Dysregulation of Plasma miR-146a and miR-155 Expression Profile in Mycosis Fungoides Is Associated with rs2910164 and rs767649 Polymorphisms
Abstract
:1. Introduction
2. Results
2.1. Demographic and Clinical Characteristics of Patients Recruited in This Study
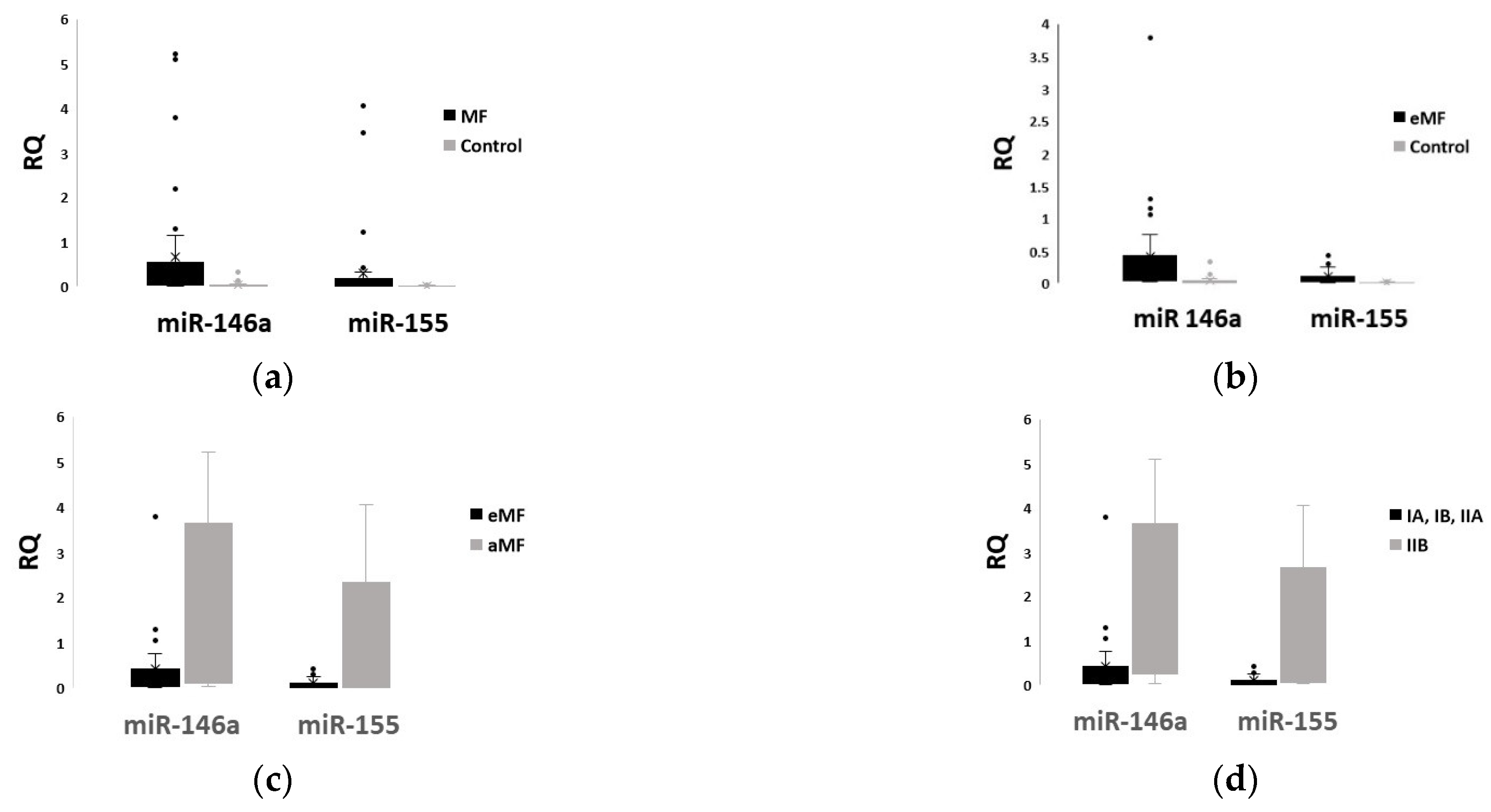
2.2. Plasma Levels of miR-146a and miR-155 Differ Significantly between Patients and Healthy Controls, and among MF Stages
2.3. Allelic and Genotypic Patterns of rs2910164 (C>G) and rs767649 (T>A) Polymorphisms Present a Statistically Significant Difference in the Distribution between MF Patients and Healthy Individuals, Displaying a Strong Association with Susceptibility to MF
2.4. rs2910164 (C>G) and rs767649 (T>A) Are Associated with Plasma Levels of miR-146a and miR-155, Respectively, and Their Concomitant Homozygous Presence Is Associated with Susceptibility to MF
2.5. rs2910164 and rs767649 Could Affect the Secondary Structure and Minimum Free Energy of miR-146a and Regulatory Region of miR-155, Respectively
3. Discussion
4. Materials and Methods
4.1. Clinical Samples
4.2. microRNAs Isolation
4.3. Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction
4.4. Genotyping
4.5. Sample Size Calculation
4.6. Statistical Analysis
- (I)
- The genotypic model, in which the wild-type homozygous genotype was used as a reference (GG for rs2910164 and TT for rs767649) to investigate distribution when compared with the other two genotypes (heterozygous genotype GC for rs2910164 and AT for rs767649, and mutant homozygous genotype CC for rs2910164 and AA for rs767649) in patients and healthy controls.
- (II)
- The dominant model, in which a wild-type homozygous genotype was used as a reference (GG for rs2910164 and TT for rs767649) to investigate the prevalence of mutant-allele-associated genotypes (CC+CG for rs2910164 and AA+AT for rs767649) within MF cases and controls.
- (III)
- The recessive model, in which wild-type-allele-associated genotypes (GC+GG for rs2910164 and AT+TT for rs767649) were used as a reference to investigate the prevalence of the mutant homozygous genotype (CC for rs2910164 and AA for rs767649) within MF and control groups.
- (IV)
- The allelic model, in which the wild-type allele (G for rs2910164 and T for rs767649) was used as a reference to investigate the prevalence of the mutant allele (C for rs2910164 and A for rs767649) within MF and control groups.
4.7. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dobos, G.; Pohrt, A.; Ram-Wolff, C.; Lebbé, C.; Bouaziz, J.-D.; Battistella, M.; Bagot, M.; De Masson, A. Epidemiology of Cutaneous T-Cell Lymphomas: A Systematic Review and Meta-Analysis of 16,953 Patients. Cancers 2020, 12, 2921. [Google Scholar] [CrossRef]
- Lovgren, M.L.; Scarisbrick, J.J. Update on Skin Directed Therapies in Mycosis Fungoides. Chin. Clin. Oncol. 2019, 8, 7. [Google Scholar] [CrossRef]
- Naraghi, Z.S.; Seirafi, H.; Valikhani, M.; Farnaghi, F.; Kavusi, S.; Dowlati, Y. Assessment of Histologic Criteria in the Diagnosis of Mycosis Fungoides. Int. J. Dermatol. 2003, 42, 45–52. [Google Scholar] [CrossRef]
- Furmanczyk, P.S.; Wolgamot, G.M.; Kussick, S.J.; Sabath, D.E.; Olerud, J.E.; Argenyi, Z.B. Diagnosis of Mycosis Fungoides with Different Algorithmic Approaches. J. Cutan. Pathol. 2010, 37, 8–14. [Google Scholar] [CrossRef]
- Massone, C.; Kodama, K.; Kerl, H.; Cerroni, L. Histopathologic Features of Early (Patch) Lesions of Mycosis Fungoides. Am. J. Surg. Pathol. 2005, 29, 550–560. [Google Scholar] [CrossRef]
- Scarisbrick, J.J.; Quaglino, P.; Prince, H.M.; Papadavid, E.; Hodak, E.; Bagot, M.; Servitje, O.; Berti, E.; Ortiz-Romero, P.; Stadler, R.; et al. The PROCLIPI International Registry of Early-Stage Mycosis Fungoides Identifies Substantial Diagnostic Delay in Most Patients. Br. J. Dermatol. 2019, 181, 350–357. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Wang, B.; Li, K.; Wang, L.; Zhao, X.; Xue, F.; Shi, R.; Zheng, J. MicroRNA Signatures in Diagnosis and Prognosis of Cutaneous T-Cell Lymphoma. J. Investog. Dermatol. 2018, 138, 2024–2032. [Google Scholar] [CrossRef] [Green Version]
- Fava, P.; Bergallo, M.; Astrua, C.; Brizio, M.; Galliano, I.; Montanari, P.; Daprà, V.; Novelli, M.; Savoia, P.; Quaglino, P.; et al. MiR-155 Expression in Primary Cutaneous T-Cell Lymphomas (CTCL). J. Eur. Acad. Dermatol. Venereol. 2017, 31, e27–e29. [Google Scholar] [CrossRef]
- Kopp, K.L.; Ralfkiaer, U.; Nielsen, B.S.; Gniadecki, R.; Woetmann, A.; Ødum, N.; Ralfkiaer, E. Expression of MiR-155 and MiR-126 in Situ in Cutaneous T-Cell Lymphoma. Apmis 2013, 121, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Di Raimondo, C.; Han, Z.; Su, C.; Wu, X.; Qin, H.; Sanchez, J.F.; Yuan, Y.C.; Martinez, X.; Abdulla, F.; Zain, J.; et al. Identification of a Distinct Mirna Regulatory Network in the Tumor Microenvironment of Transformed Mycosis Fungoides. Cancers 2021, 13, 5854. [Google Scholar] [CrossRef]
- Kopp, K.L.; Ralfkiaer, U.; Gjerdrum, L.M.R.; Helvad, R.; Pedersen, I.H.; Litman, T.; Jønson, L.; Hagedorn, P.H.; Krejsgaard, T.; Gniadecki, R.; et al. STAT5-Mediated Expression of Oncogenic MiR-155 in Cutaneous T-Cell Lymphoma. Cell Cycle 2013, 12, 1939–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manso, R.; Martinez-Magunacelaya, N.; Eraña-Tomás, I.; Monsalvez, V.; Rodriguez-Peralto, J.L.; Ortiz-Romero, P.L.; Santonja, C.; Cristóbal, I.; Piris, M.A.; Rodriguez-Pinilla, S.M. Mycosis Fungoides Progression Could Be Regulated by MicroRNAs. PLoS ONE 2018, 13, e0198477. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating MicroRNAs as Potential Cancer Biomarkers: The Advantage and Disadvantage. Clin. Epigenetics 2018, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyal, L.; Arkin, C.; Gorovitz-Haris, B.; Querfeld, C.; Rosen, S.; Knaneh, J.; Amitay-Laish, I.; Prag-Naveh, H.; Jacob-Hirsch, J.; Hodak, E. Mycosis Fungoides-Derived Exosomes Promote Cell Motility and Are Enriched with MicroRNA-155 and MicroRNA-1246, and Their Plasma-Cell-Free Expression May Serve as a Potential Biomarker for Disease Burden. Br. J. Dermatol. 2021, 185, 999–1012. [Google Scholar] [CrossRef]
- Dusílková, N.; Bašová, P.; Polívka, J.; Kodet, O.; Kulvait, V.; Pešta, M.; Trněný, M.; Stopka, T. Plasma MiR-155, Mir-203, and MiR-205 Are Biomarkers for Monitoring of Primary Cutaneous T-Cell Lymphomas. Int. J. Mol. Sci. 2017, 18, 2136. [Google Scholar] [CrossRef] [Green Version]
- Jazdzewski, K.; Murray, E.L.; Franssila, K.; Jarzab, B.; Schoenberg, D.R.; De La Chapelle, A. Common SNP in Pre-MiR-146a Decreases Mature MiR Expression and Predisposes to Papillary Thyroid Carcinoma. Proc. Natl. Acad. Sci. USA 2008, 105, 7269–7274. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, H.; Shen, J.; Zheng, Z.; Luo, X.; Gao, R.; Zhuang, X. MicroRNA-146a Rs2910164 Polymorphism and the Risk of Diffuse Large B Cell Lymphoma in the Chinese Han Population. Med. Oncol. 2014, 31, 1–5. [Google Scholar] [CrossRef]
- Wang, S.; Cao, X.; Ding, B.; Chen, J.; Cui, M.; Xu, Y.; Lu, X.; Zhang, Z.; He, A.; Jin, H. The Rs767649 Polymorphism in the Promoter of MiR-155 Contributes to the Decreased Risk for Cervical Cancer in a Chinese Population. Gene 2016, 595, 109–114. [Google Scholar] [CrossRef]
- Xie, K.; Ma, H.; Liang, C.; Wang, C.; Qin, N.; Shen, W.; Gu, Y.; Yan, C.; Zhang, K.; Dai, N.; et al. A Functional Variant in MiR-155 Regulation Region Contributes to Lung Cancer Risk and Survival. Oncotarget 2015, 6, 42781–42792. [Google Scholar] [CrossRef] [Green Version]
- Miyagaki, T. Diagnosis of Early Mycosis Fungoides. Diagnostics 2021, 11, 1721. [Google Scholar] [CrossRef]
- Cerroni, L. Mycosis Fungoides-Clinical and Histopathologic Features, Differential Diagnosis, and Treatment. Semin. Cutan. Med. Surg. 2018, 37, 2–10. [Google Scholar] [CrossRef]
- Skov, A.G.; Gniadecki, R. Delay in the Histopathologic Diagnosis of Mycosis Fungoides. Acta Derm. Venereol. 2015, 95, 472–475. [Google Scholar] [CrossRef] [Green Version]
- Gluud, M.; Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Lindahl, L.M.; Buus, T.B.; Andersen, M.H.; Bonefeld, C.M.; Krejsgaard, T.; Litvinov, I.V.; Iversen, L.; et al. MicroRNAs in the Pathogenesis, Diagnosis, Prognosis and Targeted Treatment of Cutaneous T-Cell Lymphomas. Cancers 2020, 12, 1229. [Google Scholar] [CrossRef]
- Ralfkiaer, U.; Hagedorn, P.H.; Bangsgaard, N.; Løvendorf, M.B.; Ahler, C.B.; Svensson, L.; Kopp, K.L.; Vennegaard, M.T.; Lauenborg, B.; Zibert, J.R.; et al. Diagnostic MicroRNA Profiling in Cutaneous T-Cell Lymphoma (CTCL). Blood 2011, 118, 5891–5900. [Google Scholar] [CrossRef]
- Marstrand, T.; Ahler, C.B.; Ralfkiaer, U.; Clemmensen, A.; Kopp, K.L.; Sibbesen, N.A.; Krejsgaard, T.; Litman, T.; Wasik, M.A.; Bonefeld, C.M.; et al. Validation of a Diagnostic MicroRNA Classifier in Cutaneous T-Cell Lymphomas. Leuk. Lymphoma 2014, 55, 957–958. [Google Scholar] [CrossRef]
- McGirt, L.Y.; Adams, C.M.; Baerenwald, D.A.; Zwerner, J.P.; Zic, J.A.; Eischen, C.M. MiR-223 Regulates Cell Growth and Targets Proto-Oncogenes in Mycosis Fungoides/Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2014, 134, 1101–1107. [Google Scholar] [CrossRef] [Green Version]
- Narducci, M.G.; Arcelli, D.; Picchio, M.C.; Lazzeri, C.; Pagani, E.; Sampogna, F.; Scala, E.; Fadda, P.; Cristofoletti, C.; Facchiano, A.; et al. MicroRNA Profiling Reveals That MiR-21, MiR486 and MiR-214 Are Upregulated and Involved in Cell Survival in Sézary Syndrome. Cell Death Dis. 2011, 2, e151. [Google Scholar] [CrossRef] [Green Version]
- Ito, M.; Teshima, K.; Ikeda, S.; Kitadate, A.; Watanabe, A.; Nara, M.; Yamashita, J.; Ohshima, K.; Sawada, K.; Tagawa, H. MicroRNA-150 Inhibits Tumor Invasion and Metastasis by Targeting the Chemokine Receptor CCR6, in Advanced Cutaneous T-Cell Lymphoma. Blood 2014, 123, 1499–1511. [Google Scholar] [CrossRef]
- Manfè, V.; Biskup, E.; Willumsgaard, A.; Skov, A.G.; Palmieri, D.; Gasparini, P.; Laganá, A.; Woetmann, A.; Ødum, N.; Croce, C.M.; et al. CMyc/MiR-125b-5p Signalling Determines Sensitivity to Bortezomib in Preclinical Model of Cutaneous T-Cell Lymphomas. PLoS ONE 2013, 8, e59390. [Google Scholar] [CrossRef]
- van Kester, M.S.; Ballabio, E.; Benner, M.F.; Chen, X.H.; Saunders, N.J.; van der Fits, L.; van Doorn, R.; Vermeer, M.H.; Willemze, R.; Tensen, C.P.; et al. MiRNA Expression Profiling of Mycosis Fungoides. Mol. Oncol. 2011, 5, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, L.M.; Besenbacher, S.; Rittig, A.H.; Celis, P.; Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Krejsgaard, T.; Johansen, C.; Litman, T.; Woetmann, A.; et al. Prognostic MiRNA Classifier in Early-Stage Mycosis Fungoides: Development and Validation in a Danish Nationwide Study. Blood 2018, 131, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Talaat, I.M.; Abdelmaksoud, R.E.S.; Guimei, M.; Agamia, N.F.; Nugud, A.; El-Serafi, A.T. Potential Role for MicroRNA-16 (MiR-16) and MicroRNA-93 (MiR-93) in Diagnosis and Prediction of Disease Progression in Mycosis Fungoides in Egyptian Patients. PLoS ONE 2019, 14, e0224305. [Google Scholar] [CrossRef]
- Rittig, A.H.; Lindahl, L.M.; Johansen, C.; Celis, P.; Ødum, N.; Iversen, L.; Litman, T. The MicroRNA Expression Profile Differs between Erythrodermic Mycosis Fungoides and Sézary Syndrome. Acta Derm. Venereol. 2019, 99, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- Johnson, V.E.; Vonderheid, E.C.; Hess, A.D.; Eischen, C.M.; McGirt, L.Y. Genetic Markers Associated with Progression in Early Mycosis Fungoides. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 1431–1435. [Google Scholar] [CrossRef] [Green Version]
- Doxaki, C.; Kampranis, S.C.; Eliopoulos, A.G.; Spilianakis, C.; Tsatsanis, C. Coordinated Regulation of MiR-155 and MiR-146a Genes during Induction of Endotoxin Tolerance in Macrophages. J. Immunol. 2015, 195, 5750–5761. [Google Scholar] [CrossRef] [Green Version]
- Mann, M.; Mehta, A.; Zhao, J.L.; Lee, K.; Marinov, G.K.; Garcia-Flores, Y.; Baltimore, D. An NF-ΚB-MicroRNA Regulatory Network Tunes Macrophage Inflammatory Responses. Nat. Commun. 2017, 8, 851. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H. Common Genetic Variants in Pre-MicroRNAs Were Associated with Increased Risk of Breast Cancer in Chinese Women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef]
- Kamal, A.; Elgengehy, F.T.; Elawady, Z.; Fawzy, N.A.; El Sisi, O. Role of miR-146a rs2910164 and UTS2 rs228648 Genetic Variants in Behçet’s Disease. Immunol. Investig. 2021, 51, 899–908. [Google Scholar] [CrossRef]
- Ahmadi, K.; Soleimani, A.; Soleimani Motlagh, S.; Baharvand Ahmadi, S.; Almasian, M.; Kiani, A.A. Polymorphisms of Pre-Mir-499 Rs3746444 T/C and Pre-Mir-146a Rs2910164 C/G in the Autoimmune Diseases of Rheumatoid Arthritis and Systemic Lupus Erythematosus in the West of Iran. Iran. J. Public Health 2020, 49, 782–790. [Google Scholar] [CrossRef]
- Lv, L.; Gu, H.; Chen, Z.; Tang, W.; Zhang, S.; Lin, Z. MiRNA-146a rs2910164 Confers a Susceptibility to Digestive System Cancer: A Meta-Analysis Involving 59,098 Subjects. Immunol. Investig. 2020, 51, 199–219. [Google Scholar] [CrossRef]
- Ayoub, S.E.; Shaker, O.G.; Abdelwahed, M.Y.; Ahmed, N.A.; Abdelhameed, H.G.; Bosilah, A.H.; Mohammed, S.R. Association of MicroRNA-155rs767649 Polymorphism with Susceptibility to Preeclampsia. Int. J. Mol. Cell. Med. 2019, 8, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Polina, E.R.; Oliveira, F.M.; Sbruzzi, R.C.; Crispim, D.; Canani, L.H.; Santos, K.G. Gene Polymorphism and Plasma Levels of MiR-155 in Diabetic Retinopathy. Endocr. Connect. 2019, 8, 1591–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaker, O.G.; Abdelaleem, O.O.; Fouad, N.A.; Ali, A.M.E.A.; Ahmed, T.I.; Ibrahem, E.G.; Abdelghaffar, N.K. Association between MiR-155, Its Polymorphism and Ischemia-Modified Albumin in Patients with Rheumatoid Arthritis. J. Interf. Cytokine Res. 2019, 39, 428–437. [Google Scholar] [CrossRef]
- Szlachta, K.; Thys, R.G.; Atkin, N.D.; Pierce, L.C.T.; Bekiranov, S.; Wang, Y.H. Alternative DNA Secondary Structure Formation Affects RNA Polymerase II Promoter-Proximal Pausing in Human. Genome Biol. 2018, 19, 1–19. [Google Scholar] [CrossRef]
- Timmins, M.A.; Wagner, S.D.; Ahearne, M.J. The New Biology of PTCL-NOS and AITL: Current Status and Future Clinical Impact. Br. J. Haematol. 2020, 189, 54–66. [Google Scholar] [CrossRef]
- Karambataki, M.; Malousi, A.; Tzimagiorgis, G.; Haitoglou, C.; Fragou, A.; Georgiou, E.; Papadopoulou, F.; Krassas, G.E.; Kouidou, S. Association of Two Synonymous Splicing-Associated CpG Single Nucleotide Polymorphisms in Calpain 10 and Solute Carrier Family 2 Member 2 with Type 2 Diabetes. Biomed. Rep. 2017, 6, 146–158. [Google Scholar] [CrossRef] [Green Version]
- Dezfuli, N.K.; Adcock, I.M.; Alipoor, S.D.; Seyfi, S.; Salimi, B.; Mafi Golchin, M.; Dalil Roofchayee, N.; Varhram, M.; Mortaz, E. The MiR-146a SNP Rs2910164 and MiR-155 SNP Rs767649 Are Risk Factors for Non-Small Cell Lung Cancer in the Iranian Population. Can. Respir. J. 2020, 2020, 1–8. [Google Scholar] [CrossRef]
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 Update of the WHO-EORTC Classification for Primary Cutaneous Lymphomas. Blood 2019, 133, 1703–1714. [Google Scholar] [CrossRef]
- El Maniani, M.; Rechchach, M.; El Mahfoudi, A.; El Moudane, M.; Sabbar, A. G*Power 3: A Flexible Statistical Power Analysis Program for the Social, Behavioral, and Biomedical Sciences. J. Mater. Environ. Sci. 2016, 7, 3759–3766. [Google Scholar]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Šťastný, J.; Mergny, J.L. G4Hunter Web Application: A Web Server for G-Quadruplex Prediction. Bioinformatics 2019, 35, 3493–3495. [Google Scholar] [CrossRef] [Green Version]
- Jenjaroenpun, P.; Wongsurawat, T.; Yenamandra, S.P.; Kuznetsov, V.A. QmRLFS-Finder: A Model, Web Server and Stand-Alone Tool for Prediction and Analysis of R-Loop Forming Sequences. Nucleic Acids Res. 2015, 43, W527–W534. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Churkin, A.; Retwitzer, M.D.; Reinharz, V.; Ponty, Y.; Waldispühl, J.; Barash, D. Design of RNAs: Comparing Programs for Inverse RNA Folding. Brief. Bioinform. 2018, 19, 350–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; Van Der Lee, R.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the Open-Access Database of Transcription Factor Binding Profiles and Its Web Framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed]



| Patients | Healthy Individuals | |
|---|---|---|
| n = 41 | n = 41 | |
| Demographic characteristics | ||
| Median age (range) | 59 (39–85) | 56 (34–79) |
| Male n (%) | 30 (73.17%) | 28 (68.29%) |
| Female n (%) | 11 (26.83%) | 13 (31.71%) |
| Clinical Features | n (%) | |
| Patch stage | 11 (27%) | |
| Plaque stage | 21 (51%) | |
| Tumor Stage | 6 (14%) | |
| Erythrodermic | 3 (8%) | |
| MF stage | n (%) | |
| Early-stage (IA–IIA) | 32 (78.03%) | |
| ΙA | 22 (53.65%) | |
| ΙΒ | 8 (19.51%) | |
| ΙΙA | 2 (4.87%) | |
| Advanced-stage (IIB–IV) | 9 (21.97%) | |
| ΙΙΒ | 5 (12.19%) | |
| ΙΙΙ | 3 (7.31%) | |
| ΙV | 1 (2.47%) | |
| Treatment | n (%) | |
| None | 19 (46.3%) | |
| Skin-directed | 3 (7.3%) | |
| Systematic | 12 (29.4%) | |
| Combination of skin-directed and systematic | 7 (17%) |
| miR-146a rs2910164 (C>G) | |||
| Genotypes and Alleles | MF Cases n = 33 (%) | Controls n = 26 (%) | p-Value, Unadjusted OR (95% CI) |
| GG | 23 (69.7) | 4 (15.3) | 1 (reference) |
| GC | 9 (27.3) | 6 (23) | p = 0.066, 0.261 (0.059–1.148) |
| CC | 1 (3) | 16 (61.7) | p < 0.01, 0.011 (0.001–0.107) |
| Dominant model | |||
| GG | 23 (69.7) | 4 (15.3) | 1 (reference) |
| GC+CC | 10 (30.3) | 22 (84.7) | p < 0.01, 0.079 (0.022–0.290) |
| Recessive model | |||
| GC+GG | 32 (97) | 10 (38.3) | 1 (reference) |
| CC | 1 (3) | 16 (61.7) | p < 0.01, 0.020 (0.002–0.166) |
| Allelic model | |||
| G | 55 (83.4) | 14 (26.9) | 1 (reference) |
| C | 11 (16.6) | 38 (73.1) | p < 0.01, 0.074 (0.030–0.180) |
| miR-155 rs767649 (T>A) | |||
| Genotypes and Alleles | MF Cases n = 33 (%) | Controls n = 26 (%) | p-Value, Unadjusted OR (95% CI) |
| TT | 18 (54.5) | 23 (88.3) | 1 (reference) |
| TA | 3 (9) | 1 (4) | p = 0.217, 4 (0.384–41.701) |
| AA | 12 (36.5) | 2 (7.7) | p < 0.01, 8 (1.588–40.299) |
| Dominant model | |||
| TT | 18 (54.5) | 23 (88.3) | 1 (reference) |
| ΤA+AA | 15 (45.5) | 3 (11.7) | p < 0.01, 6.667 (1.674–26.554) |
| Recessive model | |||
| ΤA+TT | 21 (63.5) | 24 (92.3) | 1 (reference) |
| AA | 12 (36.5) | 2 (7.7) | p = 0.01, 6.857 (1.374–34.217) |
| Allelic model | |||
| T | 39 (59) | 47 (90.4) | 1 (reference) |
| A | 27 (41) | 5 (9.6) | p < 0.01, 6.679 (2.347–19.003) |
| Genotypic Combinations | MF n = 33 (%) | Controls n = 26 (%) | p-Value, Unadjusted OR (95% CI) |
|---|---|---|---|
| Rest cases | 22 (67) | 26 (0) | 1 (reference) |
| GG+AA | 11 (33) | 0 (0) | p < 0.01, 1.49 (1.177–1.908) |
| Rest cases | 32 (97) | 12 (46) | 1 (reference) |
| CC+TT | 1 (3) | 14 (54) | p < 0.01, 0.025 (0.003–0.210) |
| Target | Primer Sequence 5′→3′ | Product Length | Genomic Location | Annealing Temperature |
|---|---|---|---|---|
| rs2910164 | F: CATTGGATTTACCAGGCTTTT R: CACACTCCTTATACCTTCAGAGC | 305 bp | Chr5, GRCh38.p14 (160485260, 160485564) | 58 °C |
| pre-mir-155 | F: GCATACACAAACATTTCTTTCTCTCT R: CATCCCAGTGACCAGATTATGA | 329 bp | Chr21, GRCh38.p14 (25573862, 25574191) | 58 °C |
| rs767649 | F: ACAAAAGGGGACCTGTGTGA R: TTGAAGGTAAATTGCTGGCATACT | 281 bp | Chr21, GRCh38.p14 (25572299, 25572579) | 62 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avgeros, C.; Patsatsi, A.; Dimitriadis, D.; Malousi, A.; Koletsa, T.; Papathemeli, D.; Syrnioti, A.; Avgerou, P.; Lazaridou, E.; Tzimagiorgis, G.; et al. Dysregulation of Plasma miR-146a and miR-155 Expression Profile in Mycosis Fungoides Is Associated with rs2910164 and rs767649 Polymorphisms. Int. J. Mol. Sci. 2023, 24, 271. https://doi.org/10.3390/ijms24010271
Avgeros C, Patsatsi A, Dimitriadis D, Malousi A, Koletsa T, Papathemeli D, Syrnioti A, Avgerou P, Lazaridou E, Tzimagiorgis G, et al. Dysregulation of Plasma miR-146a and miR-155 Expression Profile in Mycosis Fungoides Is Associated with rs2910164 and rs767649 Polymorphisms. International Journal of Molecular Sciences. 2023; 24(1):271. https://doi.org/10.3390/ijms24010271
Chicago/Turabian StyleAvgeros, Chrysostomos, Aikaterini Patsatsi, Dimitrios Dimitriadis, Andigoni Malousi, Triantafyllia Koletsa, Despoina Papathemeli, Antonia Syrnioti, Paraskevi Avgerou, Elizabeth Lazaridou, Georgios Tzimagiorgis, and et al. 2023. "Dysregulation of Plasma miR-146a and miR-155 Expression Profile in Mycosis Fungoides Is Associated with rs2910164 and rs767649 Polymorphisms" International Journal of Molecular Sciences 24, no. 1: 271. https://doi.org/10.3390/ijms24010271





