Effects of CYP3A43 Expression on Cell Proliferation and Migration of Lung Adenocarcinoma and Its Clinical Significance
Abstract
:1. Introduction
2. Results
2.1. Low CYP3A43 Expression Is Linked to Cancer Staging and Lymph Node Metastasis
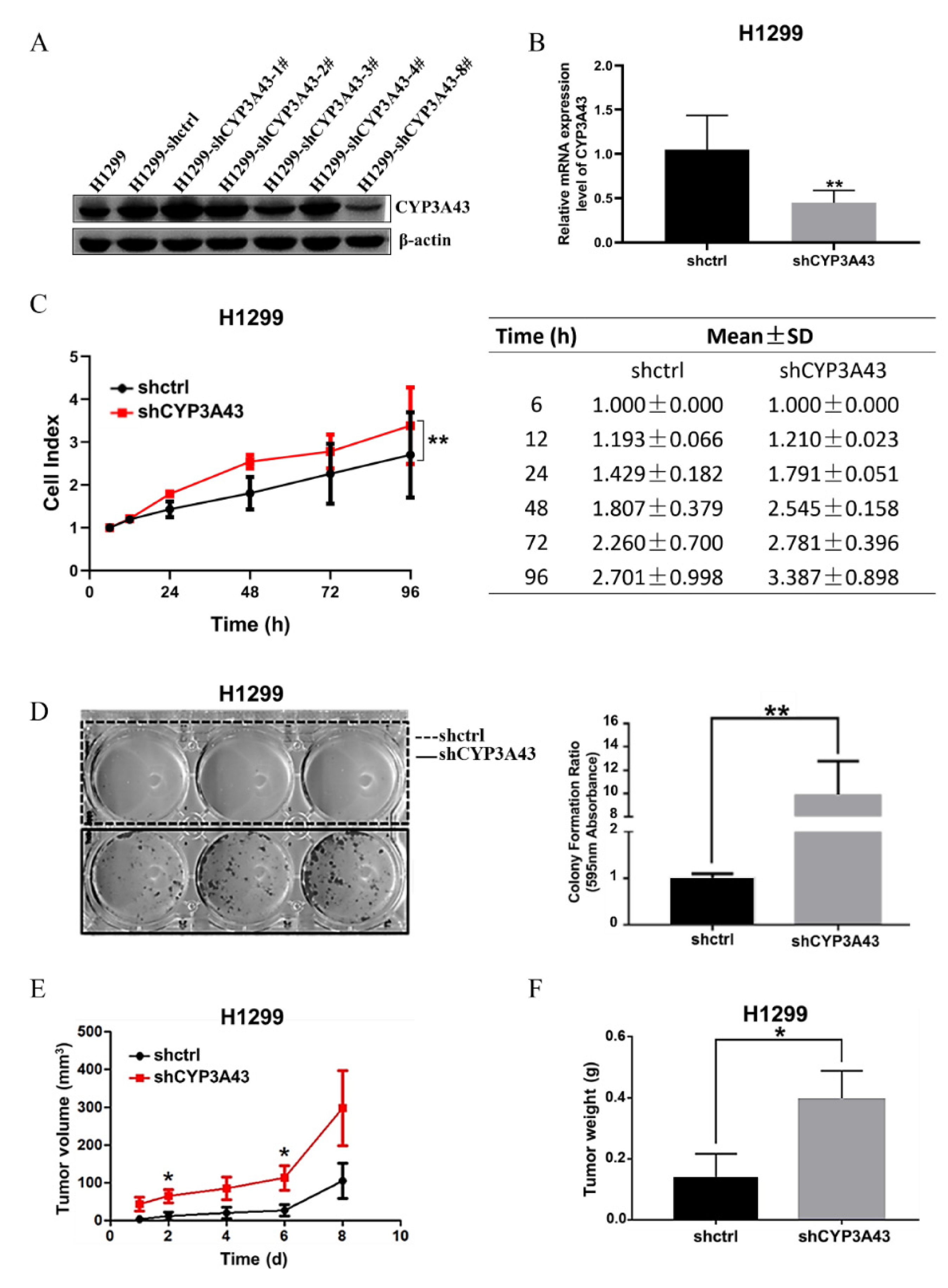
2.2. Stable CYP3A43 Knockdown Promotes Cell Proliferation In Vitro and Tumor Growth In Vivo
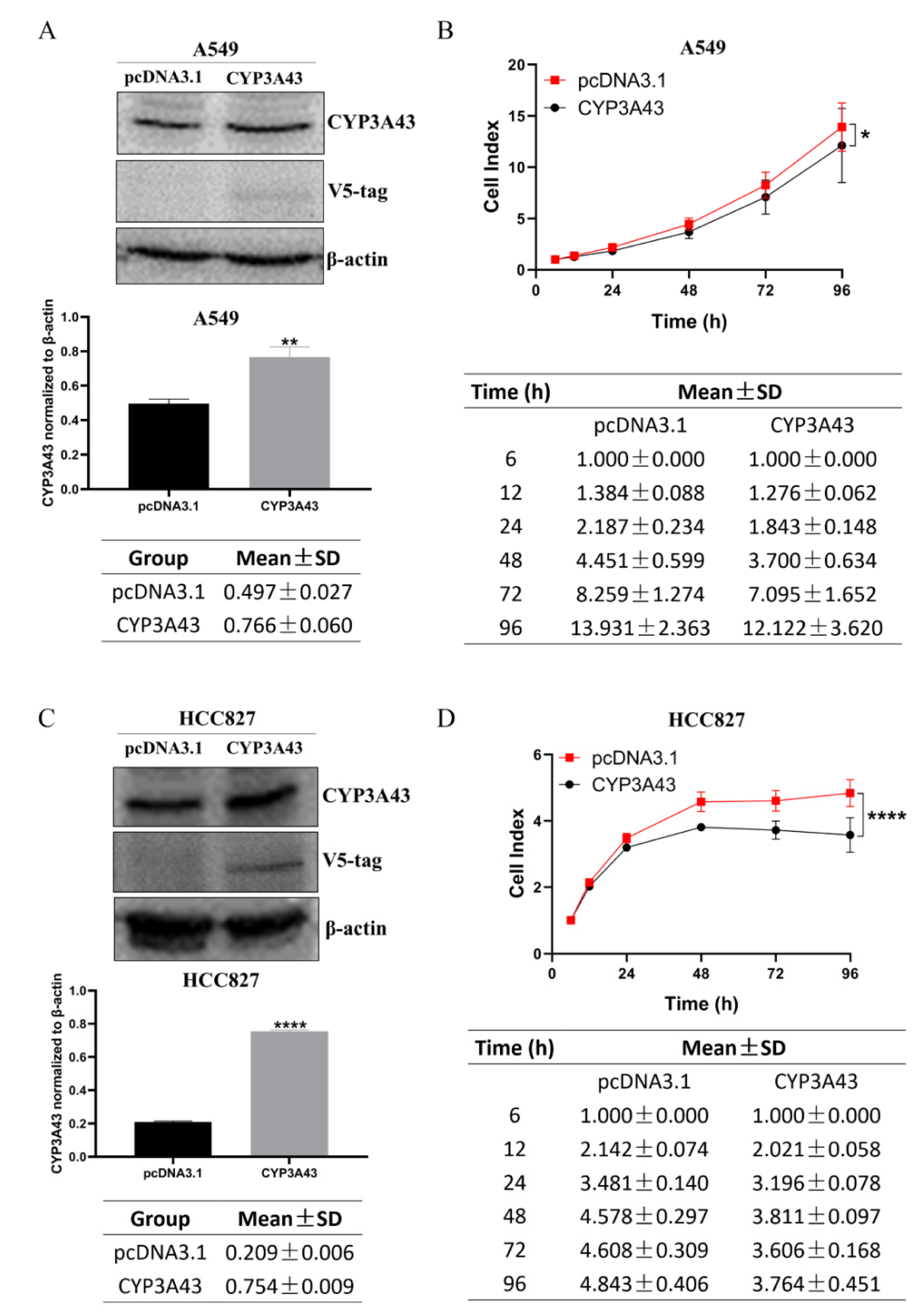
2.3. CYP3A43 Overexpression Inhibits LUAD Cell Proliferation
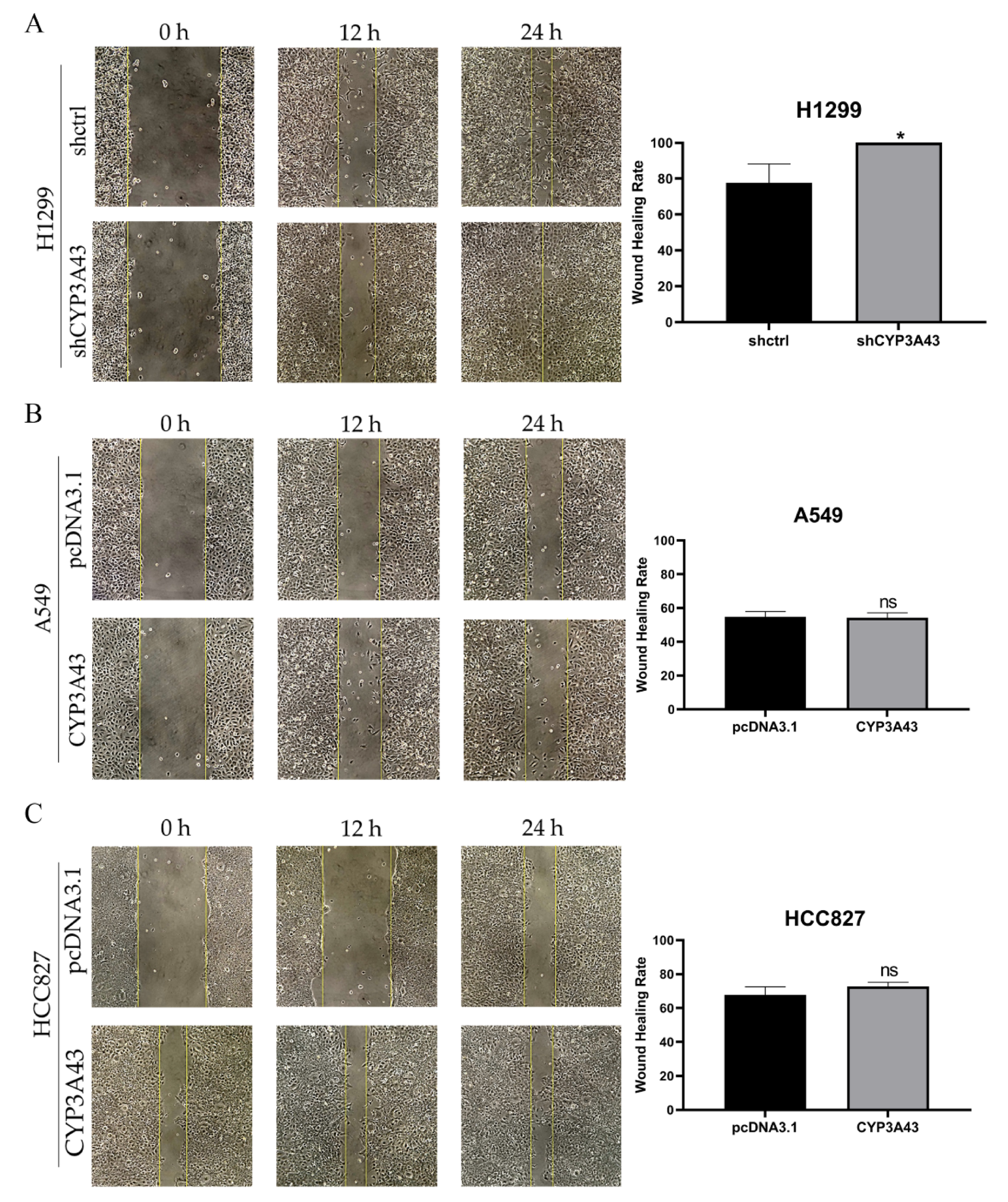
2.4. CYP3A43 Knockdown Promotes H1299 Cell Migration
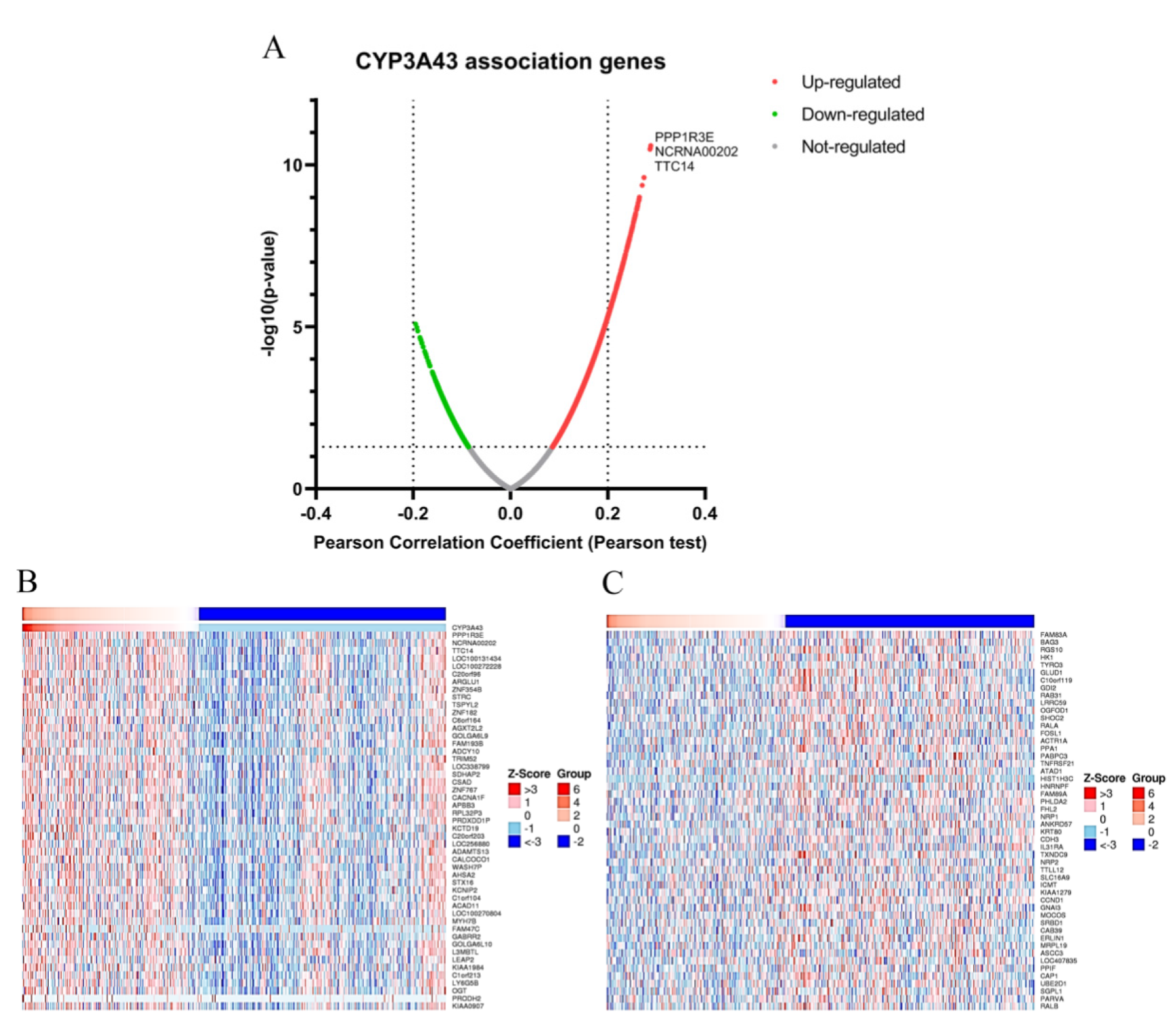
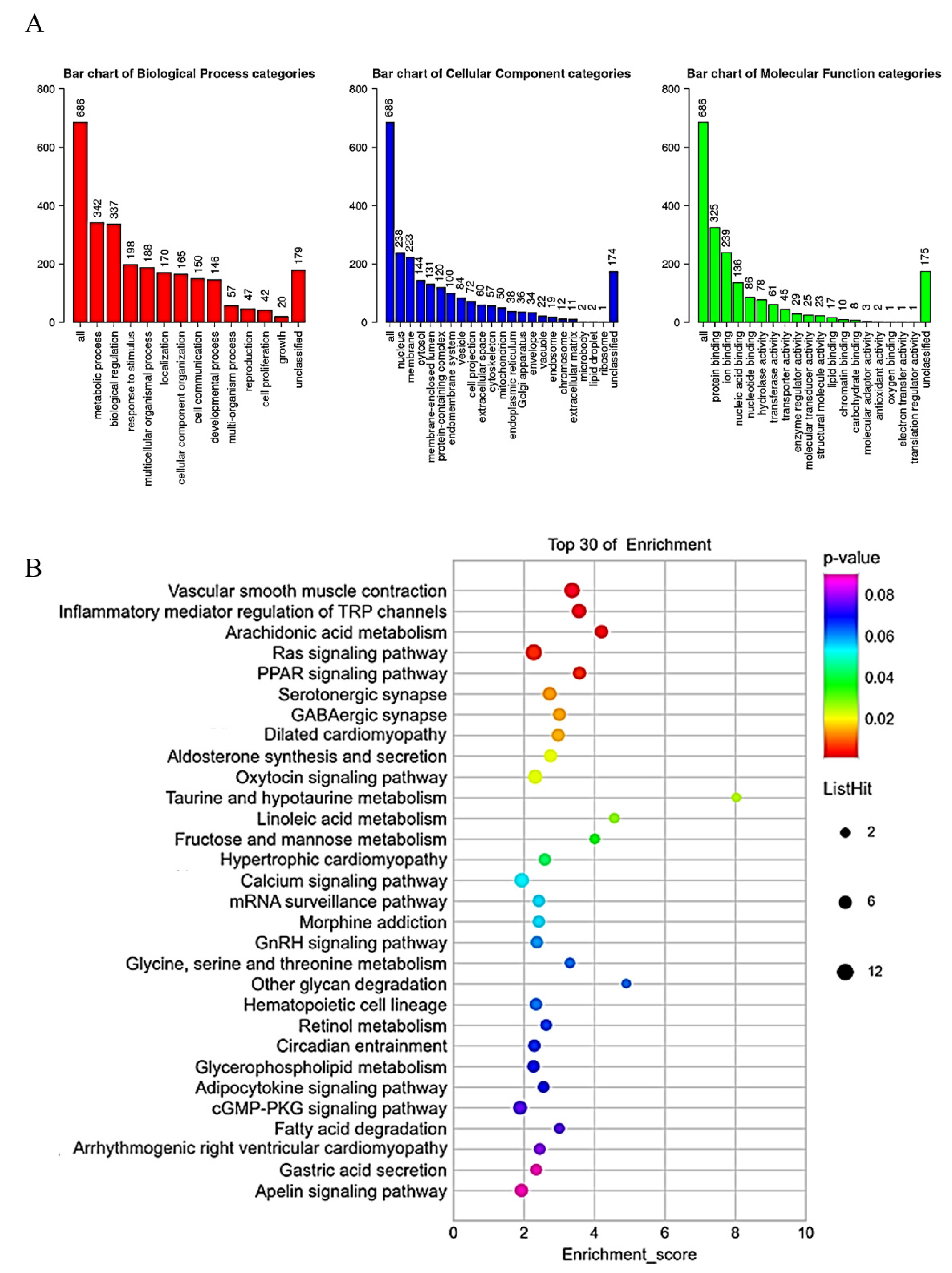
2.5. Gene Set Enrichment Analysis of CYP3A43 Co-Expressed Genes in LUAD
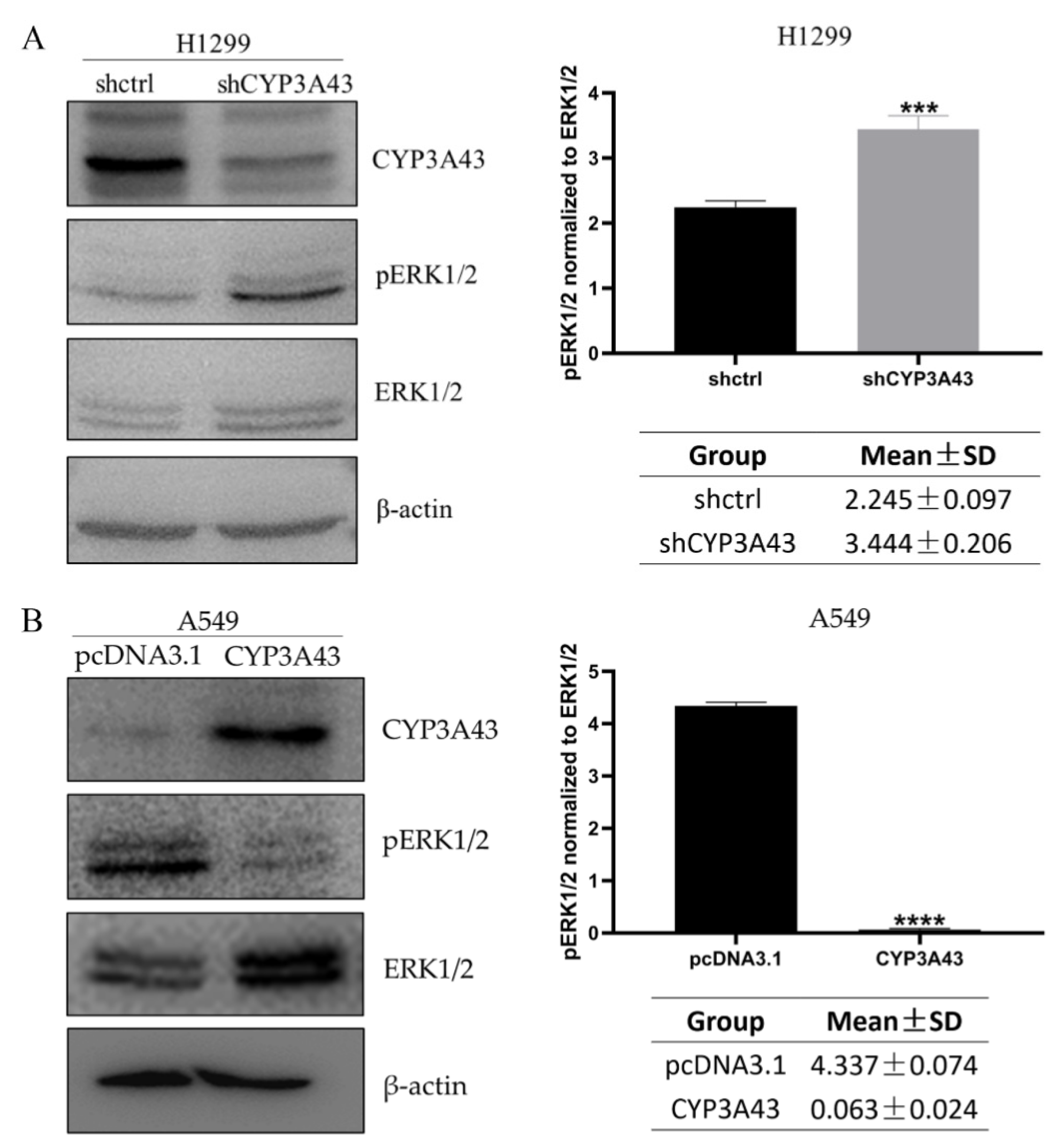
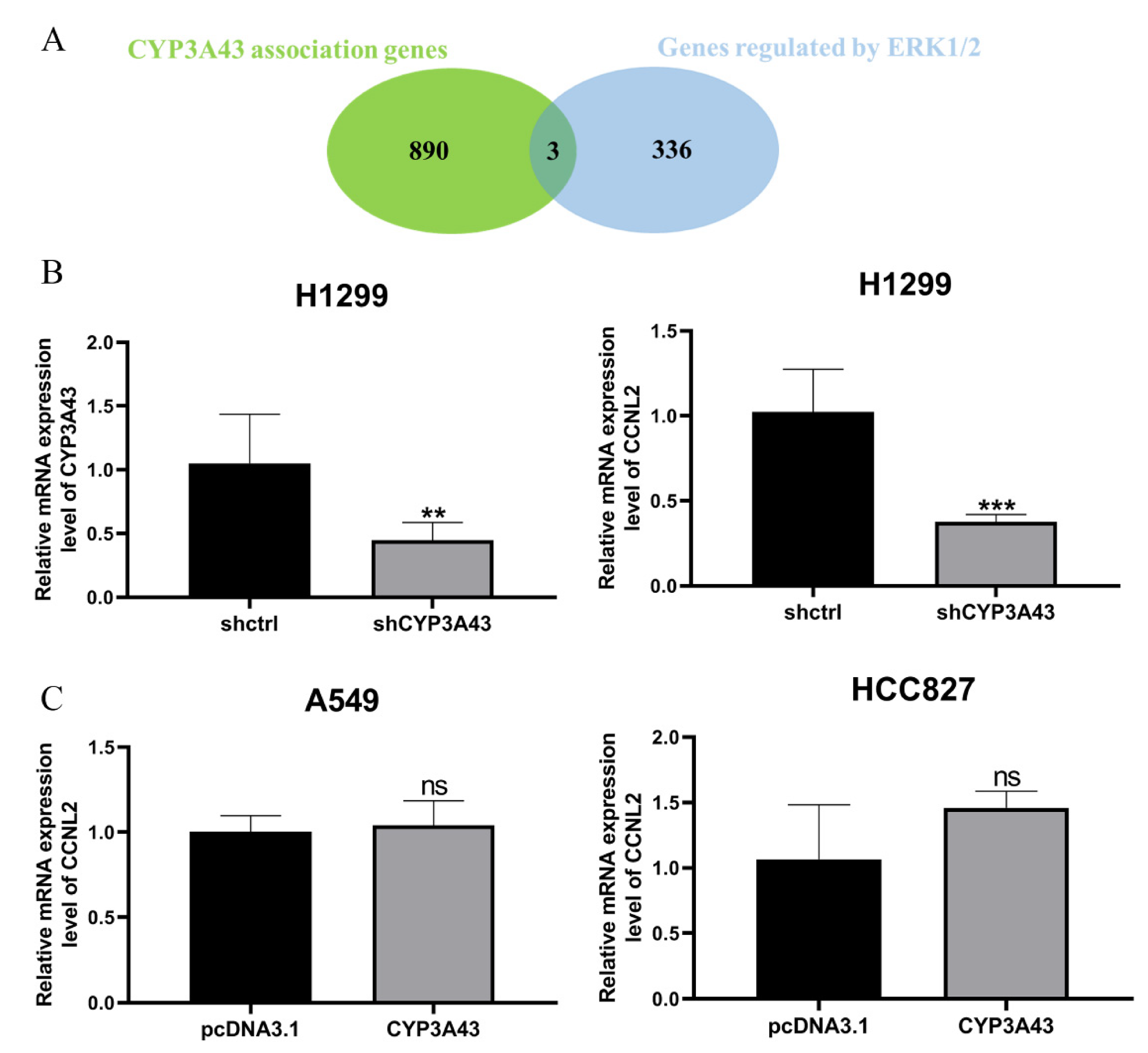
2.6. Involvement of CYP3A43 in ERK1/2 Signaling
3. Discussion
4. Materials and Methods
4.1. UALCAN Database
4.2. Cell Culture and Reagents
4.3. Plasmids, shRNA, and Transfection
4.4. Establishment of the CYP3A43-knockdown H1299 Cell Lines
4.5. Cell Index Assessment
4.6. Colony Formation Assay
4.7. Wound Healing Assay
4.8. In Vivo Xenograft Assay
4.9. LinkedOmics Database
4.10. GO and KEGG Analysis
4.11. RNA Extraction, Reverse Transcription, and RT-qPCR
4.12. Protein Extraction and Immunoblot Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Ward, E.M.; Smith, R.; Jemal, A. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 2013, 119, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
- Luk, P.P.; Selinger, C.I.; Mahar, A.; Cooper, W.A. Biomarkers for ALK and ROS1 in Lung Cancer: Immunohistochemistry and Fluorescent in Situ Hybridization. Arch. Pathol. Lab. Med. 2018, 142, 922–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerman, S.; Das, A.; Wang, S.; Julian, R.; Gandhi, L.; Wolf, J. 2017–2018 Scientific Advances in Thoracic Oncology: Small Cell Lung Cancer. J. Thorac. Oncol. 2019, 14, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Gadgeel, S.M.; Ramalingam, S.S.; Kalemkerian, G.P. Treatment of lung cancer. Radiol. Clin. North Am. 2012, 50, 961–974. [Google Scholar] [CrossRef]
- Liang, G.; Fan, W.; Luo, H.; Zhu, X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomed. Pharmacother. 2020, 128, 110255. [Google Scholar] [CrossRef]
- Ye, Z.; Huang, Y.; Ke, J.; Zhu, X.; Leng, S.; Luo, H. Breakthrough in targeted therapy for non-small cell lung cancer. Biomed. Pharmacother. 2021, 133, 111079. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef] [Green Version]
- Guengerich, F.P. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 1–17. [Google Scholar] [CrossRef]
- Evans, W.E.; Relling, M.V. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science 1999, 286, 487–491. [Google Scholar] [CrossRef]
- Danielson, P.B. The cytochrome P450 superfamily: Biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 2002, 3, 561–597. [Google Scholar] [CrossRef] [PubMed]
- Gellner, K.; Eiselt, R.; Hustert, E.; Arnold, H.; Koch, I.; Haberl, M.; Deglmann, C.J.; Burk, O.; Buntefuss, D.; Escher, S.; et al. Genomic organization of the human CYP3A locus: Identification of a new, inducible CYP3A gene. Pharmacogenetics 2001, 11, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Zeigler-Johnson, C.; Friebel, T.; Walker, A.H.; Wang, Y.; Spangler, E.; Panossian, S.; Patacsil, M.; Aplenc, R.; Wein, A.J.; Malkowicz, S.B.; et al. CYP3A4, CYP3A5, and CYP3A43 genotypes and haplotypes in the etiology and severity of prostate cancer. Cancer Res. 2004, 64, 8461–8467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, A.; Ratnasinghe, L.D.; Emerson, G.L.; Modali, R.; Lehman, T.; Runnells, G.; Carroll, A.; Carter, W.; Barnhart, S.; Rasheed, A.A.; et al. CYP3A43 Pro(340)Ala polymorphism and prostate cancer risk in African Americans and Caucasians. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 1257–1261. [Google Scholar] [CrossRef] [Green Version]
- Justenhoven, C.; Winter, S.; Hamann, U.; Haas, S.; Fischer, H.P.; Pesch, B.; Brüning, T.; Ko, Y.D.; Brauch, H. The frameshift polymorphism CYP3A43_74_delA is associated with poor differentiation of breast tumors. Cancer 2010, 116, 5358–5364. [Google Scholar] [CrossRef]
- Yu, T.; Wang, X.; Zhu, G.; Han, C.; Su, H.; Liao, X.; Yang, C.; Qin, W.; Huang, K.; Peng, T. The prognostic value of differentially expressed CYP3A subfamily members for hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 1713–1726. [Google Scholar] [CrossRef] [Green Version]
- Downie, D.; McFadyen, M.C.; Rooney, P.H.; Cruickshank, M.E.; Parkin, D.E.; Miller, I.D.; Telfer, C.; Melvin, W.T.; Murray, G.I. Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin. Cancer Res. 2005, 11, 7369–7375. [Google Scholar] [CrossRef] [Green Version]
- Elia, I.; Doglioni, G.; Fendt, S.M. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018, 28, 673–684. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [Green Version]
- Roskoski, R., Jr. ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef]
- van Eijk, M.; Boosman, R.J.; Schinkel, A.H.; Huitema, A.D.R.; Beijnen, J.H. Cytochrome P450 3A4, 3A5, and 2C8 expression in breast, prostate, lung, endometrial, and ovarian tumors: Relevance for resistance to taxanes. Cancer Chemother. Pharmacol. 2019, 84, 487–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.C.; Cheung, S.T.; Leung, K.L.; Ng, I.O.; Fan, S.T. Decreased expression of cytochrome P450 2E1 is associated with poor prognosis of hepatocellular carcinoma. Int. J. Cancer 2004, 111, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fan, X. Expression of cytochrome P450 2A13 in human non-small cell lung cancer and its clinical significance. J. Biomed. Res. 2013, 27, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvador-Barbero, B.; Alvarez-Fernández, M.; Zapatero-Solana, E.; El Bakkali, A.; Menéndez, M.D.C.; López-Casas, P.P.; Di Domenico, T.; Xie, T.; VanArsdale, T.; Shields, D.J.; et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 2020, 38, 584. [Google Scholar] [CrossRef] [PubMed]
- Tsantoulis, P.K.; Gorgoulis, V.G. Involvement of E2F transcription factor family in cancer. Eur. J. Cancer 2005, 41, 2403–2414. [Google Scholar] [CrossRef]
- Brambilla, E.; Gazdar, A. Pathogenesis of lung cancer signalling pathways: Roadmap for therapies. Eur. Respir. J. 2009, 33, 1485–1497. [Google Scholar] [CrossRef]
- Yuan, M.; Zhao, Y.; Arkenau, H.T.; Lao, T.; Chu, L.; Xu, Q. Signal pathways and precision therapy of small-cell lung cancer. Signal Transduct. Target. Ther. 2022, 7, 187. [Google Scholar] [CrossRef]
- Zhou, B.; Der, C.J.; Cox, A.D. The role of wild type RAS isoforms in cancer. Semin. Cell Dev. Biol. 2016, 58, 60–69. [Google Scholar] [CrossRef] [Green Version]
- Olson, M.F.; Marais, R. Ras protein signalling. Semin. Immunol. 2000, 12, 63–73. [Google Scholar] [CrossRef]
- Lau, A.T.Y.; Xu, Y.M. Regulation of human mitogen-activated protein kinase 15 (extracellular signal-regulated kinase 7/8) and its functions: A recent update. J. Cell. Physiol. 2019, 234, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicent, S.; López-Picazo, J.M.; Toledo, G.; Lozano, M.D.; Torre, W.; Garcia-Corchón, C.; Quero, C.; Soria, J.C.; Martín-Algarra, S.; Manzano, R.G.; et al. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br. J. Cancer 2004, 90, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, Y.; Zhu, S.; Liang, W.; Wang, Z.; Wang, Y.; Lv, T.; Yao, Y.; Yuan, D.; Song, Y. PTP1B promotes cell proliferation and metastasis through activating src and ERK1/2 in non-small cell lung cancer. Cancer Lett. 2015, 359, 218–225. [Google Scholar] [CrossRef]
- Kurgan, N.; Tsakiridis, E.; Kouvelioti, R.; Moore, J.; Klentrou, P.; Tsiani, E. Inhibition of Human Lung Cancer Cell Proliferation and Survival by Post-Exercise Serum Is Associated with the Inhibition of Akt, mTOR, p70 S6K, and Erk1/2. Cancers 2017, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Huang, S. Fisetin inhibits the growth and migration in the A549 human lung cancer cell line via the ERK1/2 pathway. Exp. Ther. Med. 2018, 15, 2667–2673. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Li, N.; Wang, C.; Yu, Y.; Yuan, L.; Zhang, M.; Cao, X. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J. Biol. Chem. 2004, 279, 11639–11648. [Google Scholar] [CrossRef] [Green Version]
- Li, H.L.; Wang, T.S.; Li, X.Y.; Li, N.; Huang, D.Z.; Chen, Q.; Ba, Y. Overexpression of cyclin L2 induces apoptosis and cell-cycle arrest in human lung cancer cells. Chin. Med. J. 2007, 120, 905–909. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Q.-Y.; Lau, A.T.Y.; Mo, H.-Y.; Zhong, Q.-H.; Zhao, X.-Y.; Yu, F.-Y.; Han, J.; Wu, Y.-Y.; Xu, Y.-M. Effects of CYP3A43 Expression on Cell Proliferation and Migration of Lung Adenocarcinoma and Its Clinical Significance. Int. J. Mol. Sci. 2023, 24, 113. https://doi.org/10.3390/ijms24010113
Wei Q-Y, Lau ATY, Mo H-Y, Zhong Q-H, Zhao X-Y, Yu F-Y, Han J, Wu Y-Y, Xu Y-M. Effects of CYP3A43 Expression on Cell Proliferation and Migration of Lung Adenocarcinoma and Its Clinical Significance. International Journal of Molecular Sciences. 2023; 24(1):113. https://doi.org/10.3390/ijms24010113
Chicago/Turabian StyleWei, Qi-Yao, Andy T. Y. Lau, Hai-Ying Mo, Qiu-Hua Zhong, Xiao-Yun Zhao, Fei-Yuan Yu, Jin Han, Yu-Yao Wu, and Yan-Ming Xu. 2023. "Effects of CYP3A43 Expression on Cell Proliferation and Migration of Lung Adenocarcinoma and Its Clinical Significance" International Journal of Molecular Sciences 24, no. 1: 113. https://doi.org/10.3390/ijms24010113




