Overexpression of Soybean GmWRI1a Stably Increases the Seed Oil Content in Soybean
Abstract
:1. Introduction
2. Results
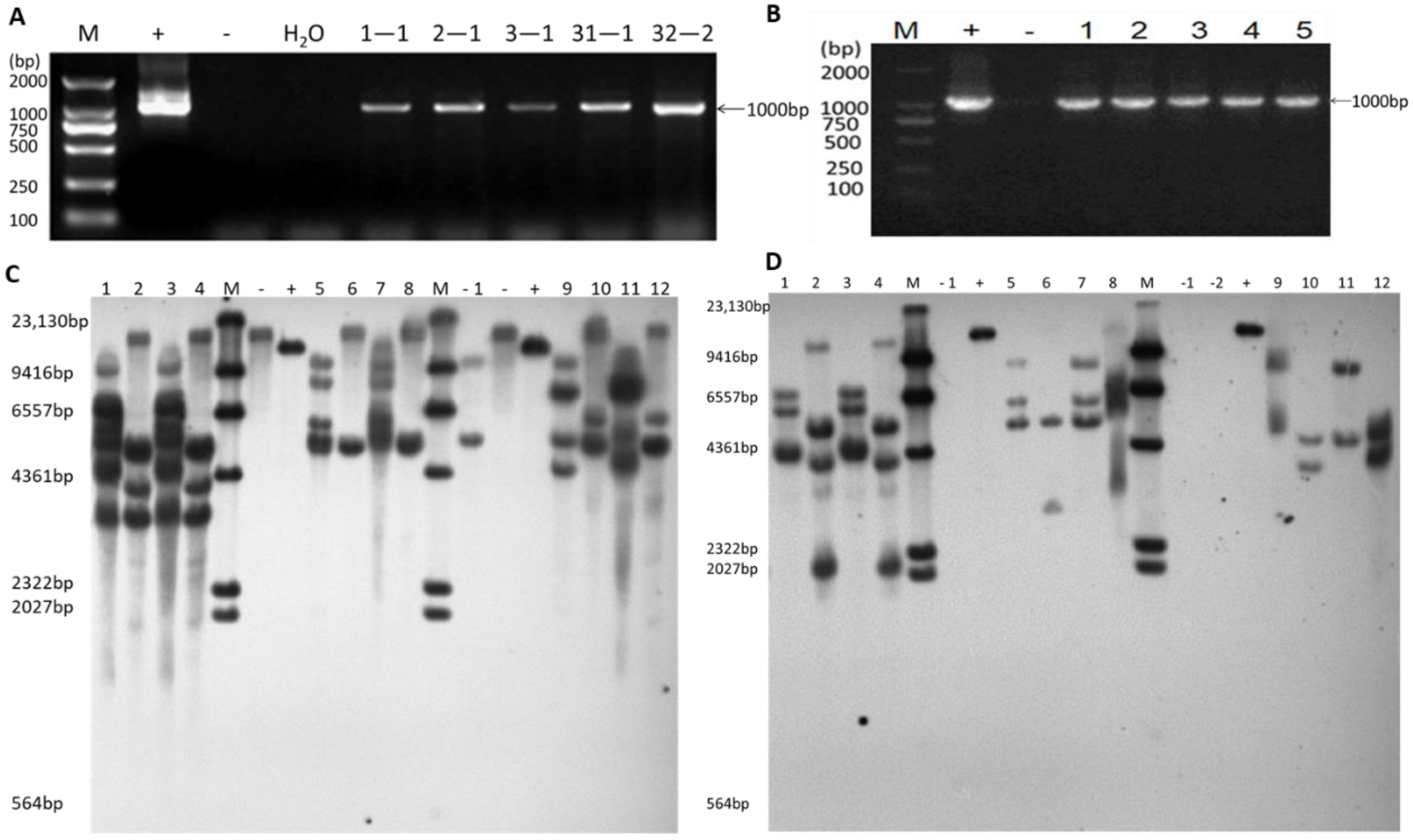
2.1. Identification of Transgene-Positive Plants and Quantification of Inserted Copy Number
2.2. Identification of Integration Sites in GmWRI1a-OE Lines 3-1 and 31-1
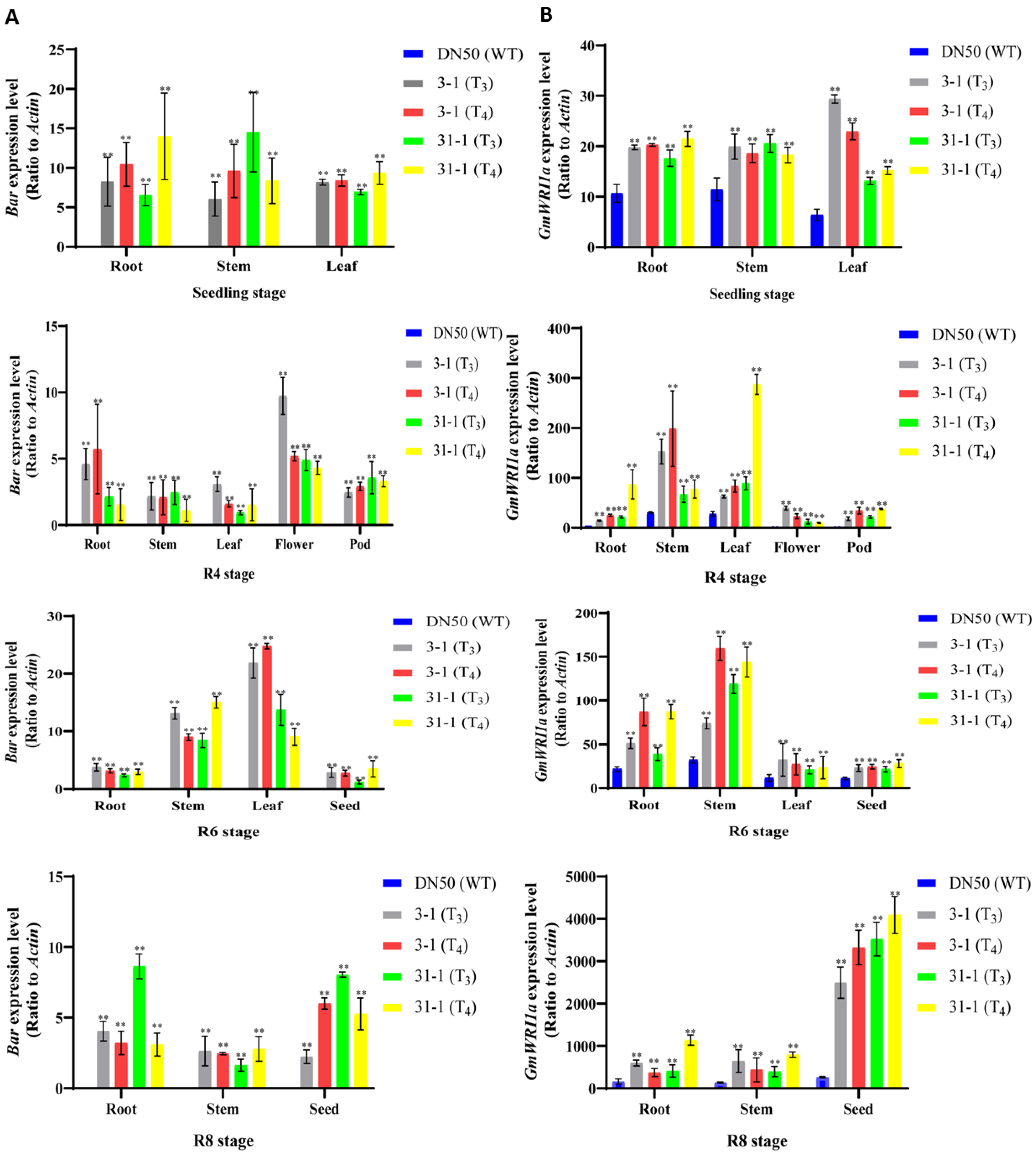
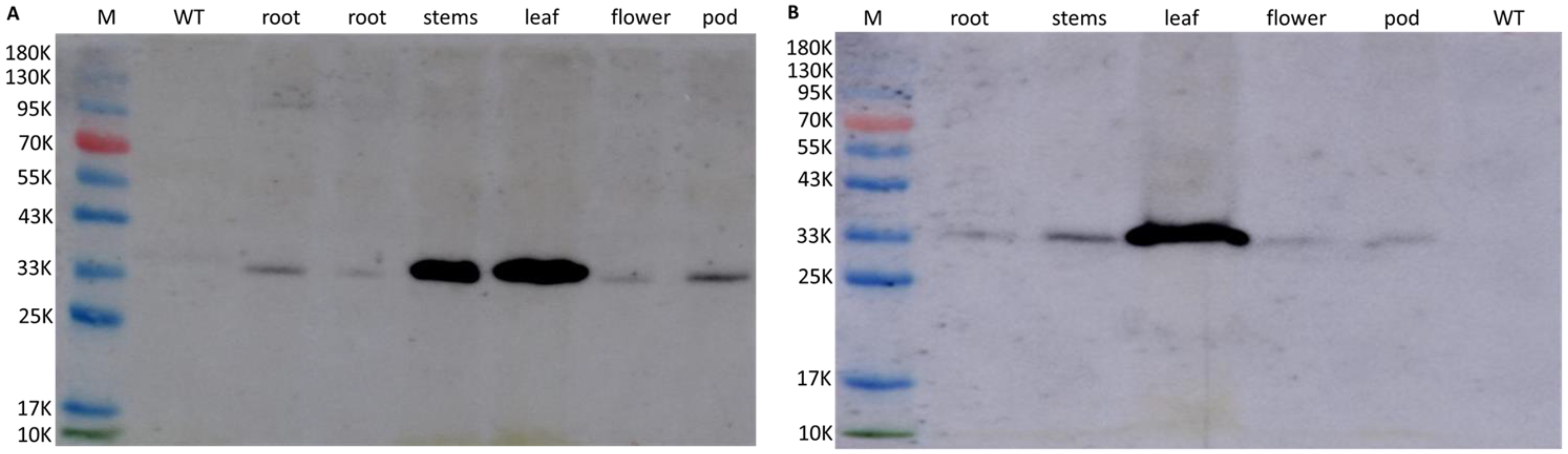
2.3. Expression Level of GmWRI1a and Bar in Transgenic Soybean Plants
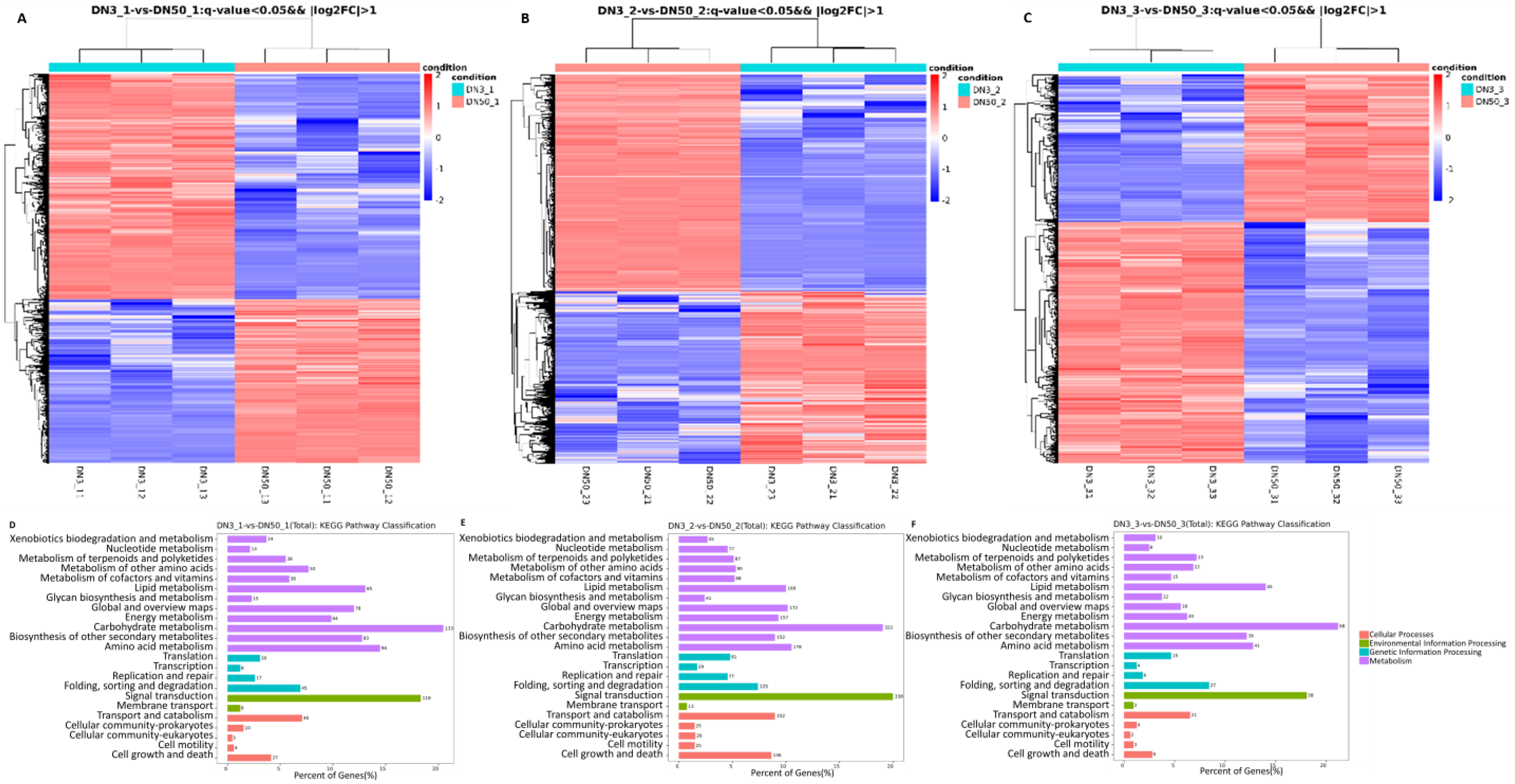
2.4. Transcriptome Sequencing Analysis
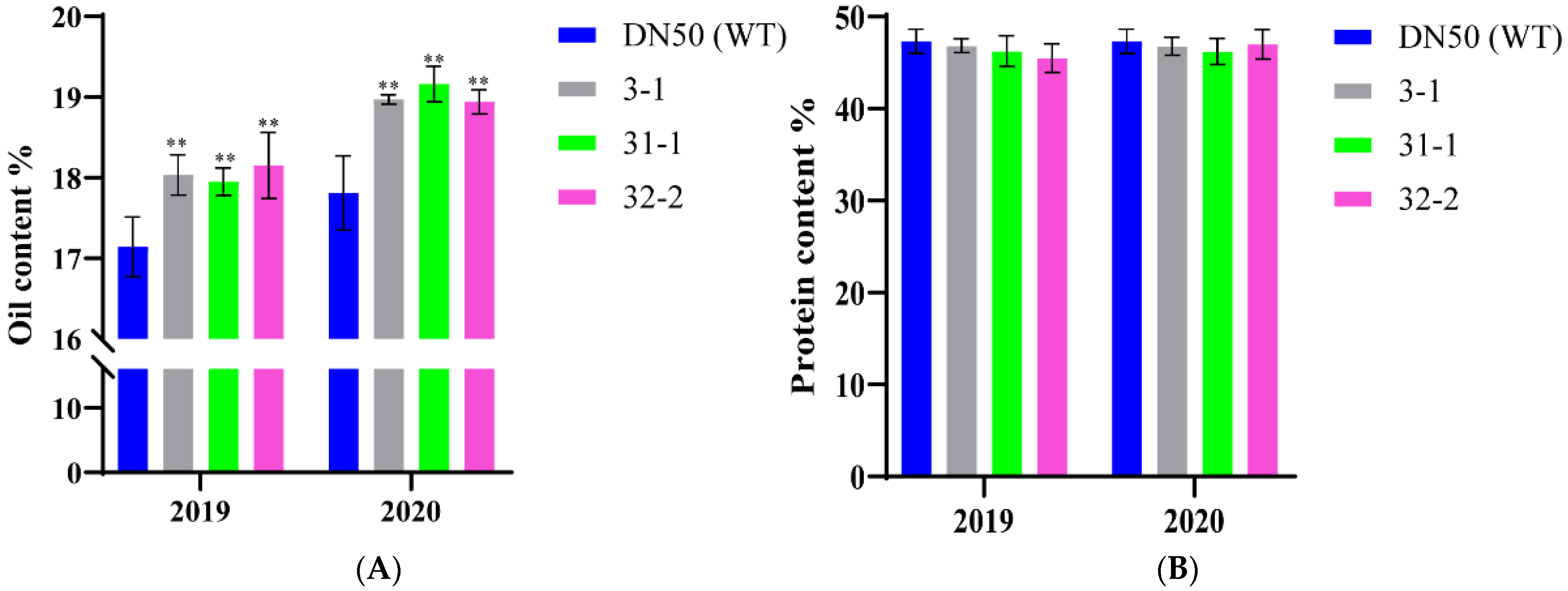
2.5. Seed Oil Content of Transgenic Plants Overexpressing GmWRI1a
2.6. Agronomic Parameters under Field Conditions
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Transformation and Identification of Transgene-Positive Plants
4.3. Confirmation of Insertion Sites and Flanking Sequences
4.4. Analysis of Gene Expression by Quantitative RT-PCR and Western Blotting
4.5. Transcriptome Sequencing and Data Analysis
4.6. Field Trial Methods
4.7. Analysis of Seed Oil and Protein Contents
4.8. Fatty Acid Composition
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


Appendix B
| GmWRI1a-P1 | 5′-ACTGCTGGATCCATGAAGAGGTCTCCAGCATC-3′ |
| GmWRI1a-P2 | 5′-CGTGCGACTAGTTCATAGATCTAGAGCATAGTCAC-3′ |
| 35s+bar-F | 5′-TGTGATAACATGGTGGAGCAC-3′ |
| 35s+bar-R | 5′-AAATCTCGGTGACGGGCA-3′ |
| GmWRI1a-probe-F | 5′-GCCTAAGCATCCAAGGAGGA-3′ |
| GmWRI1a-probe-R | 5′-GATGATGCCTAGCAACCCCA-3′ |
| BAR-target-F1 | 5′-GCGGTACCGGCAGGCTGAAG-3′ |
| BAR-target-R1 | 5′-CCGCAGGAACCGCAGGAGTG-3′ |
| Actin4-F | 5′-GTGTCAGCCATACTGTCCCCATTT-3′ |
| Actin4-R | 5′-GTTTCAAGCTCTTGCTCGTAATCA-3′ |
| QRTpcr-WRI1-F | 5′-CACCACAGCAGCACCAAGTTC-3′ |
| QRTpcr-WRI1-R | 5′-TGCCCACCAAGTCATCATC-3′ |
| QRTpcr-Bar-F | 5′-GTCCAGTCGTAGGCGTTGC-3′ |
| QRTpcr-Bar-R | 5′-GTCTGCACCATCGTCAACCA-3′ |
References
- Samarth, N.B.; Mahanwar, M.P. Modified vegetable oil based additives as a future polymeric material-review. Open J. Org. Polym. Mater. 2015, 5, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Savadi, S.; Lambani, N.; Kashyap, P.; Bisht, D.S. Genetic engineering approaches to enhance oil content in oilseed crops. Plant Growth Regul. 2017, 83, 207–222. [Google Scholar] [CrossRef]
- Chen, J.E.; Smith, A.G. A look at diacylglycerol acyltransferases (DGATs) in algae. J. Biotechnol. 2012, 162, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.G. Seed composition. In Soybeans: Improvement, Production, and Uses; Boerma, H.R., Specht, J.E., Eds.; American Society of Agronomy; Crop Science Society of America; Soil Science Society of America: Madison, WI, USA, 2004; Volume 3, pp. 621–677. [Google Scholar]
- Gupta, P.K. Molecular Biology and Genetic Engineering; Deep and Deep Publications: New Delhi, India, 2018. [Google Scholar]
- Ruuska, S.A.; Girke, T.; Benning, C.; Ohlrogge, J.B. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 2002, 14, 1191–1206. [Google Scholar] [CrossRef] [Green Version]
- Yen, C.E.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid. Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.Y.; Huang, J.X.; Chen, F.; Xu, F.; Ni, X.Y.; Xu, H.M.; Wang, Y.L.; Jiang, C.C.; Wang, H.; Xu, A.X.; et al. Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor. Appl. Genet. 2012, 124, 407–421. [Google Scholar] [CrossRef]
- Wang, Z.K.; Huang, W.J.; Chang, J.M.; Sebastian, A.; Li, Y.G.; Li, H.Y.; Wu, X.X.; Zhang, B.B.; Meng, F.L.; Li, W.B. Overexpression of SiDGAT1, a gene encoding acyl-CoA: Diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ Cult. 2014, 119, 399–410. [Google Scholar] [CrossRef]
- Lardizabal, K.; Effertz, R.; Levering, C.; Mai, J.; Pedroso, M.C.; Jury, T.; Aasen, E.; Gruys, K.; Bennett, K. Expression of Umvelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol. 2008, 148, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Santos-Mendoza, M.; Dubreucq, B.; Baud, S.; Parcy, F.; Caboche, M.; Lepiniec, L. Deciphering the regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008, 54, 608–620. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grotewold, E. Transcription factors for predictive plant metabolic engineering: Are we there yet? Curr. Opin. Plant Biol. 2008, 19, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Yang, X.H.; Zhang, F.X.; Zheng, X.; Qu, C.M.; Mu, J.Y.; Fu, F.Y.; Li, J.N.; Guan, R.Z.; Zhang, H.S.; et al. Enhance seed oil production in canola by conditional expression of B. napus LEAFY COTYLEDON1 (BnLEC1) and LEC1-LIKE (BnL1) in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef] [Green Version]
- Zafar, S.; Li, Y.L.; Li, N.N.; Zhu, K.M.; Tan, X.L. Recent advances in enhancement of oil content in oilseed crops. J. Biotechnol. 2019, 301, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoet, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of gene involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. wrinkeld1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hua, W.; Zhan, G.M.; Wei, F.; Wang, X.M.; Liu, G.H.; Wang, H.Z. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef]
- Shen, B.; Allen, W.B.; Zheng, P.Z.; Li, C.J.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef] [Green Version]
- Sanjaya Durrett, T.P.; Weise, S.E.; Benning, C. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 2011, 9, 874–883. [Google Scholar] [CrossRef]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendro, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An D and Suh: MC Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 2015, 9, 137–148. [CrossRef]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sanjaya Sedbrook, J.; Benning, C. Ectopic expression of WRINKLED1 affects fatty acid homeostasis in brachypodium distachyon vegetative tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [PubMed] [Green Version]
- Chen, L.; Zheng, Y.H.; Dong, Z.M.; Meng, F.F.; Sun, X.M.; Fan, X.H.; Zhang, Y.F.; Wang, M.L.; Wang, S.M. Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genom. 2018, 293, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Chen, L.M.; Chen, H.F.; Yang, H.L.; You, Q.B.; Bao, A.L.; Chen, S.L.; Han, Q.N.; Huang, Y.; Qiu, D.; et al. Overexpression of GmWRI1b in soybean stably improves plant architecture and associated yield parameters, and increases total seed oil production under field conditions. Plant Biotechnol. J. 2020, 18, 1639–1641. [Google Scholar] [CrossRef]
- Chen, B.B.; Zhang, G.Y.; Li, P.H.; Yang, J.H.; Guo, L.; Beening, C.; Wang, X.M.; Zhao, J. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastid glycolysis, lipid biosynthesis and homone signalling in soybean (Glycine max). Plant Biotechnol. J. 2020, 18, 155–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhercke, T.; El Tahchy, A.; Shrestha, P.; Zhou, X.R.; Singh, S.P.; Petrie, J.R. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013, 587, 364–369. [Google Scholar] [CrossRef] [Green Version]
- Vanhercke, T.; Dyer, J.M.; Mullen, R.T.; Kilaru, A.; Rahman, M.M.; Petrie, J.R.; Green, A.G.; Yurchenko, O.; Singh, S.P. Metabolic engineering for enhanced oil in biomass. Prog. Lipid. Res. 2019, 74, 103–129. [Google Scholar] [CrossRef]
- Kong, Q.; Yang, Y.; Guo, L.; Yuan, L.; Ma, W. Molecular basis of plant oil biosynthesis: Insights gained from studying the WRINKLED1 transcription factor. Front. Plant Sci. 2020, 11, 24. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Yuan, L.; Ma, W. WRINKLED1, a “Master regular” in transcriptional control of plant oil biosynthesis. Plants 2019, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Ye, R.; Gao, L.; Zhang, L.; Wang, R.; Mao, T.; Zheng, Y.; Li, D.; Lin, Y. Characterization and ectopic expression OF CoWRI1, an AP2/EREBP domain-containing transcription factor from coconut (Cocos nucifera L.) Endosperm, changes the seeds oil content in transgenic Arabidopsis thatliana and rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Wang, C.; Sun, Y.; Qu, J.; Mao, H.; Chua, N.H. Overexpression of a transcription factor increases lipid content in a woody Perennial Jatropha Curcas. Front. Plant Sci. 2018, 9, 1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohli, A.; Twyman, R.M.; Abranches, R.; Wegel, E.; Stoger, E.; Christou, P. Transgene integration, organization and interaction in plants. Plant Mol. Biol. 2003, 52, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Olhoft, P.M.; Flagel, L.E.; Somers, D.A. T-DNA locus structure in a large population of soybean plants transformed using the agrobacterium-mediated cotyledonary-node method. Plant Biotechnol. J. 2004, 2, 289–300. [Google Scholar] [CrossRef]
- Oltmanns, H.; Frame, B.; Lee, L.Y.; Johnson, S.; Li, B.; Wang, K.; Gelvin, S.B. Generation of back-bone-free, low transgene copy plants by launching T-DNA from the agrobacterium chromosome. Plant Physiol. 2010, 152, 1158–1166. [Google Scholar] [CrossRef] [Green Version]
- Honna, P.T.; Fuganti-Pagliarini, R.; Ferreira, L.C.; Molinari, M.D.C.; Marin, S.R.R.; Oliveira, M.C.N.; Farias, J.B.R.; Neumaier, N.; Mertz-Henning, L.M.; Kanamori, N.; et al. Molecular, physiological, and agronomical characterization, in greenhouse and in field conditions, of soybean plants genetically modified with AtGolS2 gene for drought tolerance. Mol. Breed. 2016, 36, 157. [Google Scholar] [CrossRef] [Green Version]
- Vogel, P.A.; Noyer, S.B.; Park, H.; Nguyen, H.; Hou, L.; Changa, T.; Khang, H.L.; Ciftci, O.N.; Wang, T.; Cahoon, E.B.; et al. Expression of the Arabidopsis WRINKLED 1 transcription factor leads to higher accumulation of palmitate in soybean seed. Plant Biotechnol. J. 2019, 17, 1369–1379. [Google Scholar] [CrossRef]
- Kang, N.K.; Kim, E.K.; Kim, Y.U.; Lee, B.; Jeong, W.J.; Jeong, B.; Chang, Y.K. Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol. Biofuels 2017, 10, 231–245. [Google Scholar] [CrossRef]
- Liu, M.; Li, D.M.; Wang, Z.K.; Meng, F.L.; Li, Y.; Wu, X.X.; Teng, W.L.; Han, Y.P.; Li, W.B. Transgenic expression of ThIPK2 gene in soybean improves stress tolerance, oleic acid content and seed size. Plant Cell Tissue Organ Cult. 2012, 111, 277–289. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Leng, J.T.; Chen, Y.Z.; Wang, Y.; Wu, C.X. Character analysis of newly-developed soybean varieties and breeding objectives in different regions of China. Soybean Sci. 2007, 26, 293–299. [Google Scholar]
- Zhang, Y.J.; Gao, H.M.; Jiang, C.Z.; Hu, M.Y.; Liu, B.Q.; Li, H. Fast analysis on fatty acids of soybean seed by gas chromatography. Soybean Sci. 2008, 5, 859–862. [Google Scholar]
- Panthee, D.R.; Pantalone, V.R.; Saxton, A.M. Modifier QTL for fatty acid composition in soybean oil. Euphytica 2006, 152, 67–73. [Google Scholar] [CrossRef]
- Xie, D.W.; Han, Y.P.; Zeng, Y.H.; Chang, W.; Teng, W.L.; Li, W.L. SSR- and SNP-related QTL underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Mol. Breed. 2012, 30, 169–179. [Google Scholar] [CrossRef]



| Lines | C16:0 | C18:0 | C18:1 | C18:2 | C18:3 |
|---|---|---|---|---|---|
| DN50 | 11.08 ± 0.02 | 3.46 ± 0.01 | 23.68 ± 0.09 | 49.71 ± 0.13 | 9.18 ± 0.03 |
| 3-1 | 10.95 ± 0.01 ** | 3.36 ± 0.02 ** | 20.13 ± 0.09 ** | 52.71 ± 0.08 ** | 9.85 ± 0.04 ** |
| 31-1 | 10.82 ± 0.02 ** | 3.49 ± 0.03 | 23.58 ± 0.12 | 50.06 ± 0.08 ** | 8.96 ± 0.06 ** |
| 32-2 | 10.40 ± 0.02 ** | 3.06 ± 0.01 ** | 22.90 ± 0.02 ** | 50.90 ± 0.06 ** | 9.70 ± 0.04 ** |
| Genotype | Plant Height (cm) | Number of Primary Internode | Number of Primary Branches | Pod Number Per Plant | Seed Number Per Plant | Seed Weight Per 100 Seeds (g) |
|---|---|---|---|---|---|---|
| DN50 (WT) | 84.02 ± 12.84 | 18.59 ± 2.14 | 3.31 ± 2.24 | 117.84 ± 63.38 | 257.79 ± 26.97 | 7.56 ± 0.77 |
| 3-1 (T3) | 88.67 ± 5.70 | 18.59 ± 2.17 | 2.58 ± 2.17 | 90.33 ± 15.60 | 259.81 ± 18.42 | 7.72 ± 1.26 |
| 3-1 (T4) | 88.92 ± 8.53 | 18.79 ± 1.53 | 3.29 ± 2.16 | 104.50 ± 14.76 | 245.89 ± 11.12 | 8.45 ± 0.86 |
| 31-1 (T3) | 86.00 ± 7.10 | 18.74 ± 1.71 | 2.59 ± 2.02 | 98.71 ± 47.10 | 206.25 ± 21.89 | 8.02 ± 1.23 |
| 31-1 (T4) | 87.12 ± 5.04 | 19.00 ± 1.22 | 2.74 ± 1.29 | 91.12 ± 25.48 | 207.73 ± 18.69 | 9.70 ± 1.06 ** |
| 32-2 (T3) | 85.33 ± 8.93 | 19.00 ± 1.05 | 3.40 ± 2.24 | 113.83 ± 43.54 | 256.06 ± 25.90 | 8.62 ± 3.64 |
| 32-2 (T4) | 70.10 ± 8.38 ** | 18.87 ± 1.41 | 2.10 ± 1.69 | 74.73 ± 21.98 ** | 157.61 ± 10.87 ** | 8.43 ± 1.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Wang, Y.; Shang, P.; Yang, C.; Yang, M.; Huang, J.; Ren, B.; Zuo, Z.; Zhang, Q.; Li, W.; et al. Overexpression of Soybean GmWRI1a Stably Increases the Seed Oil Content in Soybean. Int. J. Mol. Sci. 2022, 23, 5084. https://doi.org/10.3390/ijms23095084
Wang Z, Wang Y, Shang P, Yang C, Yang M, Huang J, Ren B, Zuo Z, Zhang Q, Li W, et al. Overexpression of Soybean GmWRI1a Stably Increases the Seed Oil Content in Soybean. International Journal of Molecular Sciences. 2022; 23(9):5084. https://doi.org/10.3390/ijms23095084
Chicago/Turabian StyleWang, Zhikun, Yuanzhuo Wang, Ping Shang, Chao Yang, Mingming Yang, Jinxiu Huang, Baizheng Ren, Zhaohui Zuo, Qingyan Zhang, Wenbin Li, and et al. 2022. "Overexpression of Soybean GmWRI1a Stably Increases the Seed Oil Content in Soybean" International Journal of Molecular Sciences 23, no. 9: 5084. https://doi.org/10.3390/ijms23095084
APA StyleWang, Z., Wang, Y., Shang, P., Yang, C., Yang, M., Huang, J., Ren, B., Zuo, Z., Zhang, Q., Li, W., & Song, B. (2022). Overexpression of Soybean GmWRI1a Stably Increases the Seed Oil Content in Soybean. International Journal of Molecular Sciences, 23(9), 5084. https://doi.org/10.3390/ijms23095084





