Integrated Multiomics Analysis of Salivary Exosomes to Identify Biomarkers Associated with Changes in Mood States and Fatigue
Abstract
1. Introduction
2. Results
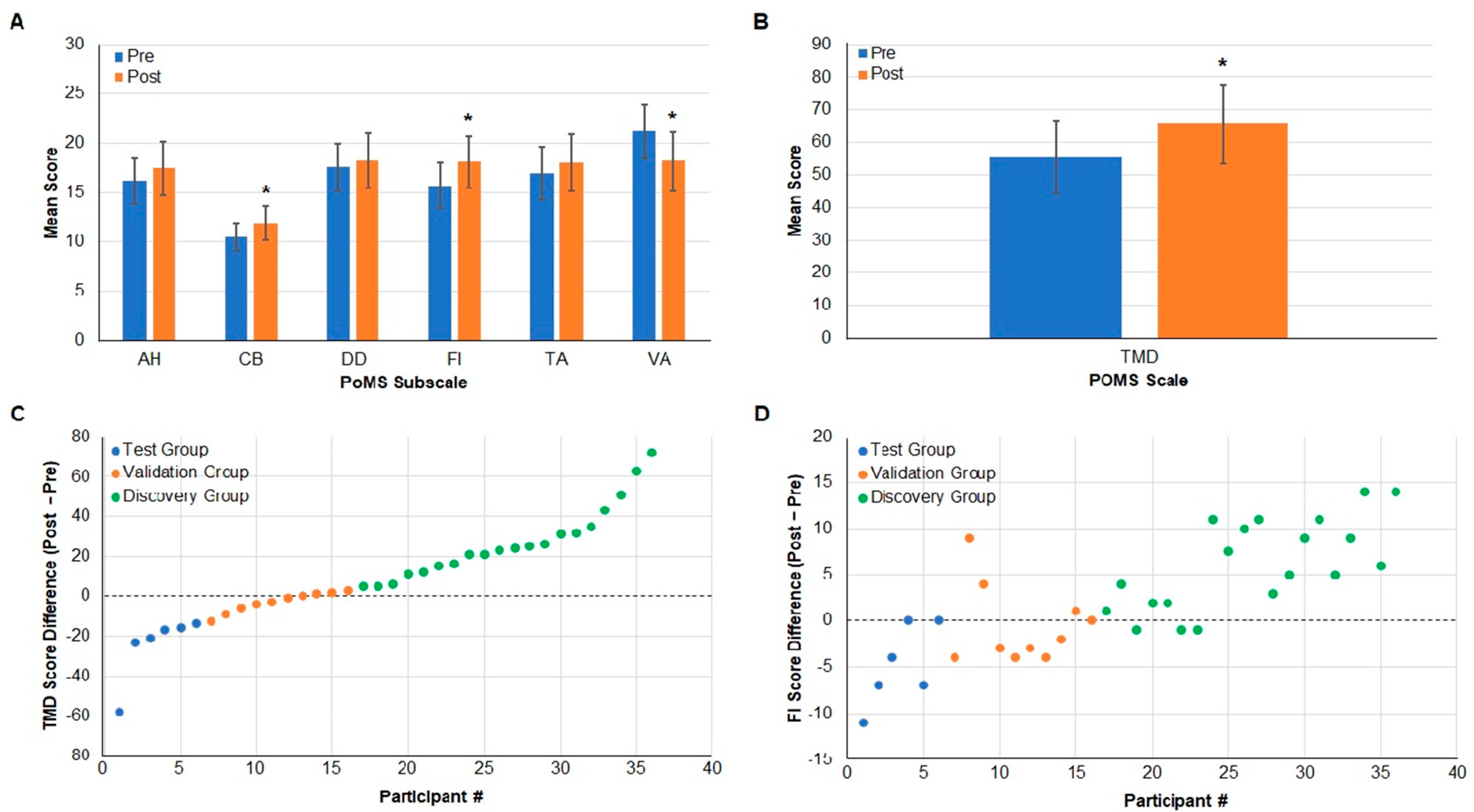
PoMS Analysis and Assignment to Study Groups
3. Multiomics Analysis of Test Group Salivary Exosomes
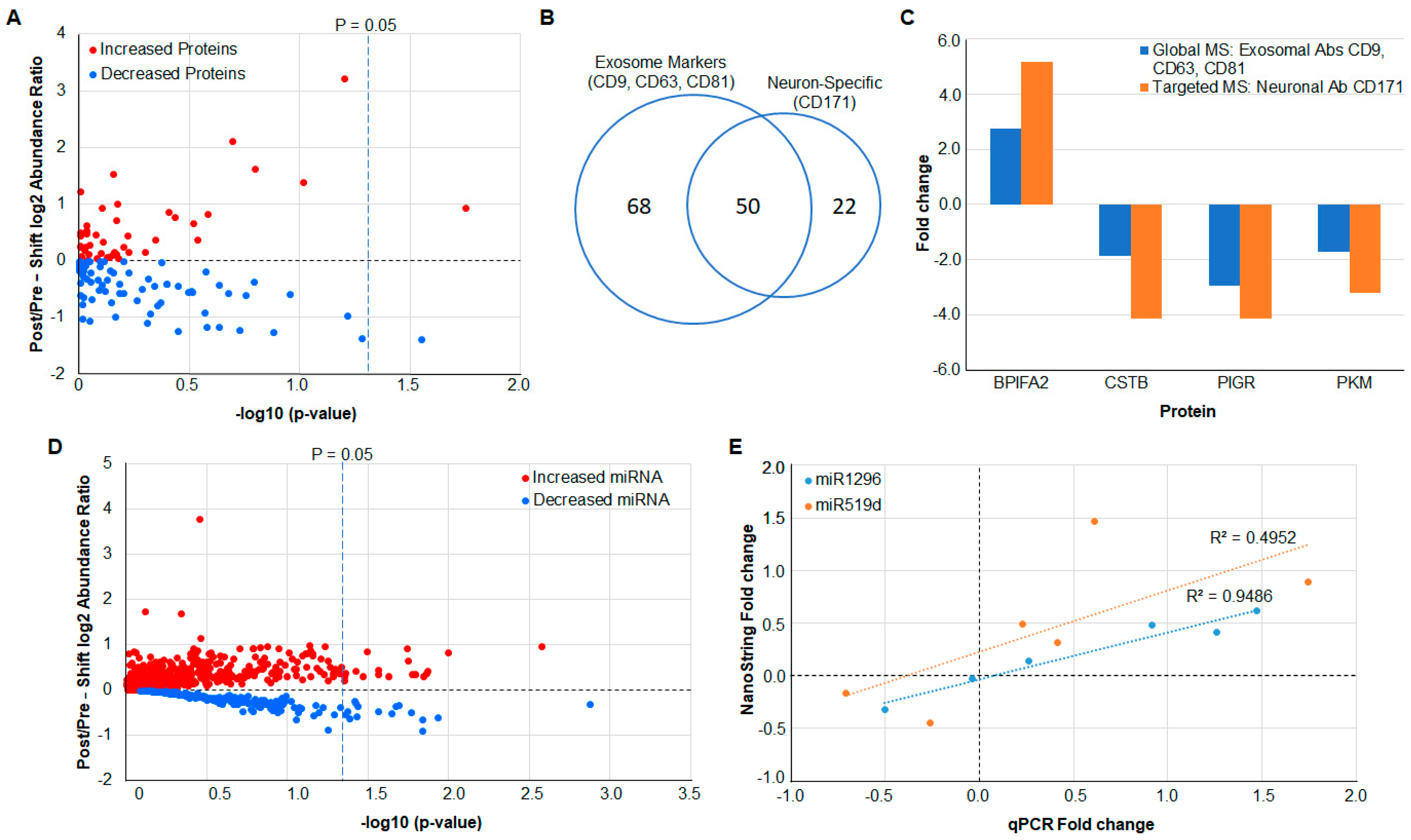
3.1. Quantitative Global Proteomics on Salivary Exosomes
3.2. Verification of Protein Measurements in Neuron-Derived Exosomes by Targeted MS
3.3. MicroRNA Analysis Using NanoString
3.4. Verification of miRNA Measurements Using qPCR
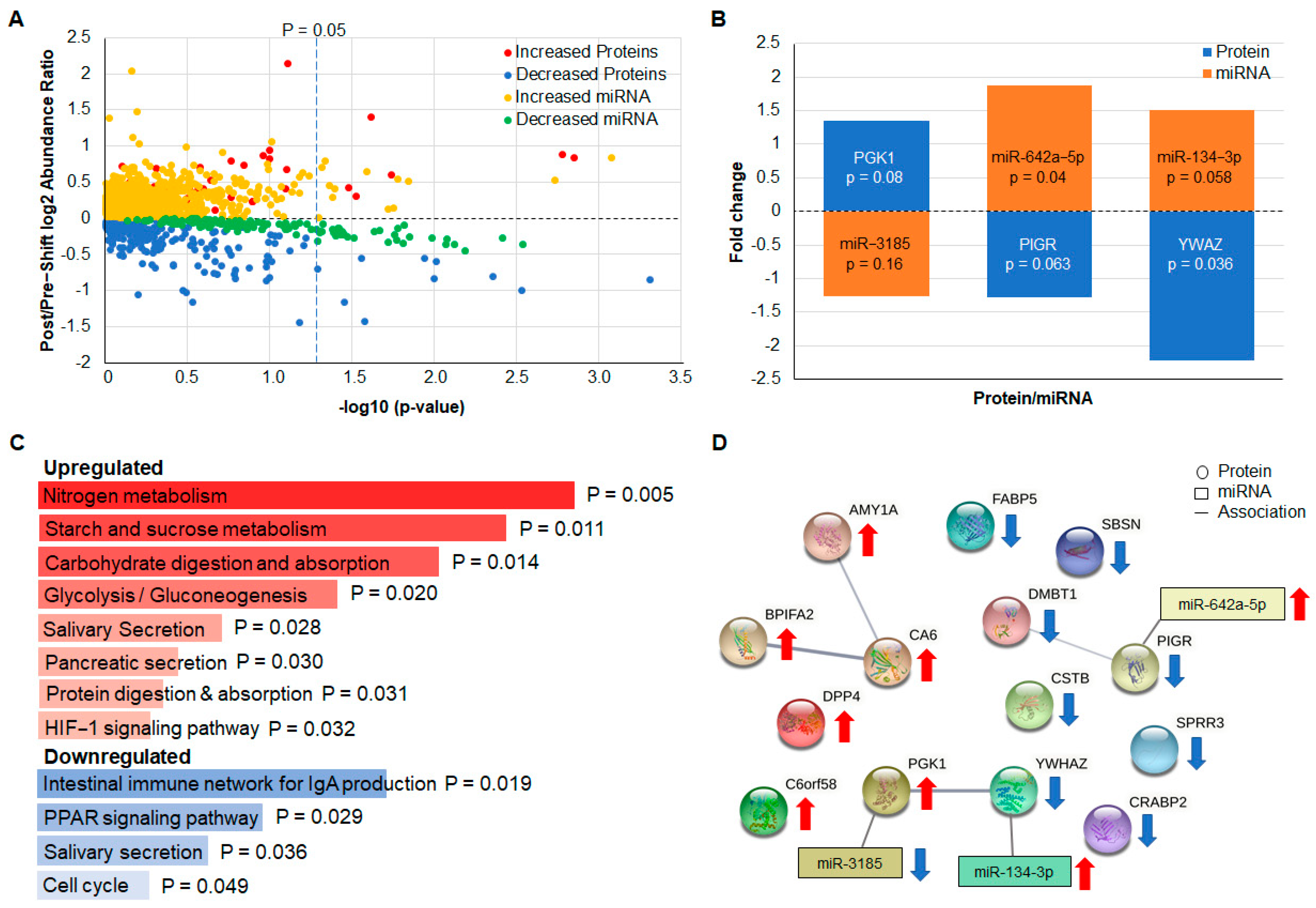
4. Biomarker Identification in Discovery Group Saliva
Identification of Additional Significantly Altered Protein and miRNA
5. Confirmation of Biomarkers and Fold Change in Validation Group Salivary Exosomes
5.1. Validation of Protein Biomarkers
5.2. Validation of miRNA Biomarkers
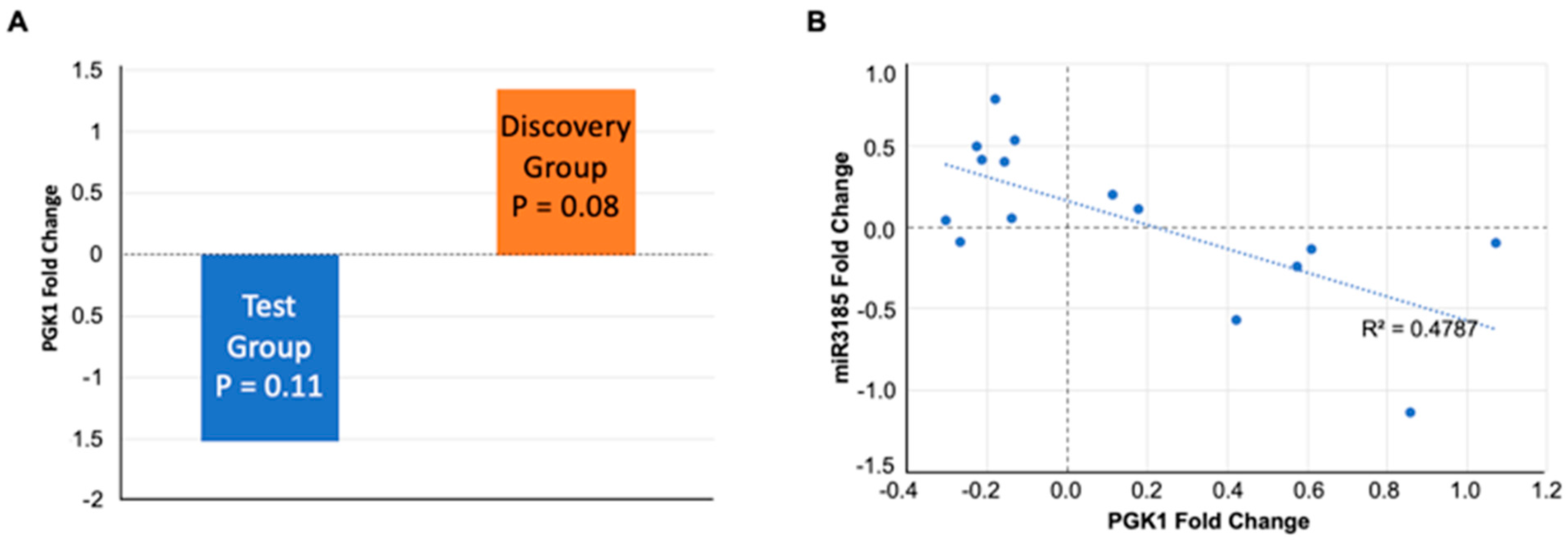
5.3. PGK1 Protein and miR3185 in Saliva as Potential Biomarkers of Fatigue
5.4. Integration of Test, Discovery, and Validation Data Reveals Proteins Associated with Work
6. Discussion
7. Methods
7.1. Participants
7.2. Whole Saliva Collection
7.3. The Profile of Mood States (PoMS) Questionnaire
7.4. Separation of Saliva Samples by PoMS TMD Score
7.5. EV Isolation and Enrichment for Exosomes
7.6. Quantitative Global Proteomics Analysis
7.7. Proteomics Analysis
7.8. Protein Bioinformatics Analysis
7.9. Targeted LC-MS/MS Protein Quantification
7.10. Global miRNA Analysis
7.11. Targeted miRNA Analysis
7.12. Identification of miRNA Target Genes
7.13. Statistical Analysis
7.14. Study Design
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorist, M.M.; Boksem, M.A.; Ridderinkhof, K.R. Impaired cognitive control and reduced cingulate activity during mental fatigue. Cogn. Brain Res. 2005, 24, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Fogt, D.L.; Kalns, J.E.; Michael, D.J. A comparison of cognitive performance decreases during acute, progressive fatigue arising from different concurrent stressors. Mil. Med. 2010, 175, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Holtzer, R.; Shuman, M.; Mahoney, J.R.; Lipton, R.; Verghese, J. Cognitive fatigue defined in the context of attention networks. Neuropsychology, development, and cognition. Aging Neuropsychol. Cogn. 2011, 18, 108–128. [Google Scholar] [CrossRef]
- Hudson, A.N.; Van Dongen, H.P.A.; Honn, K.A. Sleep deprivation, vigilant attention, and brain function: A review. Neuropsychopharmacology 2020, 45, 21–30. [Google Scholar] [CrossRef]
- Meeusen, R.; Van Cutsem, J.; Roelands, B. Endurance exercise-induced mental fatigue and the brain. Exp. Physiol. 2021, 106, 2294–2298. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- Multhoff, G.; Hightower, L.E. Distinguishing integral and receptor-bound heat shock protein 70 (Hsp70) on the cell surface by Hsp70-specific antibodies. Cell Stress Chaperones 2011, 16, 251–255. [Google Scholar] [CrossRef]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Irwin, M.R.; Wang, M.; Campomayor, C.O.; Collado-Hidalgo, A.; Cole, S. Sleep deprivation and activation of morning levels of cellular and genomic markers of inflammation. Arch. Intern. Med. 2006, 166, 1756–1762. [Google Scholar] [CrossRef]
- Carroll, J.E.; Cole, S.W.; Seeman, T.E.; Breen, E.C.; Witarama, T.; Arevalo, J.M.; Ma, J.; Irwin, M.R. Partial sleep deprivation activates the DNA damage response (DDR) and the senescence-associated secretory phenotype (SASP) in aged adult humans. Brain Behav. Immun. 2016, 51, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Dong, Y.; Xu, Z.; Gompf, H.S.; Ward, S.A.; Xue, Z.; Miao, C.; Zhang, Y.; Chamberlin, N.L.; Xie, Z. Sleep disturbance induces neuroinflammation and impairment of learning and memory. Neurobiol. Dis. 2012, 48, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Gottshall, J.L.; Guedes, V.A.; Pucci, J.U.; Brooks, D.; Watson, N.; Sheth, P.; Gabriel, A.; Mithani, S.; Leete, J.L.; Lai, C.; et al. Poor Sleep Quality is linked to elevated extracellular vesicle-associated inflammatory cytokines in warfighters with chronic mild traumatic brain injuries. Front. Pharmacol. 2022, 12, 762077. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.N.; Hoffman, M.P. Interactions between developing nerves and salivary glands. Organogenesis 2013, 9, 199–205. [Google Scholar] [CrossRef]
- Dominy, S.S.; Brown, J.N.; Ryder, M.I.; Gritsenko, M.; Jacobs, J.M.; Smith, R.D. Proteomic analysis of saliva in HIV-positive heroin addicts reveals proteins correlated with cognition. PLoS ONE 2014, 9, e89366. [Google Scholar] [CrossRef]
- Di Pietro, V.D.; Porto, E.; Ragusa, M.; Barbagallo, C.; Davies, D.; Forcione, M.; Logan, A.; Di Pietro, C.; Purrello, M.; Grey, M.; et al. Salivary MicroRNAS: Diagnostic Markers of Mild Traumatic Brain Injury in Contact-Sport. Front. Mol. Neurosci. 2018, 11, 290. [Google Scholar] [CrossRef]
- Ciregia, F.; Giusti, L.; Valle, Y.D.; Donadio, E.; Consensi, A.; Giacomelli, C.; Sernissi, F.; Scarpellini, P.; Maggi, F.; Lucacchini, A.; et al. A multidisciplinary approach to study a couple of monozygotic twins discordant for the chronic fatigue syndrome: A focus on potential salivary biomarkers. J. Transl. Med. 2013, 11, 243–257. [Google Scholar] [CrossRef]
- Bermejo-Pareja, F.; Antequera, D.; Vargas, T.; Molina, J.; Carro, E. Saliva levels of Abeta1-42 as potential biomarker of Alzheimer’s disease: A pilot study. BMC Neurol. 2010, 10, 108. [Google Scholar] [CrossRef]
- Shi, M.; Sui, Y.T.; Peskind, E.R.; Li, G.; Hwang, H.; Devic, I.; Ginghina, C.; Edgar, J.S.; Pan, C.; Goodlett, D.R.; et al. Salivary tau species are potential biomarkers of Alzheimer’s disease. J. Alzheimers Dis. 2011, 27, 299–305. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015, 11, 600–607. [Google Scholar] [CrossRef]
- Tanaka, M.; Mizuno, K.; Yamaguti, K.; Kuratsune, H.; Fujii, A.; Baba, H.; Matsuda, K.; Nishimae, A.; Takesaka, T.; Watanabe, Y. Autonomic nervous alterations associated with daily level of fatigue. Behav. Brain Funct. 2011, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Proctor, G.B.; Carpenter, G.H. Regulation of salivary gland function by autonomic nerves. Auton. Neurosci. 2007, 133, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Pihur, V.; Datta, S.; Datta, S. Meta analysis of Chronic Fatigue Syndrome through integration of clinical, gene expression, SNP and proteomic data. Bioinformation 2011, 6, 120–124. [Google Scholar] [CrossRef][Green Version]
- Casado, B. and Baraniuk, J.N. Abstract: Decreased Mucosal Protein Secretion in the Nonallergic Rhinitis of Chronic Fatigue Syndrome. J. Allergy Clin. Immunol. 2009, 123, S260. [Google Scholar] [CrossRef]
- Maes, M.; Twist, F.N.; Kubera, M.; Ringel, K. Evidence for inflammation and activation of cell-mediated immunity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): Increased interleukin-1, tumor necrosis factor-α, PMN-elastase, lysozyme and neopterin. J. Affect. Disord. 2012, 136, 933–939. [Google Scholar] [CrossRef]
- Savitski, M.M.; Mathieson, T.; Zinn, N.; Sweetman, G.; Doce, C.; Becher, I.; Pachl, F.; Kuster, B.; Bantscheff, M. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 2013, 12, 3586–3598. [Google Scholar] [CrossRef]
- Nie, H.; Ju, H.; Fan, J.; Shi, X.; Cheng, Y.; Cang, X.; Zheng, Z.; Duan, X.; Yi, W. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 2020, 11, 36. [Google Scholar] [CrossRef]
- Barber, T.M.; Bhatti, A.A.; Elder, P.J.D.; Ball, S.P.; Calvez, R.; Ramsden, D.B.; Cuthbertson, D.J.; Pfeiffer, A.F.; Burnett, D.; Weickert, M.O. AMY1 Gene Copy Number Correlates with Glucose Absorption and Visceral Fat Volume, but Not with Insulin Resistance. J. Clin. Endocrinol. Metab. 2020, 105, dgaa473. [Google Scholar] [CrossRef] [PubMed]
- Love, K.M.; Liu, Z. DPP4 Activity, Hyperinsulinemia, and Atherosclerosis. J. Clin. Endocrinol. Metab. 2021, 106, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Mitra, B.; Rau, T.F.; Surendran, N.; Brennan, J.H.; Thaveenthiran, P.; Sorich, E.; Fitzgerald, M.C.; Rosenfeld, J.V.; Patel, S.A. Plasma micro-RNA biomarkers for diagnosis and prognosis after traumatic brain injury: A pilot study. J. Clin. Neurosci. 2017, 38, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Vissing, J.; Akman, H.O.; Aasly, J.; Kahler, S.G.; Bacino, C.A.; DiMauro, S.; Haller, R.G. Level of residual enzyme activity modulates the phenotype in phosphoglycerate kinase deficiency. Neurology 2018, 91, e1077–e1082. [Google Scholar] [CrossRef] [PubMed]
- David, D.; Almeida, L.S.; Maggi, M.; Araújo, C.; Imreh, S.; Valentini, G.; Fekete, G.; Haltrich, I. Clinical Severity of PGK1 Deficiency Due To a Novel p.E120K Substitution Is Exacerbated by Co-inheritance of a Subclinical Translocation t(3;14)(q26.33;q12), Disrupting NUBPL Gene. JIMD Rep. 2015, 23, 55–65. [Google Scholar]
- Boyd, P.J.; Tu, W.-Y.; Shorrock, H.K.; Groen, E.J.N.; Carter, R.N.; Powis, R.A.; Thomson, S.R.; Thomson, D.; Graham, L.C.; Motyl, A.A.L.; et al. Bioenergetic status modulates motor neuron vulnerability and pathogenesis in a zebrafish model of spinal muscular atrophy. PLoS Genet. 2017, 13, e1006744. [Google Scholar] [CrossRef]
- Du, Z.-Q.; Yang, C.-X.; Rothschild, M.F.; Ross, J.W. Novel microRNA families expanded in the human genome. BMC Genom. 2013, 14, 98. [Google Scholar] [CrossRef]
- Han, L.; Zhang, H.; Zeng, Y.; Lv, Y.; Tao, L.; Ma, J.; Xu, H.; Ma, K.; Shi, Q.; Xiao, B.; et al. Identification of the miRNA-3185/CYP4A11 axis in cardiac tissue as a biomarker for mechanical asphyxia. Forensic Sci. Int. 2020, 311, 110293. [Google Scholar] [CrossRef]
- Pascut, D.; Pratama, M.Y.; Gilardi, F.; Giuffrè, M.; Crocè, L.S.; Tiribelli, C. Weighted miRNA co-expression networks analysis identifies circulating miRNA predicting overall survival in hepatocellular carcinoma patients. Sci. Rep. 2020, 10, 18967. [Google Scholar] [CrossRef]
- McNair, D.; Lorr, M.; Droppleman, L. Manual for the Profile of Mood States; Educational and Industrial Testing Service: San Diego, CA, USA, 1971. [Google Scholar]
- McNair, D.M. Profile of Mood States; Educational and Industrial Testing Service: San Diego, CA, USA, 1992. [Google Scholar]
- Heuchert, J.P.; McNair, D.M. Profile of Mood States 2. Multi-Health Systems.APA Psych Tests Database. 2012. Available online: https://psycnet.apa.org/record/9999-05057-000?doi=1 (accessed on 8 February 2022).
- Albrecht, R.R.; Ewing, S.J. Standardizing the administration of the Profile of Mood States (POMS): Development of alternative word lists. J. Pers. Assess 1989, 53, 31–39. [Google Scholar] [CrossRef]
- Heinzelman, P.; Powers, D.; Wohlschlegel, J.; John, V. Shotgun Proteomic Profiling of Bloodborne Nanoscale Extracellular Vesicles. Methods Mol. Biol. 2019, 1897, 403–416. [Google Scholar]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.T.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from Human Saliva as a Source of microRNA Biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef]


| Gene Symbol | Protein Description | Protein Fold Change: Post-/Pre-Work | Protein p-Value: Post-/Pre-Work | miRNA | miRNA Fold Change: Post-/Pre-Work | miRNA p-Value: Post-/Pre-Work |
|---|---|---|---|---|---|---|
| LEG1 | Liver-enriched gene 1 protein | 4.46 | 0.079 | |||
| AMY1A | Alpha-amylase 1 n,c | 1.85 | 0.002 | |||
| BPIFA2 | BPI fold-containing family A member n | 1.64 | 0.006 | |||
| CA6 | Carbonic anhydrase 6 n | 1.5 | 0.083 | |||
| PGK1 | Phosphoglycerate kinase 1 n | 1.34 | 0.08 | hsa-miR-3185 | 0.79 | 0.016 |
| DPP4 | Dipeptidyl peptidase 4 | 1.24 | 0.03 | |||
| SBSN | Suprabasin | −1.23 | 0.06 | |||
| PIGR | Polymeric immunoglobulin receptor m,n,c | −1.28 | 0.063 | hsa-miR-642a-5p | 1.9 | 0.04 |
| CSTB | Cystatin-B n,c | −1.34 | 0.057 | |||
| SPRR3 | Small proline-rich protein 3 | −1.49 | 0.01 | |||
| CRABP2 | Cellular retinoic acid-binding protein 2 | −1.49 | 0.098 | |||
| FABP5 | Fatty acid-binding protein 5 | −1.72 | 0.004 | |||
| YWHAZ | 14-3-3 protein zeta/delta n,c | −2.22 | 0.036 | hsa-miR-134-3p | 1.5 | 0.058 |
| DMBT1 | Deleted in malignant brain tumors 1 n,c | −2.63 | 0.027 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohn, W.; Zhu, C.; Campagna, J.; Bilousova, T.; Spilman, P.; Teter, B.; Li, F.; Guo, R.; Elashoff, D.; Cole, G.M.; et al. Integrated Multiomics Analysis of Salivary Exosomes to Identify Biomarkers Associated with Changes in Mood States and Fatigue. Int. J. Mol. Sci. 2022, 23, 5257. https://doi.org/10.3390/ijms23095257
Cohn W, Zhu C, Campagna J, Bilousova T, Spilman P, Teter B, Li F, Guo R, Elashoff D, Cole GM, et al. Integrated Multiomics Analysis of Salivary Exosomes to Identify Biomarkers Associated with Changes in Mood States and Fatigue. International Journal of Molecular Sciences. 2022; 23(9):5257. https://doi.org/10.3390/ijms23095257
Chicago/Turabian StyleCohn, Whitaker, Chunni Zhu, Jesus Campagna, Tina Bilousova, Patricia Spilman, Bruce Teter, Feng Li, Rong Guo, David Elashoff, Greg M. Cole, and et al. 2022. "Integrated Multiomics Analysis of Salivary Exosomes to Identify Biomarkers Associated with Changes in Mood States and Fatigue" International Journal of Molecular Sciences 23, no. 9: 5257. https://doi.org/10.3390/ijms23095257
APA StyleCohn, W., Zhu, C., Campagna, J., Bilousova, T., Spilman, P., Teter, B., Li, F., Guo, R., Elashoff, D., Cole, G. M., Avidan, A., Faull, K. F., Whitelegge, J., Wong, D. T. W., & John, V. (2022). Integrated Multiomics Analysis of Salivary Exosomes to Identify Biomarkers Associated with Changes in Mood States and Fatigue. International Journal of Molecular Sciences, 23(9), 5257. https://doi.org/10.3390/ijms23095257






