Identification of Epitopes on Rhinovirus 89 Capsid Proteins Capable of Inducing Neutralizing Antibodies
Abstract
:1. Introduction
2. Results
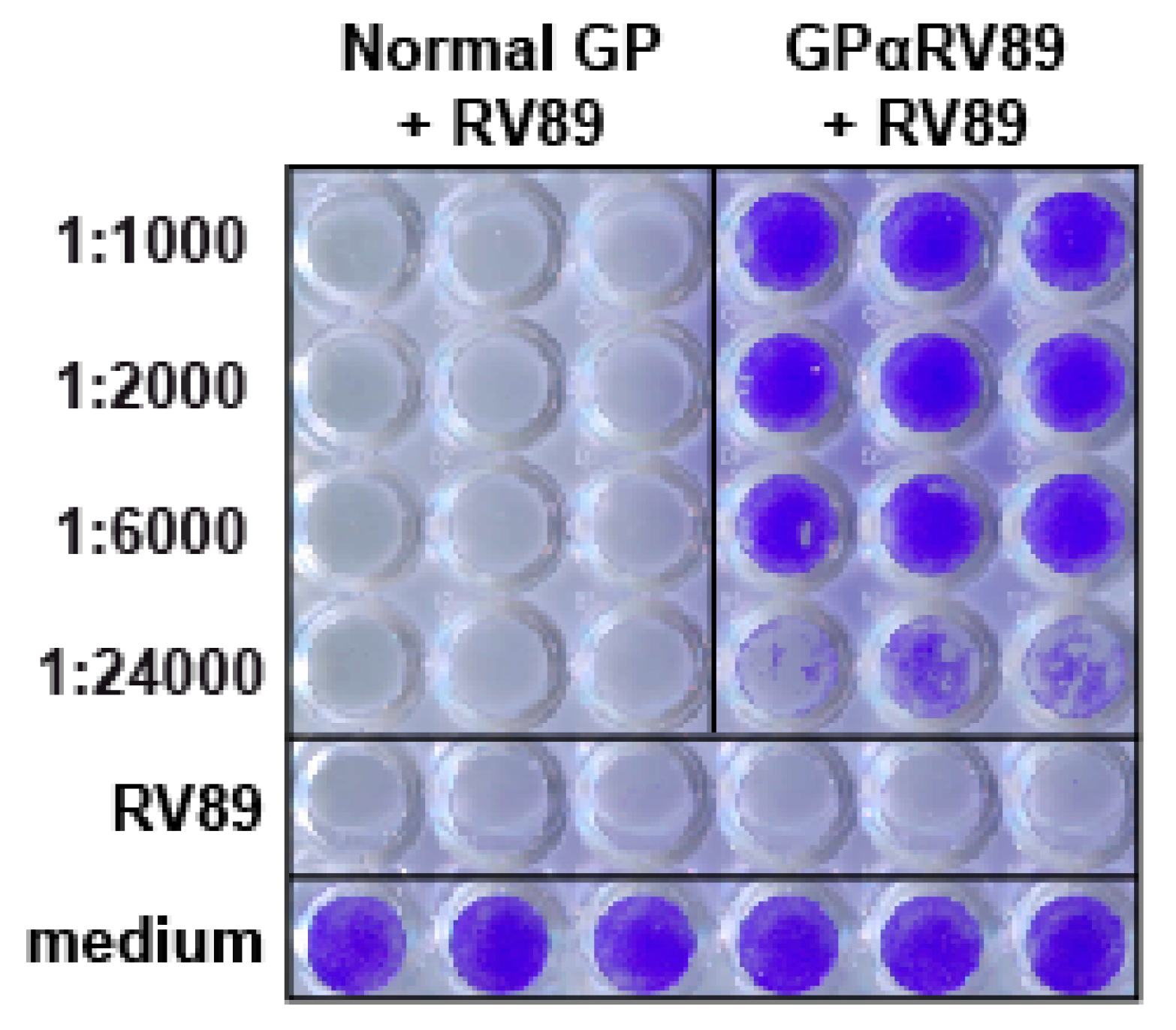
2.1. Identification of an Antiserum with High Titers of RV89-Neutralizing Antibodies
2.2. Neutralizing Antibodies React with Three Protein Fragments of VP1 and VP2 Capsid Proteins but Neither with VP3 Nor with VP4
2.3. GPαRV89 Virus-Neutralizing Antibodies Recognize Peptide Epitopes in VP1 and VP2
2.4. Recombinant PreS-Fusion Proteins Containing VP1 and VP2 Epitopes Recognized by GPαRV89 Induce Protective Antibody Responses upon Immunization
3. Discussion
4. Materials and Methods
4.1. Antisera and Detection Antibodies
4.2. RV89-Derived Recombinant Capsid Proteins, Protein Fragments and Synthetic Peptides
4.3. SDS-PAGE and Immunoblotting
4.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. In Vitro Virus Neutralization Assays
4.6. Construction of Recombinant PreS-Carrier-Based Fusion Proteins, 89RX1 and 89RX2
4.7. Expression, Purification and Characterization of Recombinant PreS-Carrier-Based Fusion Proteins
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bramley, T.J.; Lerner, D.; Sames, M. Productivity losses related to the common cold. J. Occup. Environ. Med. 2002, 44, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Gangnon, R.E.; Evans, M.D.; Roberg, K.A.; Anderson, E.L.; Pappas, T.E.; Printz, M.C.; Lee, W.M.; Shult, P.A.; Reisdorf, E.; et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 2008, 178, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K.G.; Kent, J.; Ireland, D.C. Respiratory viruses and exacerbations of asthma in adults. BMJ 1993, 307, 982–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansel, T.T.; Johnston, S.L.; Openshaw, P.J. Microbes and mucosal immune responses in asthma. Lancet 2013, 381, 861–867. [Google Scholar] [CrossRef]
- Heymann, P.W.; Platts-Mills, T.A.; Johnston, S.L. Role of viral infections, atopy and antiviral immunity in the etiology of wheezing exacerbations among children and young adults. Pediatr. Infect. Dis. J. 2005, 24, 217–222. [Google Scholar] [CrossRef]
- Papi, A.; Bellettato, C.M.; Braccioni, F.; Romagnoli, M.; Casolari, P.; Caramori, G.; Fabbri, L.M.; Johnston, S.L. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Respir. Crit. Care Med. 2006, 173, 1114–1121. [Google Scholar] [CrossRef]
- Donaldson, G.C.; Seemungal, T.A.; Patel, I.S.; Lloyd-Owen, S.J.; Wilkinson, T.M.; Wedzicha, J.A. Longitudinal changes in the nature, severity and frequency of COPD exacerbations. Eur. Respir. J. 2003, 22, 931–936. [Google Scholar] [CrossRef]
- Heymann, P.W.; Kennedy, J.L. Rhinovirus-induced asthma exacerbations during childhood: The importance of understanding the atopic status of the host. J. Allergy Clin. Immunol. 2012, 130, 1315–1316. [Google Scholar] [CrossRef] [Green Version]
- Kloepfer, K.M.; Lee, W.M.; Pappas, T.E.; Kang, T.J.; Vrtis, R.F.; Evans, M.D.; Gangnon, R.E.; Bochkov, Y.A.; Jackson, D.J.; Lemanske, R.F., Jr.; et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J. Allergy Clin. Immunol. 2014, 133, 1301–1307. [Google Scholar] [CrossRef] [Green Version]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser-Liggett, C.M.; Liggett, S.B. Sequencing and analyses of all know human rhinovirus genomes reveal structure and evolution. Science 2009, 324, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Rossmann, M.G.; Arnold, E.; Erickson, J.W.; Frankenberger, E.A.; Griffith, J.P.; Hecht, H.J.; Johnson, J.E.; Kamer, G.; Luo, M.; Mosser, A.G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 1985, 317, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Conant, R.M.; Hamparian, V.V. Rhinoviruses: Basis for a numbering system. II. Serologic characterization of prototype strain. J. Immunol. 1968, 100, 114–119. [Google Scholar] [PubMed]
- Schieble, J.H.; Fox, V.L.; Lester, F.; Lennette, E.H. Rhinoviruses: An antigenic study of the prototype virus strains. Proc. Soc. Exp. Biol. Med. 1974, 147, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Aab, A.; Wirz, O.; van de Veen, W.; Söllner, S.; Stanic, B.; Rückert, B.; Aniscenko, J.; Edwards, M.R.; Johnston, S.L.; Papadopoulos, N.G.; et al. Human rhinoviruses enter and induce proliferation of B lymphocytes. Allergy 2017, 72, 232–243. [Google Scholar] [CrossRef]
- Nikonova, A.; Khaitov, M.; Jackson, D.J.; Traub, S.; Trujillo-Torralbo, M.B.; Kudlay, D.A.; Dvornikov, A.S.; Del-Rosario, A.; Valenta, R.; Stanciu, L.A.; et al. M1-like macrophages are potent producers of anti-viral interferons and M1-associated marker-positive lung macrophages are decreased during rhinovirus-induced asthma exacerbations. EBioMedicine 2020, 54, 102734. [Google Scholar] [CrossRef]
- Greve, J.M.; Davis, G.; Meyer, A.M.; Forte, C.P.; Yost, S.C.; Marlor, C.W.; Kamarck, M.E.; McClelland, A. The major human rhinovirus receptor is ICAM-1. Cell 1989, 56, 839–847. [Google Scholar] [CrossRef]
- Hofer, F.; Gruenberger, M.; Kowalski, H.; Machat, H.; Huettinger, M.; Kuechler, E.; Blaas, D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 1994, 91, 1839–1842. [Google Scholar] [CrossRef] [Green Version]
- Bochkov, Y.A.; Watters, K.; Ashraf, S.; Griggs, T.F.; Devries, M.K.; Jackson, D.J.; Palmenberg, A.C.; Gern, J.E. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. USA 2015, 28, 5485–5490. [Google Scholar] [CrossRef] [Green Version]
- McLean, G.R. Developing a vaccine for human rhinoviruses. J. Vaccines Immun. 2014, 2, 16–20. [Google Scholar]
- Glanville, N.; Johnston, S.L. Challenges in developing a cross-serotype rhinovirus vaccine. Curr. Opin. Virol. 2015, 11, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Niespodziana, K.; Stenberg-Hammar, K.; Megremis, S.; Cabauatan, C.R.; Napora-Wijata, K.; Vacal, P.C.; Gallerano, D.; Lupinek, C.; Ebner, D.; Schlederer, T.; et al. PreDicta chip-based high resolution diagnosis of rhinovirus-induced wheeze. Nat. Commun. 2018, 9, 2382. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.C.; Tucker, D.N.; Knopf, H.L.; Wenzel, R.P.; Kapikian, A.Z.; Chanock, R.M. Comparison of protective effect of neutralizing antibody in serum and nasal secretions in experimental rhinovirus type 13 illness. Am. J. Epidemiol. 1969, 90, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.C.; Tucker, D.N.; Knope, H.L.; Wenzel, R.P.; Hornick, R.B.; Kapikian, A.Z.; Chanock, R.M. Evidence for protective effect of an inactivated rhinovirus vaccine administered by the nasal route. Am. J. Epidemiol. 1969, 90, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Buscho, R.F.; Perkins, J.C.; Knopf, H.L.; Kapikian, A.Z.; Chanock, R.M. Further characterization of the local respiratory tract antibody response induced by intranasal instillation of inactivated rhinovirus 13 vaccine. J. Immunol. 1972, 108, 169–177. [Google Scholar] [PubMed]
- Hamory, B.H.; Hamparian, V.V.; Conant, R.M.; Gwaltney, J.M., Jr. Human responses to two decavalent rhinovirus vaccines. J. Infect. Dis. 1975, 132, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Cooney, M.K.; Wise, J.A.; Kenny, G.E.; Fox, J.P. Broad antigenic relationships among rhinovirus serotypes revealed by cross-immunization of rabbits with different serotypes. J. Immunol. 1975, 114, 635–639. [Google Scholar] [PubMed]
- Fox, J.P. Is a rhinovirus vaccine possible? Am. J. Epidemiol. 1976, 103, 345–354. [Google Scholar] [CrossRef]
- Francis, M.J.; Hastings, G.Z.; Sangar, D.V.; Clark, R.P.; Syred, A.; Clarke, B.E.; Rowlands, D.J.; Brown, F. A synthetic peptide which elicits neutralizing antibody against human rhinovirus type 2. J. Gen. Virol. 1987, 68, 2687–2691. [Google Scholar] [CrossRef]
- McCray, J.; Werner, G. Different rhinovirus serotypes neutralized by antipeptide antibodies. Nature 1987, 329, 736–738. [Google Scholar] [CrossRef]
- Hastings, G.Z.; Speller, S.A.; Francis, M.J. Neutralizing antibodies to human rhinovirus produced in laboratory animals and humans that recognize a linear sequence from VP2. J. Gen. Virol. 1990, 71, 3055–3059. [Google Scholar] [CrossRef]
- Appleyard, G.; Russell, S.M.; Clarke, B.E.; Speller, S.A.; Trowbridge, M.; Vadolas, J. Neutralization epitopes of human rhinovirus type 2. J. Gen. Virol. 1990, 71, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Skern, T.; Neubauer, C.; Frasel, L.; Gründler, P.; Sommergruber, W.; Zorn, M.; Kuechler, E.; Blaas, D. A neutralizing epitope on human rhinovirus type 2 includes amino acid residues between 153 and 164 of virus capsid protein VP2. J. Gen. Virol. 1987, 68, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Sherry, B.; Mosser, A.G.; Colonno, R.J.; Rueckert, R.R. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J. Virol. 1986, 57, 246–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalnin, K.; Yan, Y.; Kleanthous, H. Neutralizining Immunogen (NImIV) of Rhinovirus and Its Use for Vaccine Applications. U.S. Patent 8,652,486, 18 February 2014. [Google Scholar]
- Katpally, U.; Fu, T.M.; Freed, D.C.; Casimiro, D.R.; Smith, T.J. Antibodies to the buried N terminus of rhinovirus VP4 exhibit cross-serotypic neutralization. J. Virol. 2009, 83, 7040–7048. [Google Scholar] [CrossRef] [Green Version]
- Glanville, N.; McLean, G.R.; Guy, B.; Lecouturier, V.; Berry, C.; Girerd, Y.; Gregoire, C.; Walton, R.P.; Pearson, R.M.; Kebadze, T.; et al. Cross-serotype immunity induced by immunization with a conserved rhinovirus capsid protein. PLoS Pathog. 2013, 9, e1003669. [Google Scholar] [CrossRef] [Green Version]
- Edlmayr, J.; Niespodziana, K.; Popow-Kraupp, T.; Krzyzanek, V.; Focke-Tejkl, M.; Blaas, D.; Grote, M.; Valenta, R. Antibodies induced with recombinant VP1 from human rhinovirus exhibit cross-neutralisation. Eur. Respir. J. 2011, 37, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Stenberg-Hammar, K.; Niespodziana, K.; Söderhäll, C.; James, A.; Cabauatan, C.R.; Konradsen, J.R.; Melén, E.; van Hage, M.; Valenta, R.; Hedlin, G. Rhinovirus-specific antibody responses in preschool children with acute wheeze reflect severity of respiratory symptoms. Allergy 2016, 71, 1728–1735. [Google Scholar] [CrossRef]
- Niespodziana, K.; Napora, K.; Cabauatan, C.; Focke-Tejkl, M.; Keller, W.; Niederberger, V.; Tsolia, M.; Christodoulou, I.; Papadopoulos, N.G.; Valenta, R. Misdirected antibody responses against an N-terminal epitope on human rhinovirus VP1 as explanation for recurrent RV infections. FASEB J. 2012, 26, 1001–1008. [Google Scholar] [CrossRef]
- Conant, R.M.; Hamparian, V.V. Rhinoviruses: Basis for a Numbering System: 1. HeLa Cells for Propagation and Serologic Procedures. J. Immunol. 1968, 100, 107–113. [Google Scholar]
- Pazderova, P.; Waltl, E.E.; Niederberger-Leppin, V.; Flicker, S.; Valenta, R.; Niespodziana, K. ELISA-Based Assay for Studying Major and Minor Group Rhinovirus-Receptor Interactions. Vaccines 2020, 8, 315. [Google Scholar] [CrossRef]
- Bartlett, N.W.; Walton, R.P.; Edwards, M.R.; Aniscenko, J.; Caramori, G.; Zhu, J.; Glanville, N.; Choy, K.J.; Jourdan, P.; Burnet, J.; et al. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat. Med. 2008, 14, 199–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niespodziana, K.; Focke-Tejkl, M.; Linhart, B.; Civaj, V.; Blatt, K.; Valent, P.; van Hage, M.; Grönlund, H.; Valenta, R. A hypoallergenic cat vaccine based on Fel d 1-derived peptides fused to hepatitis B PreS. J. Allergy Clin. Immunol. 2011, 127, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Focke-Tejkl, M.; Weber, M.; Niespodziana, K.; Neubauer, A.; Huber, H.; Henning, R.; Stegfellner, G.; Maderegger, B.; Hauer, M.; Stolz, F.; et al. Development and characterization of a recombinant, hypoallergenic, peptide-based vaccine for grass pollen allergy. J. Allergy Clin. Immunol. 2015, 135, 1207–1217.e11. [Google Scholar] [CrossRef] [Green Version]
- Gallerano, D.; Devanaboyina, S.C.; Swoboda, I.; Linhart, B.; Mittermann, I.; Keller, W.; Valenta, R. Biophysical characterization of recombinant HIV-1 subtype C virus infectivity factor. Amino Acids 2011, 40, 981–989. [Google Scholar] [CrossRef] [PubMed]






| RV Strain | Identified Epitope/Antigen | AA Residue/s (AA Position) | Methodology | Ref. |
|---|---|---|---|---|
| RV2 Group A minor | VP1 | TL (85-6) E (92) K (264) DRS (267-9) T (276) P (278) | RV2 escape mutants (n = 51) were identified using 14 neutralizing monoclonal antibodies. Amino acid substitutions among the 51 mutants were identified. | [31] |
| VP2 | NP (163-4) A (236) N (238) | |||
| VP2 | GREVKAETRLNPD (153-64) | Deletions at the 3’-end gene of VP2 using Bal-31 nuclease were created. The polypeptides were probed using a neutralizing monoclonal antibody (mAb 8F5). | [32] | |
| VP2/NIm-II | VKAETRLNPDLQPTE (156-70) | Rabbit anti-peptide antibodies (at 1:4 dilution) were used to determine neutralizing epitopes. | [28] | |
| RV-14 Group B major | VP1 | MYVPPGAPNP (151-60) | Amino acid sequences were postulated as potential receptor-binding sites. | [11] |
| VP1 | PPGA (154-7) | A rabbit polyclonal anti-peptide antiserum was used to neutralize RV14. | [29] | |
| VP1/NIm-IA | D (91) E (95) | Approach was the same as Appleyard et al. (1990) (30). Thirty-five RV14-neutralizing monoclonal antibodies were used to identify 62 escape mutants. | [33] | |
| VP1/NIm-IB | Q (83) K (85) DS (138-9) | |||
| VP1/NIM-II | E (210) | |||
| VP1/NIm-III | K (287) | |||
| VP2/NIm-II | SA (158-9) EV (161-2) E (136) | |||
| VP1/NIm-IV | PVIKKRK (275-85) | Residues were discovered via a molecular evolution experiment/VP1 gene shuffling. | [34] | |
| VP1/NIm-IV | NTEPVIKKRKGDIKSY (272-91) | Residues were discovered via a molecular evolution experiment/VP1 gene shuffling. | [34] | |
| VP4 | GAQVSTQKSGSHENQNILTNGSNQTFTVINY (1-31) | Antibodies generated to N-terminal VP4 fragment of RV14 showed cross-serotypic neutralization. | [35] | |
| RV16 Group A major | VP0 | Consensus sequence for VP4 and VP2 | Recombinant VP0 (VP4 + VP2) was used for the immunization of mice. Antibodies induced by VP0 immunogen enhanced neutralizing antibody responses to heterologous virus infection. | [36] |
| RV89 Group Amajor | VP1 | GenBank: AY355270 | Recombinant VP1 proteins of RV89 and RV14 were used to induce polyclonal antibodies, which exhibited cross-neutralization. | [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niespodziana, K.; Cabauatan, C.R.; Pazderova, P.; Vacal, P.C.; Wortmann, J.; Keller, W.; Errhalt, P.; Valenta, R. Identification of Epitopes on Rhinovirus 89 Capsid Proteins Capable of Inducing Neutralizing Antibodies. Int. J. Mol. Sci. 2022, 23, 5113. https://doi.org/10.3390/ijms23095113
Niespodziana K, Cabauatan CR, Pazderova P, Vacal PC, Wortmann J, Keller W, Errhalt P, Valenta R. Identification of Epitopes on Rhinovirus 89 Capsid Proteins Capable of Inducing Neutralizing Antibodies. International Journal of Molecular Sciences. 2022; 23(9):5113. https://doi.org/10.3390/ijms23095113
Chicago/Turabian StyleNiespodziana, Katarzyna, Clarissa R. Cabauatan, Petra Pazderova, Phyllis C. Vacal, Judith Wortmann, Walter Keller, Peter Errhalt, and Rudolf Valenta. 2022. "Identification of Epitopes on Rhinovirus 89 Capsid Proteins Capable of Inducing Neutralizing Antibodies" International Journal of Molecular Sciences 23, no. 9: 5113. https://doi.org/10.3390/ijms23095113
APA StyleNiespodziana, K., Cabauatan, C. R., Pazderova, P., Vacal, P. C., Wortmann, J., Keller, W., Errhalt, P., & Valenta, R. (2022). Identification of Epitopes on Rhinovirus 89 Capsid Proteins Capable of Inducing Neutralizing Antibodies. International Journal of Molecular Sciences, 23(9), 5113. https://doi.org/10.3390/ijms23095113






