Quality Control of Human Pluripotent Stem Cell Colonies by Computational Image Analysis Using Convolutional Neural Networks
Abstract
:1. Introduction
2. Results
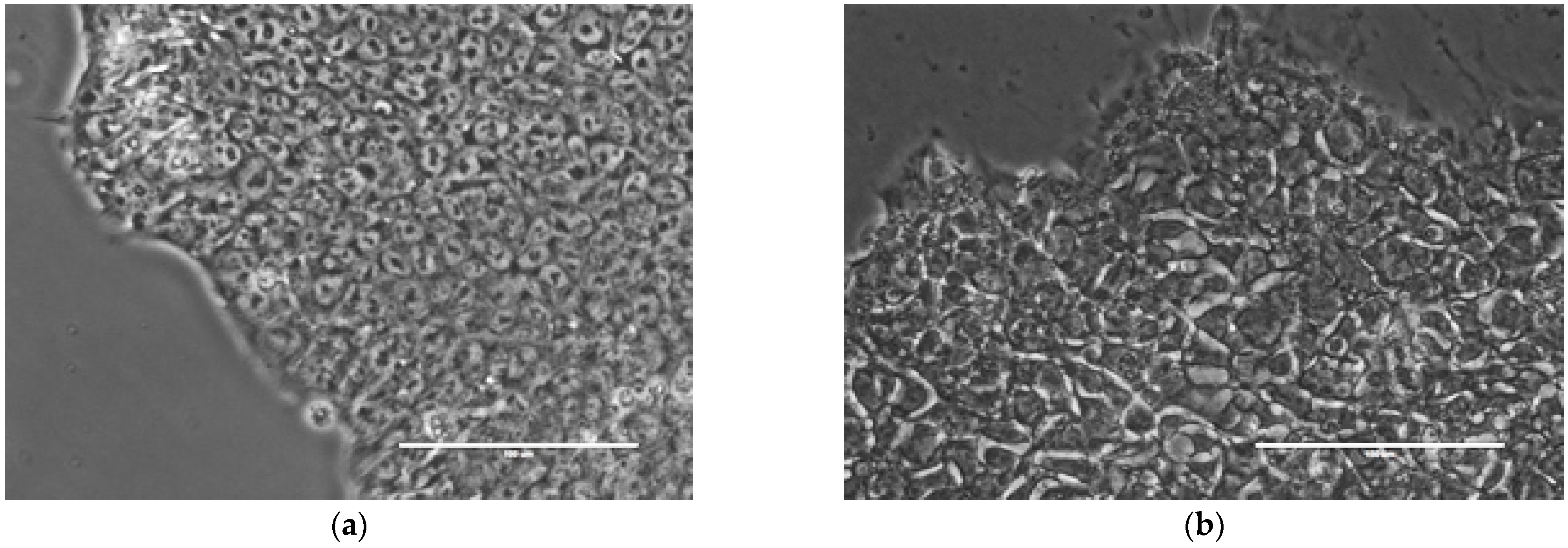
2.1. Image Acquisition and Phenotyping
2.2. CNN-Based Automated Classification of hPSC Colonies according to Their Phenotype
2.3. Characteristic Spatial Scale for Assessing the Morphological Phenotype
2.4. Proteome Analysis in H9 Cells with Good and Bad Phenotype
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Image Acquisition, and Colony Phenotyping
4.2. Image Preprocessing and Augmentation
4.3. CNN Model Selection and Training
4.4. Proteome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soldner, F.; Jaenisch, R. Stem cells, genome editing, and the path to translational medicine. Cell 2018, 175, 615–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, S.; Blau, H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010, 465, 704–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradi, S.; Mahdizadeh, H.; Šarić, T.; Kim, J.; Harati, J.; Shahsavarani, H.; Greber, B.; Moore, J.B. 4th. Research and therapy with induced pluripotent stem cells (iPSCs): Social, legal, and ethical considerations. Stem Cell Res. Ther. 2019, 10, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Pu, J. Differentiation and application of human pluripotent stem cells derived cardiovascular cells for treatment of heart diseases: Promises and challenges. Front. Cell Dev. Biol. 2021, 9, 658088. [Google Scholar] [CrossRef] [PubMed]
- Allegrucci, C.; Young, L.E. Differences between human embryonic stem cell lines. Hum. Reprod. Update 2007, 13, 103–120. [Google Scholar] [CrossRef]
- Allegrucci, C.; Wu, Y.Z.; Thurston, A.; Denning, C.N.; Priddle, H.; Mummery, C.L.; Ward-van Oostwaard, D.; Andrews, P.W.; Stojkovic, M.; Smith, N.; et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum. Mol. Genet. 2007, 16, 1253–1268. [Google Scholar] [CrossRef] [Green Version]
- Daniszewski, M.; Crombie, D.E.; Henderson, R.; Liang, H.H.; Wong, R.C.B.; Hewitt, A.W.; Pébay, A. Automated cell culture systems and their applications to human pluripotent stem cell studies. SLAS Technol. 2018, 23, 315–325. [Google Scholar] [CrossRef] [Green Version]
- Shariatzadeh, M.; Chandra, A.; Wilson, S.L.; McCall, M.J.; Morizur, L.; Lesueur, L.; Chose, O.; Gepp, M.M.; Schulz, A.; Neubauer, J.C.; et al. Distributed automated manufacturing of pluripotent stem cell products. Int. J. Adv. Manuf. Technol. 2020, 106, 1085–1103. [Google Scholar] [CrossRef] [Green Version]
- Elanzew, A.; Nießing, B.; Langendoerfer, D.; Rippel, O.; Piotrowski, T.; Schenk, F.; Kulik, M.; Peitz, M.; Breitkreuz, Y.; Jung, S.; et al. The StemCellFactory: A modular system integration for automated generation and expansion of human induced pluripotent stem cells. Front. Bioeng. Biotechnol. 2020, 8, 580352. [Google Scholar] [CrossRef]
- Tristan, C.A.; Ormanoglu, P.; Slamecka, J.; Malley, C.; Chu, P.H.; Jovanovic, V.M.; Gedik, Y.; Jethmalani, Y.; Bonney, C.; Barnaeva, E.; et al. Robotic high-throughput biomanufacturing and functional differentiation of human pluripotent stem cells. Stem Cell Rep. 2021, 16, 3076–3092. [Google Scholar] [CrossRef]
- Krasnova, O.A.; Gursky, V.V.; Chabina, A.S.; Kulakova, K.A.; Alekseenko, L.L.; Panova, A.V.; Kiselev, S.L.; Neganova, I.E. Prognostic analysis of human pluripotent stem cells based on their morphological portrait and expression of pluripotent markers. Int. J. Mol. Sci. 2022, 23, 12902. [Google Scholar] [CrossRef] [PubMed]
- Eliceiri, K.W.; Berthold, M.R.; Goldberg, I.G.; Ibáñez, L.; Manjunath, B.S.; Martone, M.E.; Murphy, R.F.; Peng, H.; Plant, A.L.; Roysam, B.; et al. Biological imaging software tools. Nat. Methods 2012, 9, 697–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronnello, C.; Francipane, M.G. Moving towards induced pluripotent stem cell-based therapies with artificial intelligence and machine learning. Stem Cell Rev. Rep. 2022, 18, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.V.; Murphy, R.F. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics 2001, 17, 1213–1223. [Google Scholar] [CrossRef] [Green Version]
- Orlov, N.; Shamir, L.; Macura, T.; Johnston, J.; Eckley, D.M.; Goldberg, I.G. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognit. Lett. 2008, 29, 1684–1693. [Google Scholar] [CrossRef] [Green Version]
- Ponomarev, G.V.; Arlazarov, V.L.; Gelfand, M.S.; Kazanov, M.D. ANA HEp-2 cells image classification using number, size, shape and localization of targeted cell regions. Pattern Recognit. 2014, 47, 2360–2366. [Google Scholar] [CrossRef] [Green Version]
- Shamir, L.; Orlov, N.; Eckley, D.M.; Macura, T.; Johnston, J.; Goldberg, I.G. Wndchrm—An open source utility for biological image analysis. Source Code Biol. Med. 2008, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Tokunaga, K.; Saitoh, N.; Goldberg, I.G.; Sakamoto, C.; Yasuda, Y.; Yoshida, Y.; Yamanaka, S.; Nakao, M. Computational image analysis of colony and nuclear morphology to evaluate human induced pluripotent stem cells. Sci. Rep. 2014, 4, 6996. [Google Scholar] [CrossRef] [Green Version]
- Kato, R.; Matsumoto, M.; Sasaki, H.; Joto, R.; Okada, M.; Ikeda, Y.; Kanie, K.; Suga, M.; Kinehara, M.; Yanagihara, K.; et al. Parametric analysis of colony morphology of non-labelled live human pluripotent stem cells for cell quality control. Sci. Rep. 2016, 6, 34009. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Zhang, S.; Zhang, Y.; Lu, J.; Holcombe, M.; Zhang, X. A machine learning assisted, label-free, non-invasive approach for somatic reprogramming in induced pluripotent stem cell colony formation detection and prediction. Sci. Rep. 2017, 7, 13496. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wang, L.; Zhou, L.; Zhang, J. HEp-2 cell image classification with deep convolutional neural networks. IEEE J. Biomed. Health Inform. 2017, 21, 416–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Harrison, P.J.; Wieslander, H.; Pielawski, N.; Kartasalo, K.; Partel, G.; Solorzano, L.; Suveer, A.; Klemm, A.H.; Spjuth, O.; et al. Deep learning in image cytometry: A review. Cytom. A 2019, 95, 366–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kensert, A.; Harrison, P.J.; Spjuth, O. Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov. 2019, 24, 466–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowski, T.; Rippel, O.; Elanzew, A.; Nießing, B.; Stucken, S.; Jung, S.; König, N.; Haupt, S.; Stappert, L.; Brüstle, O.; et al. Deep-learning-based multi-class segmentation for automated, non-invasive routine assessment of human pluripotent stem cell culture status. Comput. Bio.l Med. 2021, 129, 104172. [Google Scholar] [CrossRef] [PubMed]
- Fischbacher, B.; Hedaya, S.; Hartley, B.J.; Wang, Z.; Lallos, G.; Hutson, D.; Zimmer, M.; Brammer, J.; Paull, D.; The NYSCF Global Stem Cell Array Team. Modular deep learning enables automated identification of monoclonal cell lines. Nat. Mach. Intell. 2021, 3, 632–640. [Google Scholar] [CrossRef]
- Shifat-E-Rabbi, M.; Yin, X.; Fitzgerald, C.E.; Rohde, G.K. Cell image classification: A comparative overview. Cytom. A 2020, 97, 347–362. [Google Scholar] [CrossRef] [Green Version]
- Bjørlykke, Y.; Søviknes, A.M.; Hoareau, L.; Vethe, H.; Mathisen, A.F.; Chera, S.; Vaudel, M.; Ghila, L.M.; Ræder, H. Reprogrammed cells display distinct proteomic signatures associated with colony morphology variability. Stem Cells Int. 2019, 2019, 8036035. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Barbaric, I.; Biga, V.; Gokhale, P.J.; Jones, M.; Stavish, D.; Glen, A.; Coca, D.; Andrews, P.W. Time-lapse analysis of human embryonic stem cells reveals multiple bottlenecks restricting colony formation and their relief upon culture adaptation. Stem Cell Rep. 2014, 3, 142–155. [Google Scholar] [CrossRef]
- Maddah, M.; Shoukat-Mumtaz, U.; Nassirpour, S.; Loewke, K. A system for automated, noninvasive, morphology-based evaluation of induced pluripotent stem cell cultures. J. Lab. Autom. 2014, 19, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Konagaya, S.; Ando, T.; Yamauchi, T.; Suemori, H.; Iwata, H. Long-term maintenance of human induced pluripotent stem cells by automated cell culture system. Sci. Rep. 2015, 5, 16647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paull, D.; Sevilla, A.; Zhou, H.; Hahn, A.K.; Kim, H.; Napolitano, C.; Tsankov, A.; Shang, L.; Krumholz, K.; Jagadeesan, P.; et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods 2015, 12, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Archibald, P.R.; Chandra, A.; Thomas, D.; Chose, O.; Massouridès, E.; Laâbi, Y.; Williams, D.J. Comparability of automated human induced pluripotent stem cell culture: A pilot study. Bioprocess Biosyst. Eng. 2016, 39, 1847–1858. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, T.E.; Bergendahl, V.; Levenstein, M.E.; Yu, J.; Probasco, M.D.; Thomson, J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods 2006, 3, 637–646. [Google Scholar] [CrossRef]
- Chen, G.; Gulbranson, D.R.; Hou, Z.; Bolin, J.M.; Ruotti, V.; Probasco, M.D.; Smuga-Otto, K.; Howden, S.E.; Diol, N.R.; Propson, N.E.; et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 2011, 8, 424–429. [Google Scholar] [CrossRef] [Green Version]
- Harkness, L.; Chen, X.; Gillard, M.; Gray, P.P.; Davies, A.M. Media composition modulates human embryonic stem cell morphology and may influence preferential lineage differentiation potential. PLoS ONE 2019, 14, e0213678. [Google Scholar] [CrossRef] [Green Version]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 2014, 14, 1006-0. [Google Scholar] [CrossRef]
- Novikova, S.; Tolstova, T.; Kurbatov, L.; Farafonova, T.; Tikhonova, O.; Soloveva, N.; Rusanov, A.; Archakov, A.; Zgoda, V. Nuclear proteomics of induced leukemia cell differentiation. Cells 2022, 11, 3221. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]



| Model Configuration | Quality Measures | ||||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | AUC | |
| VGG13 | 0.83 | 0.85 | 0.81 | 0.83 | 0.99 |
| VGG13–FirstPool4 | 0.80 | 0.88 | 0.74 | 0.81 | 0.99 |
| VGG12 | 0.74 | 0.81 | 0.70 | 0.75 | 0.98 |
| VGG12–FirstPool4 | 0.69 | 0.92 | 0.62 | 0.74 | 0.95 |
| Res + VGG13 | 0.80 | 0.85 | 0.76 | 0.80 | 0.98 |
| Preprocessing Method | Quality Measures | ||||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | AUC | |
| no preprocessing | 0.83 | 0.85 | 0.81 | 0.83 | 0.99 |
| gray level transform | 0.80 | 0.92 | 0.73 | 0.81 | 0.99 |
| binarization | 0.70 | 0.93 | 0.63 | 0.76 | 0.98 |
| normalization | 0.80 | 0.85 | 0.76 | 0.80 | 0.99 |
| histogram equalization | 0.84 | 0.93 | 0.77 | 0.84 | 0.99 |
| Augmentation Method | Quality Measures | ||||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | AUC | |
| no augmentation | 0.84 | 0.93 | 0.77 | 0.84 | 0.99 |
| rotations | 0.85 | 0.85 | 0.85 | 0.85 | 0.98 |
| cropping | 0.85 | 0.92 | 0.80 | 0.86 | 0.99 |
| rotations + cropping | 0.89 | 0.93 | 0.86 | 0.89 | 0.99 |
| Predicted: Good | Predicted: Bad | |
|---|---|---|
| Actual: Good | 24 | 2 |
| Actual: Bad | 4 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamaeva, A.; Krasnova, O.; Khvorova, I.; Kozlov, K.; Gursky, V.; Samsonova, M.; Tikhonova, O.; Neganova, I. Quality Control of Human Pluripotent Stem Cell Colonies by Computational Image Analysis Using Convolutional Neural Networks. Int. J. Mol. Sci. 2023, 24, 140. https://doi.org/10.3390/ijms24010140
Mamaeva A, Krasnova O, Khvorova I, Kozlov K, Gursky V, Samsonova M, Tikhonova O, Neganova I. Quality Control of Human Pluripotent Stem Cell Colonies by Computational Image Analysis Using Convolutional Neural Networks. International Journal of Molecular Sciences. 2023; 24(1):140. https://doi.org/10.3390/ijms24010140
Chicago/Turabian StyleMamaeva, Anastasiya, Olga Krasnova, Irina Khvorova, Konstantin Kozlov, Vitaly Gursky, Maria Samsonova, Olga Tikhonova, and Irina Neganova. 2023. "Quality Control of Human Pluripotent Stem Cell Colonies by Computational Image Analysis Using Convolutional Neural Networks" International Journal of Molecular Sciences 24, no. 1: 140. https://doi.org/10.3390/ijms24010140
APA StyleMamaeva, A., Krasnova, O., Khvorova, I., Kozlov, K., Gursky, V., Samsonova, M., Tikhonova, O., & Neganova, I. (2023). Quality Control of Human Pluripotent Stem Cell Colonies by Computational Image Analysis Using Convolutional Neural Networks. International Journal of Molecular Sciences, 24(1), 140. https://doi.org/10.3390/ijms24010140








