Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Properties of the Catalytic Domain of SARS-CoV-2 3CLpro
2.2. Enzymatic Properties of the Monomeric Form of the Catalytic Domain
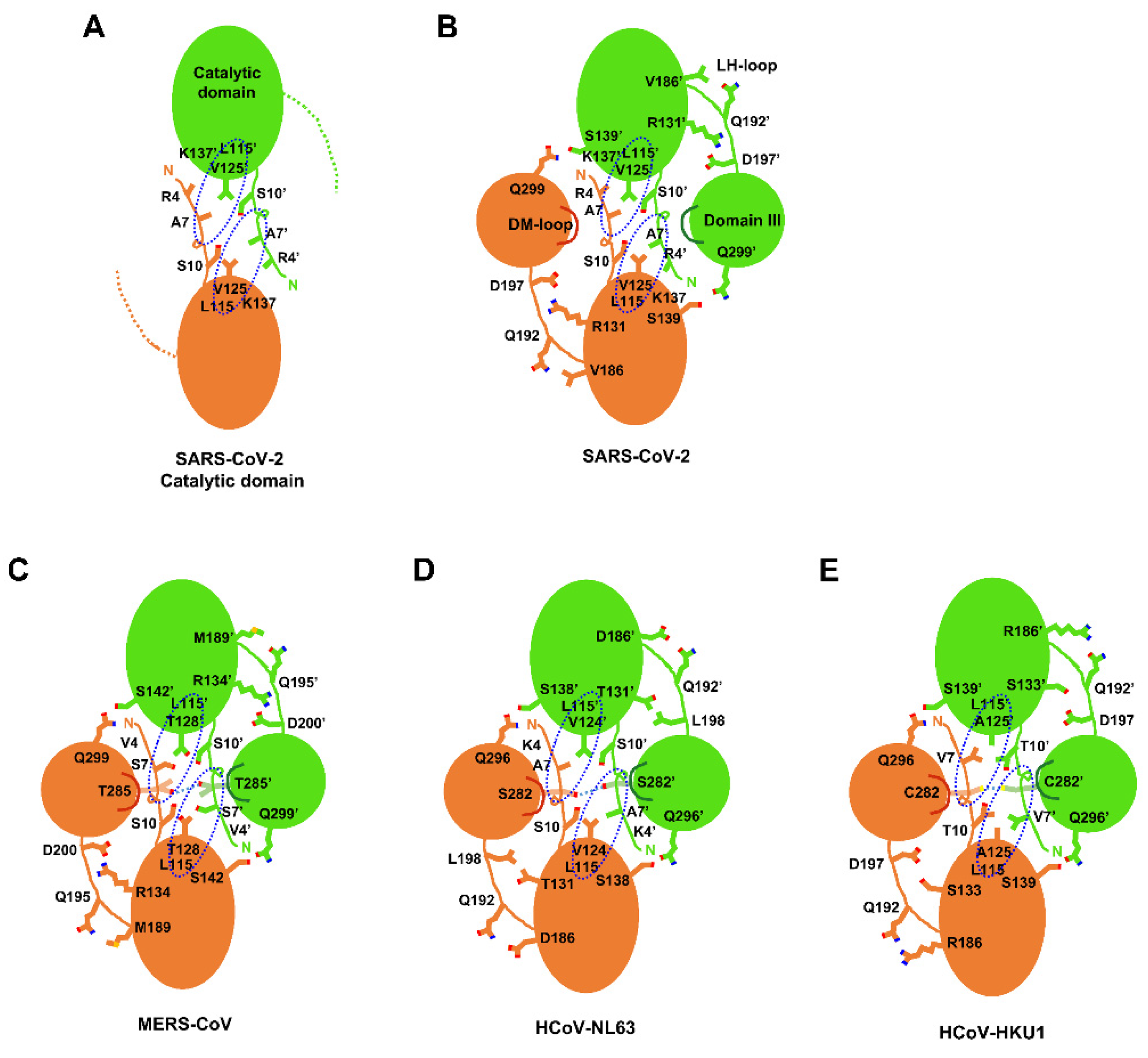
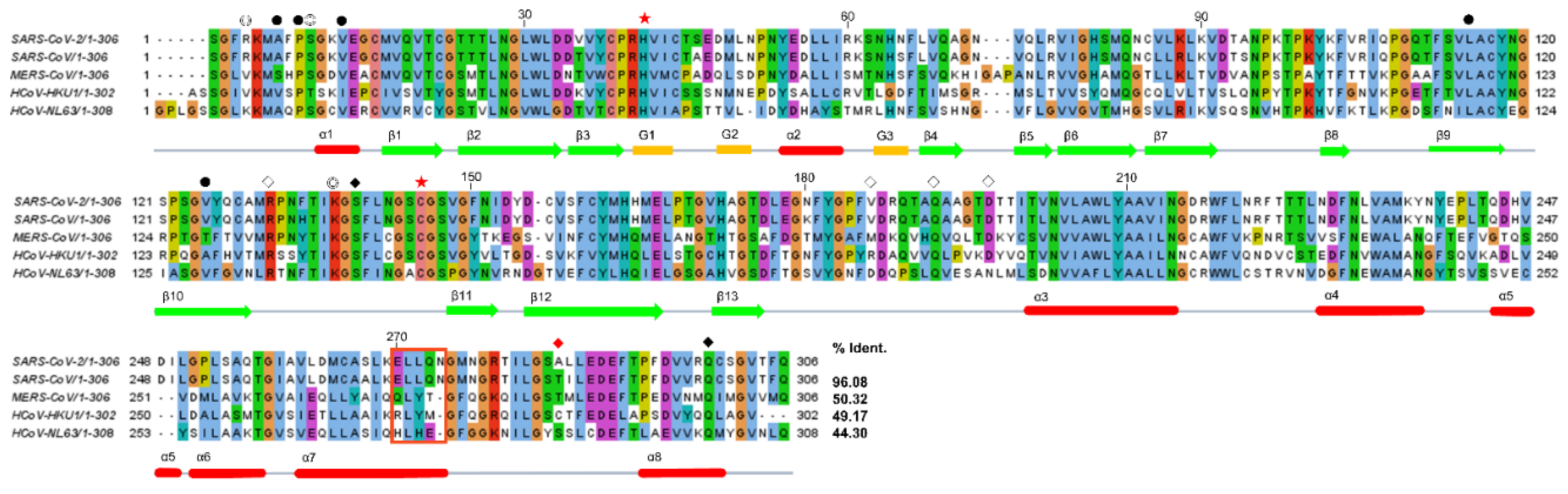
2.3. Structural Comparison among Various 3CLpros
3. Materials and Methods
3.1. Protein Expression and Purification of SARS-CoV-2 3CLpro and its Catalytic Domain
3.2. Enzymatic Kinetic Studies with SARS-CoV-2 3CLpro
3.3. Size-Exlusion Chromatography (SEC)
3.4. Crystallization
3.5. Structure Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.J.; Chen, M.-Y.; Liu, Y.-L.; Witschi, M.; Ardoin, A.; Calba, C.; Mathieu, P.; Masserey, V.; Maraglino, F.; Marro, S.; et al. Early introduction of severe acute respiratory syndrome coronavirus 2 into Europe. Emerg. Infect. Dis. 2020, 26, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Vena, A.; Giacobbe, D.R. The novel Chinese coronavirus (2019-nCoV) infections: Challenges for fighting the storm. Eur. J. Clin. Investig. 2020, 50, e13209. [Google Scholar] [CrossRef]
- Wang, W.; Tang, J.; Wei, F. Updated understanding of the outbreak of 2019 novel coronavirus (2019–nCoV) in Wuhan, China. J. Med. Virol. 2020, 92, 441–447. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Epidemiology Working Group for NCIP Epidemic Response; Chinese Center for Disease Control and Prevention. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020, 10, 145–151. [Google Scholar] [CrossRef]
- Wu, P.; Hao, X.; Lau, E.H.Y.; Wong, J.Y.; Leung, K.S.M.; Wu, J.T.; Cowling, B.J.; Leung, G.M. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Eurosurveillance 2020, 25, 2000044. [Google Scholar] [CrossRef]
- Biggerstaff, M.; Cauchemez, S.; Reed, C.; Gambhir, M.; Finelli, L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: A systematic review of the literature. BMC Infect. Dis. 2014, 14, 480. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, Q.; Ran, J.; Musa, S.S.; Yang, G.; Wang, W.; Lou, Y.; Gao, D.; Yang, L.; He, D.; et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020, 92, 214–217. [Google Scholar] [CrossRef]
- Cao, Y.-C.; Deng, Q.-X.; Dai, S.-X. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med. Infect. Dis. 2020, 35, 101647. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Alshrari, A.S.; Asdaq, S.M.B. Trends in the development of remdesivir based inventions against COVID-19 and other disorders: A patent review. J. Infect. Public Health 2021, 14, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Kelleni, M. Paxlovid and Molnupiravir Approved to Manage COVID-19: A Countdown for SARS CoV-2 Variant Apocalypse. OSF Prepr. 2021, in press. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Kim, D.Y.; Kim, M.-S.; Shin, D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzym. Inhib. Med. Chem. 2020, 35, 1539–1544. [Google Scholar] [CrossRef]
- Zhong, N.; Zhang, S.; Xue, F.; Kang, X.; Zou, P.; Chen, J.; Liang, C.; Rao, Z.; Jin, C.; Lou, Z.; et al. C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer. Protein Sci. 2009, 18, 839–844. [Google Scholar] [CrossRef]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef]
- Shi, J.; Wei, Z.; Song, J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: Defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004, 279, 24765–24773. [Google Scholar] [CrossRef]
- Liang, J.; Karagiannis, C.; Pitsillou, E.; Darmawan, K.K.; Ng, K.; Hung, A.; Karagiannis, T.C. Site mapping and small molecule blind docking reveal a possible target site on the SARS-CoV-2 main protease dimer interface. Comput. Biol. Chem. 2020, 89, 107372. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T.J.B. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Dampalla, C.S.; Zheng, J.; Perera, K.D.; Wong, L.-Y.R.; Meyerholz, D.K.; Nguyen, H.N.; Kashipathy, M.M.; Battaile, K.P.; Lovell, S.; Kim, Y.; et al. Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2101555118. [Google Scholar] [CrossRef] [PubMed]
- Su, H.-X.; Yao, S.; Zhao, W.-F.; Li, M.-J.; Liu, J.; Shang, W.-J.; Xie, H.; Ke, C.-Q.; Hu, H.-C.; Gao, M.-N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Nie, T.; Shao, Q.; Li, M.; Su, H.; Xu, Y. In silico screening-based discovery of novel covalent inhibitors of the SARS-CoV-2 3CL protease. Eur. J. Med. Chem. 2022, 231, 114130. [Google Scholar] [CrossRef] [PubMed]
- Bacha, U.; Barrila, J.; Velazquez-Campoy, A.; Leavitt, A.S.A.; Freire, E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry 2004, 43, 4906–4912. [Google Scholar] [CrossRef]
- Lin, P.-Y.; Chou, C.-Y.; Chang, H.-C.; Hsu, W.-C.; Chang, G.-G. Correlation between dissociation and catalysis of SARS-CoV main protease. Arch. Biochem. Biophys. 2008, 472, 34–42. [Google Scholar] [CrossRef]
- Cheng, S.-C.; Chang, G.-G.; Chou, C.-Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010, 98, 1327–1336. [Google Scholar] [CrossRef]
- Tomar, S.; Johnston, M.L.; John, S.E.S.; Osswald, H.L.; Nyalapatla, P.R.; Paul, L.N.; Ghosh, A.K.; Denison, M.R.; Mesecar, A.D. Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): Implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015, 290, 19403–19422. [Google Scholar] [CrossRef]
- Johansen-Leete, J.; Ullrich, S.; Fry, S.E.; Frkic, R.; Bedding, M.J.; Aggarwal, A.; Ashhurst, A.S.; Ekanayake, K.B.; Mahawaththa, M.C.; Sasi, V.M.; et al. Antiviral cyclic peptides targeting the main protease of SARS-CoV-2. Chem. Sci. 2022, 13, 3826–3836. [Google Scholar] [CrossRef]
- Tripathi, P.K.; Upadhyay, S.; Singh, M.; Raghavendhar, S.; Bhardwaj, M.; Sharma, P.; Patel, A.K. Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int. J. Biol. Macromol. 2020, 164, 2622–2631. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Goyal, B.; Goyal, D. Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy. ACS Comb. Sci. 2020, 22, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Luttens, A.; Gullberg, H.; Abdurakhmanov, E.; Vo, D.D.; Akaberi, D.; Talibov, V.O.; Nekhotiaeva, N.; Vangeel, L.; De Jonghe, S.; Jochmans, D.; et al. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc. 2022, 144, 2905–2920. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Arutyunova, E.; Khan, M.B.; Lu, J.; Joyce, M.A.; Saffran, H.A.; Shields, J.A.; Kandadai, A.S.; Belovodskiy, A.; Hena, M.; et al. Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors. RSC Med. Chem. 2021, 12, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, S.; Xue, F.; Zou, Y.; Chen, C.; Bartlam, M.; Rao, Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J. Virol. 2008, 82, 8647–8655. [Google Scholar] [CrossRef][Green Version]
- Wang, F.; Chen, C.; Tan, W.; Yang, K.; Yang, H. Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design. Sci. Rep. 2016, 6, 22677. [Google Scholar] [CrossRef]
- Kuo, C.J.; Chi, Y.H.; Hsu, J.T.A.; Liang, P.H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004, 318, 862–867. [Google Scholar] [CrossRef]
- Wu, A.; Wang, Y.; Zeng, C.; Huang, X.; Xu, S.; Su, C.; Wang, M.; Chen, Y.; Guo, D. Prediction and biochemical analysis of putative cleavage sites of the 3C-like protease of Middle East respiratory syndrome coronavirus. Virus Res. 2015, 208, 56–65. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Matthews, B. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]


| PDB Code | 7WHC |
|---|---|
| Data collection statistics | |
| Wavelength | 1.0000 Å |
| Resolution | 50.0–2.27 (2.31–2.27) Å |
| Redundancy | 3.4 (3.1) * |
| Unique reflections | 32201 (1203) |
| Completeness | 89.0 (63.2) % |
| I/σ | 17.03 (1.67) |
| Rmergy † | 5.7 (63.2) % |
| Crystal parameters and refinement statistics | |
| Resolution | 31.99–2.27 (2.31–2.27) Å |
| Space group | P21 |
| Cell dimensions | 52.47 Å × 121.09 Å × 62.70 Å |
| Volume fraction of solvent | 48.13% |
| Vm (Å3/Dalton) | 2.37 |
| Total number of residues | 5628 |
| Total non-H atoms | 5778 |
| Number of water molecules | 150 |
| Average temperature factors | |
| Protein | 48.90 Å2 |
| Solvent | 45.70 Å2 |
| R-factor | 20.16% |
| Free R-factor | 26.41% |
| Stereochemical ideality: | |
| Bond | 0.009 Å |
| Angle | 1.297° |
| Chirality | 0.049° |
| Planarity | 0.006° |
| Dihedral | 13.954° |
| Ramachandran plot | |
| Residues in favored regions | 92.78% |
| Residues in allowed regions | 6.94% |
| Residues in disallowed regions | 0.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.; Kim, H.Y.; Shin, D.H.; Kim, M.-S. Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro. Int. J. Mol. Sci. 2022, 23, 5268. https://doi.org/10.3390/ijms23095268
Jo S, Kim HY, Shin DH, Kim M-S. Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro. International Journal of Molecular Sciences. 2022; 23(9):5268. https://doi.org/10.3390/ijms23095268
Chicago/Turabian StyleJo, Seri, Hwa Young Kim, Dong Hae Shin, and Mi-Sun Kim. 2022. "Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro" International Journal of Molecular Sciences 23, no. 9: 5268. https://doi.org/10.3390/ijms23095268
APA StyleJo, S., Kim, H. Y., Shin, D. H., & Kim, M.-S. (2022). Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro. International Journal of Molecular Sciences, 23(9), 5268. https://doi.org/10.3390/ijms23095268





