Lineage Reconstruction of In Vitro Identified Antigen-Specific Autoreactive B Cells from Adaptive Immune Receptor Repertoires
Abstract
:1. Introduction
1.1. Formation of B Cell Lineages Is a Constant Somatic Microevolution
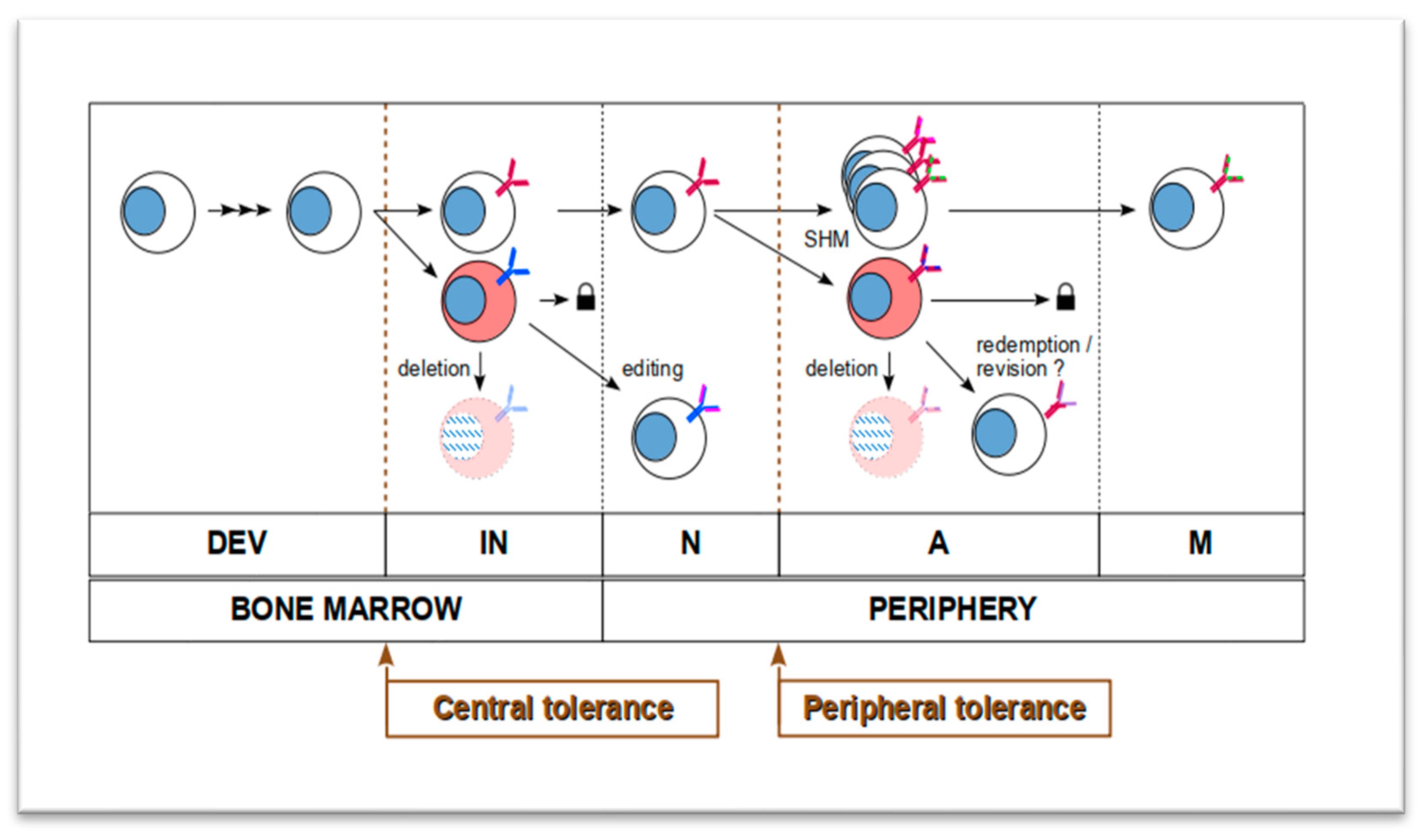
1.2. Humoral Autoimmunity Is Interpreted as Evolving Autoreactive B Cell Lineages
1.3. Autoreactive B-Cell Lineages Leave Their Footprints in BCR Repertoires
2. Results
2.1. PR/AR IgH-Related Sequences Were Extracted from AIRRs of pRD Patients
2.2. Expansion of PR/AR-Related Clonotypes Were Detected in pRD Patients
2.3. Diversification and Class-Switching in PR/AR Lineages Imply Early Activation
2.4. Phylogeny Reveals Paths of PR/AR Antibody Maturation in pRD Patients
2.5. 9G4+ B Cells Also Expand and Thrive in SLE
3. Discussion
4. Materials and Methods
4.1. Preparation of Input IgH Sequences
4.2. ImmChainTracer Pipeline
4.3. Reconstruction of Phylogenetic Tree
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Christie, S.M.; Fijen, C.; Rothenberg, E. V(D)J Recombination: Recent Insights in Formation of the Recombinase Complex and Recruitment of DNA Repair Machinery. Front. Cell Dev. Biol. 2022, 10, 886718. [Google Scholar] [CrossRef]
- Yu, K.; Lieber, M.R. Current Insights into the Mechanism of Mammalian Immunoglobulin Class Switch Recombination. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Pilzecker, B.; Jacobs, H. Mutating for Good: DNA Damage Responses During Somatic Hypermutation. Front. Immunol. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.C.; de Bouteiller, O.; Liu, Y.J.; Potter, K.; Banchereau, J.; Capra, J.D.; Pascual, V. Somatic Hypermutation Introduces Insertions and Deletions into Immunoglobulin V Genes. J. Exp. Med. 1998, 187, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupo, C.; Spisak, N.; Walczak, A.M.; Mora, T. Learning the Statistics and Landscape of Somatic Mutation-Induced Insertions and Deletions in Antibodies. PLoS Comput. Biol. 2022, 18, e1010167. [Google Scholar] [CrossRef] [PubMed]
- Nemazee, D. Mechanisms of Central Tolerance for B Cells. Nat. Rev. Immunol. 2017, 17, 281–294. [Google Scholar] [CrossRef]
- Tan, C.; Noviski, M.; Huizar, J.; Zikherman, J. Self-Reactivity on a Spectrum: A Sliding Scale of Peripheral B Cell Tolerance. Immunol. Rev. 2019, 292, 37–60. [Google Scholar] [CrossRef]
- Lee, S.; Ko, Y.; Kim, T.J. Homeostasis and Regulation of Autoreactive B Cells. Cell. Mol. Immunol. 2020, 17, 561–569. [Google Scholar] [CrossRef]
- Meffre, E.; Papavasiliou, F.; Cohen, P.; de Bouteiller, O.; Bell, D.; Karasuyama, H.; Schiff, C.; Banchereau, J.; Liu, Y.J.; Nussenzweig, M.C. Antigen Receptor Engagement Turns off the V(D)J Recombination Machinery in Human Tonsil B Cells. J. Exp. Med. 1998, 188, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Hillion, S.; Rochas, C.; Youinou, P.; Jamin, C. Expression and Reexpression of Recombination Activating Genes: Relevance to the Development of Autoimmune States. Ann. N. Y. Acad. Sci. 2005, 1050, 10–18. [Google Scholar] [CrossRef]
- Da Sylva, T.R.; Fong, I.C.; Cunningham, L.A.; Wu, G.E. RAG1/2 Re-Expression Causes Receptor Revision in a Model B Cell Line. Mol. Immunol. 2007, 44, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Diamond, B. B Cell Receptor Revision Diminishes the Autoreactive B Cell Response after Antigen Activation in Mice. J. Clin. Investig. 2008, 118, 2896–2907. [Google Scholar] [CrossRef]
- Reed, J.H.; Jackson, J.; Christ, D.; Goodnow, C.C. Clonal Redemption of Autoantibodies by Somatic Hypermutation Away from Self-Reactivity during Human Immunization. J. Exp. Med. 2016, 213, 1255–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnett, D.L.; Reed, J.H.; Christ, D.; Goodnow, C.C. Clonal Redemption and Clonal Anergy as Mechanisms to Balance B Cell Tolerance and Immunity. Immunol. Rev. 2019, 292, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Hershberg, U.; Luning Prak, E.T. The Analysis of Clonal Expansions in Normal and Autoimmune B Cell Repertoires. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140239. [Google Scholar] [CrossRef] [Green Version]
- Tipton, C.M.; Fucile, C.F.; Darce, J.; Chida, A.; Ichikawa, T.; Gregoretti, I.; Schieferl, S.; Hom, J.; Jenks, S.; Feldman, R.J.; et al. Diversity, Cellular Origin and Autoreactivity of Antibody-Secreting Cell Population Expansions in Acute Systemic Lupus Erythematosus. Nat. Immunol. 2015, 16, 755–765. [Google Scholar] [CrossRef] [Green Version]
- Cowan, G.J.M.; Miles, K.; Capitani, L.; Giguere, S.S.B.; Johnsson, H.; Goodyear, C.; McInnes, I.B.; Breusch, S.; Gray, D.; Gray, M. In Human Autoimmunity, a Substantial Component of the B Cell Repertoire Consists of Polyclonal, Barely Mutated IgG+ve B Cells. Front. Immunol. 2020, 11, 395. [Google Scholar] [CrossRef]
- Csomos, K.; Ujhazi, B.; Blazso, P.; Herrera, J.L.; Tipton, C.M.; Kawai, T.; Gordon, S.; Ellison, M.; Wu, K.; Stowell, M.; et al. Partial RAG Deficiency in Humans Induces Dysregulated Peripheral Lymphocyte Development and Humoral Tolerance Defect with Accumulation of T-Bet+ B Cells. Nat. Immunol. 2022, 23, 1256–1272. [Google Scholar] [CrossRef]
- Horns, F.; Vollmers, C.; Croote, D.; Mackey, S.F.; Swan, G.E.; Dekker, C.L.; Davis, M.M.; Quake, S.R. Lineage Tracing of Human B Cells Reveals the in Vivo Landscape of Human Antibody Class Switching. eLife 2016, 5, e16578. [Google Scholar] [CrossRef]
- Chaudhary, N.; Wesemann, D.R. Analyzing Immunoglobulin Repertoires. Front. Immunol. 2018, 9, 462. [Google Scholar] [CrossRef]
- Magadán, S. Adaptive Immune Receptor Repertoires, an Overview of This Exciting Field. Immunol. Lett. 2020, 221, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Wardemann, H.; Busse, C.E. Novel Approaches to Analyze Immunoglobulin Repertoires. Trends Immunol. 2017, 38, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Bashford-Rogers, R.J.M.; Smith, K.G.C.; Thomas, D.C. Antibody Repertoire Analysis in Polygenic Autoimmune Diseases. Immunology 2018, 155, 3–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silberstein, L.E.; Jefferies, L.C.; Goldman, J.; Friedman, D.; Moore, J.S.; Nowell, P.C.; Roelcke, D.; Pruzanski, W.; Roudier, J.; Silverman, G.J. Variable Region Gene Analysis of Pathologic Human Autoantibodies to the Related i and I Red Blood Cell Antigens. Blood 1991, 78, 2372–2386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, F.K.; Longhurst, C.; Chapman, C.J.; Ehrenstein, M.; Spellerberg, M.B.; Hamblin, T.J.; Ravirajan, C.T.; Latchman, D.; Isenberg, D. Utilization of the VH4-21 Gene Segment by Anti-DNA Antibodies from Patients with Systemic Lupus Erythematosus. J. Autoimmun. 1993, 6, 809–825. [Google Scholar] [CrossRef] [PubMed]
- van Vollenhoven, R.F.; Bieber, M.M.; Powell, M.J.; Gupta, P.K.; Bhat, N.M.; Richards, K.L.; Albano, S.A.; Teng, N.N. VH4-34 Encoded Antibodies in Systemic Lupus Erythematosus: A Specific Diagnostic Marker That Correlates with Clinical Disease Characteristics. J. Rheumatol. 1999, 26, 1727–1733. [Google Scholar]
- Zikherman, J.; Parameswaran, R.; Weiss, A. Endogenous Antigen Tunes the Responsiveness of Naive B Cells but Not T Cells. Nature 2012, 489, 160–164. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.E.; Rosen, L.B.; Csomos, K.; Rosenberg, J.M.; Mathew, D.; Keszei, M.; Ujhazi, B.; Chen, K.; Lee, Y.N.; Tirosh, I.; et al. Broad-Spectrum Antibodies against Self-Antigens and Cytokines in RAG Deficiency. J. Clin. Investig. 2015, 125, 4135–4148. [Google Scholar] [CrossRef] [Green Version]
- Delmonte, O.M.; Villa, A.; Notarangelo, L.D. Immune Dysregulation in Patients with RAG Deficiency and Other Forms of Combined Immune Deficiency. Blood 2020, 135, 610–619. [Google Scholar] [CrossRef]
- Gupta, N.T.; Vander Heiden, J.A.; Uduman, M.; Gadala-Maria, D.; Yaari, G.; Kleinstein, S.H. Change-O: A Toolkit for Analyzing Large-Scale B Cell Immunoglobulin Repertoire Sequencing Data. Bioinformatics 2015, 31, 3356–3358. [Google Scholar] [CrossRef] [Green Version]
- Wardemann, H.; Yurasov, S.; Schaefer, A.; Young, J.W.; Meffre, E.; Nussenzweig, M.C. Predominant Autoantibody Production by Early Human B Cell Precursors. Science 2003, 301, 1374–1377. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-H.; Zhang, Y.; Hu, Y.-F.; Wahl, L.M.; Cisar, J.O.; Notkins, A.L. The Broad Antibacterial Activity of the Natural Antibody Repertoire Is Due to Polyreactive Antibodies. Cell Host Microbe 2007, 1, 51–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott de Bruin, L.M.; Bosticardo, M.; Barbieri, A.; Lin, S.G.; Rowe, J.H.; Poliani, P.L.; Ching, K.; Eriksson, D.; Landegren, N.; Kämpe, O.; et al. Hypomorphic Rag1 Mutations Alter the Preimmune Repertoire at Early Stages of Lymphoid Development. Blood 2018, 132, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Barak, M.; Zuckerman, N.S.; Edelman, H.; Unger, R.; Mehr, R. IgTree: Creating Immunoglobulin Variable Region Gene Lineage Trees. J. Immunol. Methods 2008, 338, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Yaari, G.; Benichou, J.I.C.; Vander Heiden, J.A.; Kleinstein, S.H.; Louzoun, Y. The Mutation Patterns in B-Cell Immunoglobulin Receptors Reflect the Influence of Selection Acting at Multiple Time-Scales. Philos. Trans. R. Soc. B 2015, 370, 20140242. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.W.; Khavrutskii, I.V.; Wallqvist, A.; Bavari, S.; Cooper, C.L.; Chaudhury, S. BRILIA: Integrated Tool for High-Throughput Annotation and Lineage Tree Assembly of B-Cell Repertoires. Front. Immunol. 2016, 7, 681. [Google Scholar] [CrossRef] [Green Version]
- Foglierini, M.; Pappas, L.; Lanzavecchia, A.; Corti, D.; Perez, L. AncesTree: An Interactive Immunoglobulin Lineage Tree Visualizer. PLoS Comput. Biol. 2020, 16, e1007731. [Google Scholar] [CrossRef]
- Yang, X.; Tipton, C.M.; Woodruff, M.C.; Zhou, E.; Lee, F.E.-H.; Sanz, I.; Qiu, P. GLaMST: Grow Lineages along Minimum Spanning Tree for b Cell Receptor Sequencing Data. BMC Genom. 2020, 21, 583. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
- Stern, J.N.H.; Yaari, G.; Vander Heiden, J.A.; Church, G.; Donahue, W.F.; Hintzen, R.Q.; Huttner, A.J.; Laman, J.D.; Nagra, R.M.; Nylander, A.; et al. B Cells Populating the Multiple Sclerosis Brain Mature in the Draining Cervical Lymph Nodes. Sci. Transl. Med. 2014, 6, 248ra107. [Google Scholar] [CrossRef] [Green Version]
- Richardson, C.; Chida, A.S.; Adlowitz, D.; Silver, L.; Fox, E.; Jenks, S.A.; Palmer, E.; Wang, Y.; Heimburg-Molinaro, J.; Li, Q.-Z.; et al. Molecular Basis of 9G4 B Cell Autoreactivity in Human Systemic Lupus Erythematosus. J. Immunol. 2013, 191, 4926–4939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipton, C.M.; Hom, J.R.; Fucile, C.F.; Rosenberg, A.F.; Sanz, I. Understanding B-Cell Activation and Autoantibody Repertoire Selection in Systemic Lupus Erythematosus: A B-Cell Immunomics Approach. Immunol. Rev. 2018, 284, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Cashman, K.S.; Jenks, S.A.; Woodruff, M.C.; Tomar, D.; Tipton, C.M.; Scharer, C.D.; Eun-Hyung Lee, F.; Boss, J.M.; Sanz, I. Understanding and Measuring Human B-Cell Tolerance and Its Breakdown in Autoimmune Disease. Immunol. Rev. 2019, 292, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Nouri, N.; Kleinstein, S.H. Optimized Threshold Inference for Partitioning of Clones From High-Throughput B Cell Repertoire Sequencing Data. Front. Immunol. 2018, 9, 1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindenbaum, O.; Nouri, N.; Kluger, Y.; Kleinstein, S.H. Alignment Free Identification of Clones in B Cell Receptor Repertoires. Nucleic Acids Res. 2021, 49, e21. [Google Scholar] [CrossRef]
- See, P.; Lum, J.; Chen, J.; Ginhoux, F. A Single-Cell Sequencing Guide for Immunologists. Front. Immunol. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Friedensohn, S.; Khan, T.A.; Reddy, S.T. Advanced Methodologies in High-Throughput Sequencing of Immune Repertoires. Trends Biotechnol. 2017, 35, 203–214. [Google Scholar] [CrossRef]
- Lindeman, I.; Stubbington, M.J.T. Antigen Receptor Sequence Reconstruction and Clonality Inference from ScRNA-Seq Data. In Computational Methods for Single-Cell Data Analysis; Yuan, G.-C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 223–249. ISBN 978-1-4939-9057-3. [Google Scholar]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous Epitope and Transcriptome Measurement in Single Cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [Green Version]
- Brochet, X.; Lefranc, M.-P.; Giudicelli, V. IMGT/V-QUEST: The Highly Customized and Integrated System for IG and TR Standardized V-J and V-D-J Sequence Analysis. Nucleic Acids Res. 2008, 36, W503–W508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H.; Grolemund, G. R for Data Science: Import, Tidy, Transform, Visualize, and Model Data, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2017; ISBN 978-1-4919-1039-9. [Google Scholar]
- Vander Heiden, J.A.; Marquez, S.; Marthandan, N.; Bukhari, S.A.C.; Busse, C.E.; Corrie, B.; Hershberg, U.; Kleinstein, S.H.; Matsen IV, F.A.; Ralph, D.K.; et al. AIRR Community Standardized Representations for Annotated Immune Repertoires. Front. Immunol. 2018, 9, 2206. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blazso, P.; Csomos, K.; Tipton, C.M.; Ujhazi, B.; Walter, J.E. Lineage Reconstruction of In Vitro Identified Antigen-Specific Autoreactive B Cells from Adaptive Immune Receptor Repertoires. Int. J. Mol. Sci. 2023, 24, 225. https://doi.org/10.3390/ijms24010225
Blazso P, Csomos K, Tipton CM, Ujhazi B, Walter JE. Lineage Reconstruction of In Vitro Identified Antigen-Specific Autoreactive B Cells from Adaptive Immune Receptor Repertoires. International Journal of Molecular Sciences. 2023; 24(1):225. https://doi.org/10.3390/ijms24010225
Chicago/Turabian StyleBlazso, Peter, Krisztian Csomos, Christopher M. Tipton, Boglarka Ujhazi, and Jolan E. Walter. 2023. "Lineage Reconstruction of In Vitro Identified Antigen-Specific Autoreactive B Cells from Adaptive Immune Receptor Repertoires" International Journal of Molecular Sciences 24, no. 1: 225. https://doi.org/10.3390/ijms24010225
APA StyleBlazso, P., Csomos, K., Tipton, C. M., Ujhazi, B., & Walter, J. E. (2023). Lineage Reconstruction of In Vitro Identified Antigen-Specific Autoreactive B Cells from Adaptive Immune Receptor Repertoires. International Journal of Molecular Sciences, 24(1), 225. https://doi.org/10.3390/ijms24010225




