The Active Site of the Enzyme 10-Formyl-THFDH in the Honey Bee Apis mellifera—A Key Player in Formic Acid Detoxification
Abstract
1. Introduction
2. Results
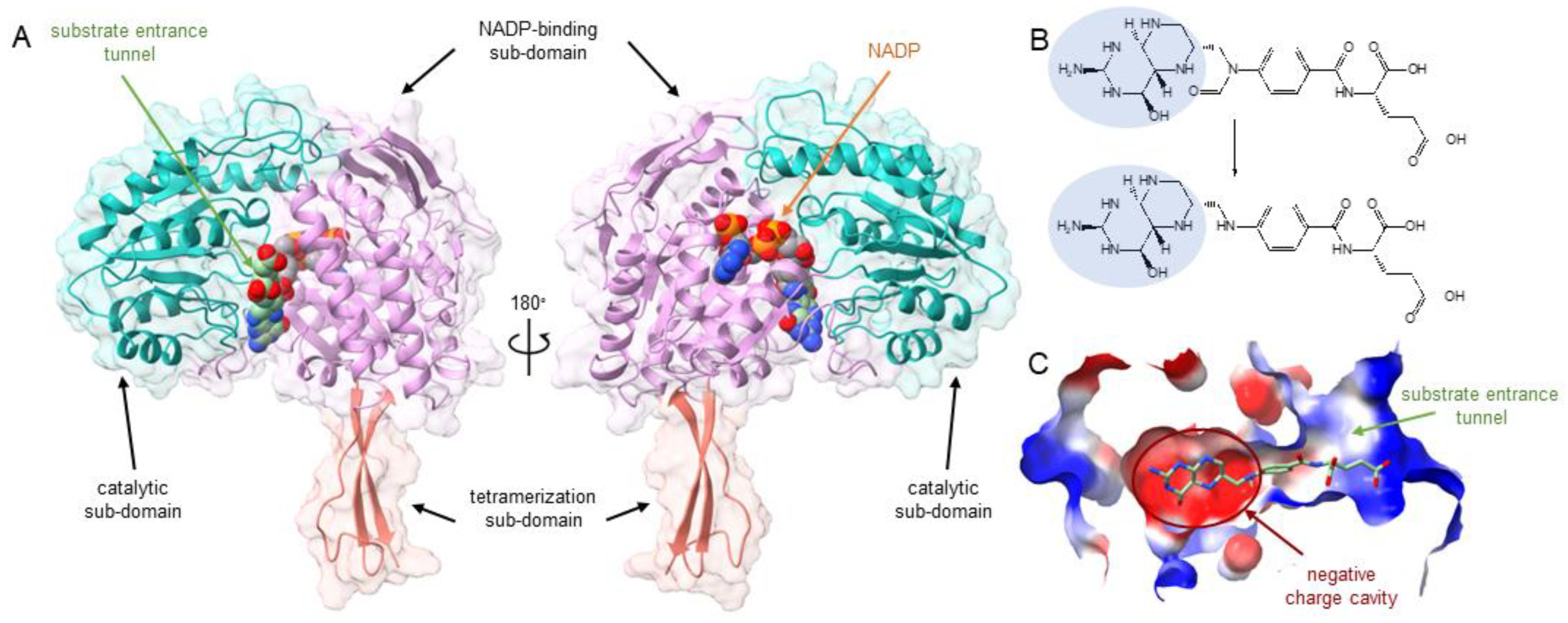
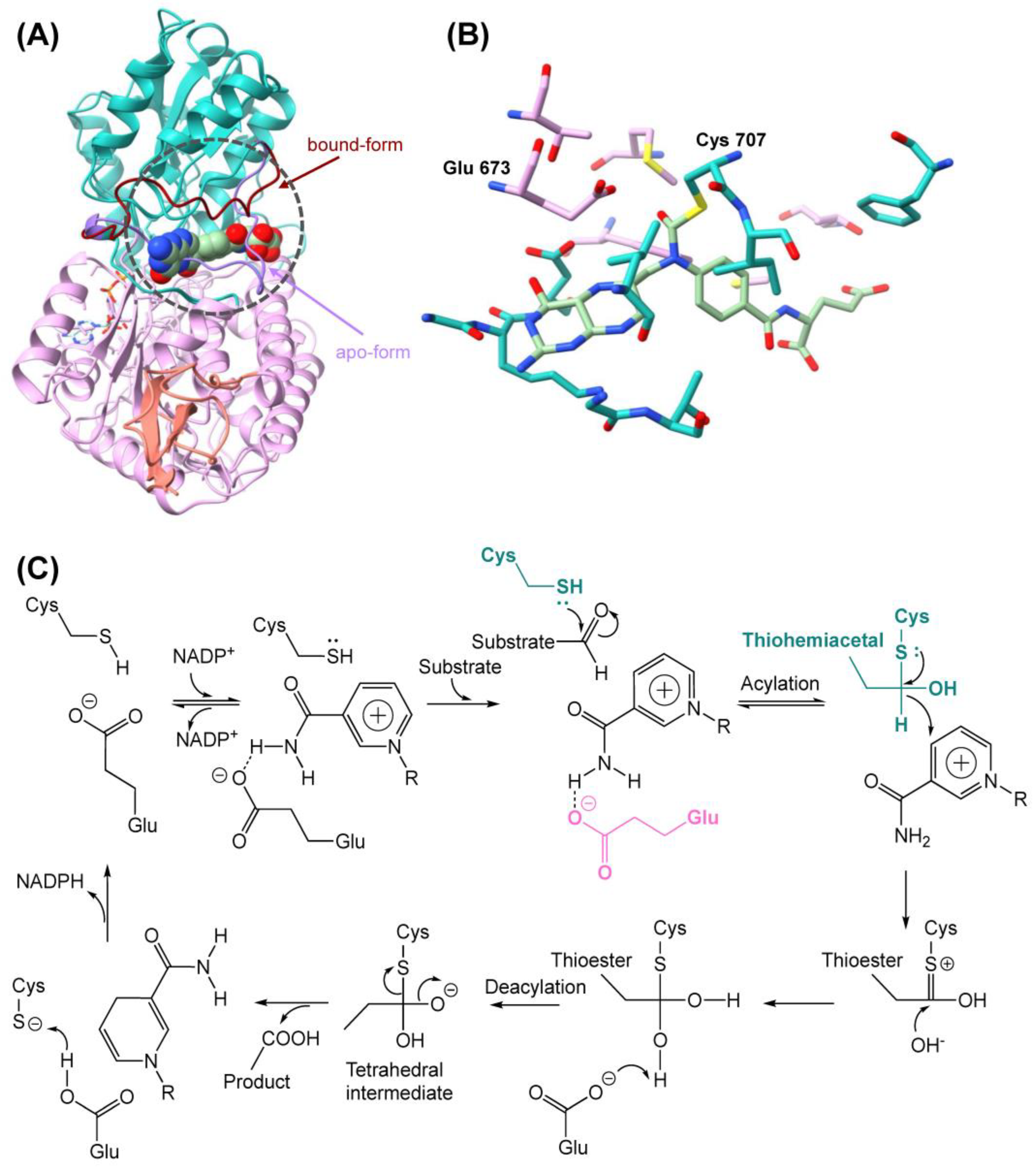
2.1. Homology Modeling, Docking, and Sequence Alignment of Wildtype Enzyme
2.2. Homology Modeling and Docking of Mutants
2.3. Comparison of AlphaFold2 Model and Homology Models
2.4. Enzyme Kinetics
3. Discussion
4. Materials and Methods
4.1. Homology Modeling
4.2. Covalent Docking
4.3. AlphaFold2 Structural Prediction
4.4. Structural Analysis
4.5. Visualization
4.6. Multiple Sequence Alignment
4.7. A. Mellifera Sampling
4.8. RNA Extraction
4.9. First-Strand cDNA Synthesis
4.10. Creation of pFBD-eGFP-Amel_10-Formyl-THFDH Expression Vector
4.11. Mutagenesis of the Expression Vector
4.12. Creation of the Recombinant Bacmid
4.13. Creation of Baculovirus
4.14. Expression and Purification of Recombinant Proteins
4.15. Synthesis of 10-Formyl-THF
4.16. Enzyme Activity Assays
4.17. Analysis of Kinetic Data
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Havens, K.; Vitt, P. The Importance of Phenological Diversity in Seed Mixes for Pollinator Restoration. Nat. Areas J. 2016, 36, 531. [Google Scholar] [CrossRef]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Boil. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef] [PubMed]
- Paudel, Y.P.; Mackereth, R.; Hanley, R.; Qin, W. Honey Bees (Apis mellifera L.) and Pollination Issues: Current status, impacts and potential drivers of decline. J. Agric. Sci. 2015, 7, 93. [Google Scholar] [CrossRef]
- Pettis, J.S.; Delaplane, K.S. Coordinated responses to honey bee decline in the USA. Apidologie 2010, 41, 256–263. [Google Scholar] [CrossRef]
- Seitz, N.; Traynor, K.S.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Ellis, J.D.; Rose, R.; Tarpy, D.R.; Sagili, R.R.; Caron, D.M.; et al. A national survey of managed honey bee 2014–2015 annual colony losses in the USA. J. Apic. Res. 2015, 54, 292–304. [Google Scholar] [CrossRef]
- Anderson, D.; Trueman, J. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 2000, 24, 165–189. [Google Scholar] [CrossRef]
- Le Conte, Y.; Ellis, M.; Ritter, W. Varroa mites and honey bee health: Can Varroa explain part of the colony losses? Apidologie 2010, 41, 353–363. [Google Scholar] [CrossRef]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef]
- Sumpter, D.J.T.; Martin, S.J. The dynamics of virus epidemics in Varroa-infested honey bee colonies. J. Anim. Ecol. 2004, 73, 51–63. [Google Scholar] [CrossRef]
- Shen, M.; Yang, X.; Cox-Foster, D.; Cui, L. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 2005, 342, 141–149. [Google Scholar] [CrossRef] [PubMed]
- De Rycke, P.H.; Joubert, J.J.; Hosseinian, S.H.; Jacobs, F.J. The possible role of Varroa destructor in the spreading of American foulbrood among apiaries. Exp. Appl. Acarol. 2002, 27, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Imdorf, A.; Charrière, J.-D.; Kilchenmann, V.; Bogdanov, S.; Fluri, P. Alternative strategy in central Europe for the control of Varroa destructor in honey bee colonies. Apiacta 2003, 38, 258–278. [Google Scholar]
- Bogdanov, S. Contaminants of bee products. Apidologie 2006, 37, 1–18. [Google Scholar] [CrossRef]
- Kraus, B.; Berg, S. Effect of a lactic acid treatment during winter in temperate climate upon Varroa jacobsoni Oud. and the bee (Apis mellifera L.) colony. Exp. Appl. Acarol. 1994, 18, 459–468. [Google Scholar] [CrossRef]
- Imdorf, A.; Charrière, J.-D.; Rosenkranz, P. Varroa control with formic acid. FAIR 1999, 24, CT97-3686. [Google Scholar]
- Gregorc, A.; Planinc, I. The Control of Varroa destructor Using Oxalic Acid. Vet. J. 2002, 163, 306–310. [Google Scholar] [CrossRef]
- Gregorc, A.; Poklukar, J. Rotenone and oxalic acid as alternative acaricidal treatments for Varroa destructor in honeybee colonies. Vet. Parasitol. 2003, 111, 351–360. [Google Scholar] [CrossRef]
- Avila-Ramos, F.; Otero-Colina, G.; Sánchez-Arroyo, H.; Santillán-Galicia, M.T.; Tecante, A. A gel formulation of formic acid for control of Varroa destructor. Trends Acarol. 2010, 545–549. [Google Scholar] [CrossRef]
- Girişgin, A.O.; Aydin, L. Efficacies of formic, oxalic and lactic acids against Varroa destructor in naturally infested honeybee (Apis mellifera L.) colonies in Turkey. Kafkas Univ. Vet. Fak. Derg. 2010, 16, 941–945. [Google Scholar]
- Genath, A.; Sharbati, S.; Buer, B.; Nauen, R.; Einspanier, R. Comparative transcriptomics indicates endogenous differences in detoxification capacity after formic acid treatment between honey bees and varroa mites. Sci. Rep. 2020, 10, 21943. [Google Scholar] [CrossRef] [PubMed]
- Mating, M.; Sharbati, S.; Einspanier, R. A Detoxification Enzyme for Apis mellifera Newly Characterized by Recombinant Expression: 10-Formyl Tetrahydrofolate Dehydrogenase. Front. Insect Sci. 2022, 2, 5. [Google Scholar] [CrossRef]
- Krupenko, N.I.; Dubard, M.E.; Strickland, K.C.; Moxley, K.M.; Oleinik, N.V.; Krupenko, S.A. ALDH1L2 Is the Mitochondrial Homolog of 10-Formyltetrahydrofolate Dehydrogenase. J. Biol. Chem. 2010, 285, 23056–23063. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, L.; Brode, E. Research on monocarbon compounds. I. The tetrahydrofolate formylase from pigeon liver. Purification and mechanism. Biochem. Z. 1961, 334, 108–132. [Google Scholar] [PubMed]
- Johlin, F.C.; Swain, E.; Smith, C.; Tephly, T.R. Studies on the mechanism of methanol poisoning: Purification and comparison of rat and human liver 10-formyltetrahydrofolate dehydrogenase. Mol. Pharmacol. 1989, 35, 745–750. [Google Scholar]
- Anguera, M.C.; Field, M.; Perry, C.; Ghandour, H.; Chiang, E.; Selhub, J.; Shane, B.; Stover, P.J. Regulation of Folate-mediated One-carbon Metabolism by 10-Formyltetrahydrofolate Dehydrogenase. J. Biol. Chem. 2006, 281, 18335–18342. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Edich, M.; Briggs, D.C.; Kippes, O.; Gao, Y.; Thorn, A. The impact of AlphaFold on experimental structure solution. Faraday Discuss. 2022, 240, 184–195. [Google Scholar] [CrossRef]
- Dembele, H.; Mating, M.; Singh, R.; Fatehi, S.; Herrera, A.I.; Park, Y.; Prakash, O. Ecdysis triggering hormone peptide in the African malaria mosquito Anopheles gambiae: The peptide structure for receptor activation. Insect Sci. 2022, 29, 1309–1317. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Venthur, H.; Mutis, A.; Zhou, J.-J.; Quiroz, A. Ligand binding and homology modelling of insect odorant-binding proteins. Physiol. Entomol. 2014, 39, 183–198. [Google Scholar] [CrossRef]
- Jindal, V.; Li, D.; Rault, L.C.; Fatehi, S.; Singh, R.; Mating, M.; Zou, Y.; Ng, H.-L.; Kaczmarek, K.; Zabrocki, J.; et al. Bee-safe peptidomimetic acaricides achieved by comparative genomics. Sci. Rep. 2022, 12, 17263. [Google Scholar] [CrossRef]
- Tsybovsky, Y.; Donato, H.; Krupenko, N.I.; Davies, C.; Krupenko, S.A. Crystal Structures of the Carboxyl Terminal Domain of Rat 10-Formyltetrahydrofolate Dehydrogenase: Implications for the Catalytic Mechanism of Aldehyde Dehydrogenases. Biochemistry 2007, 46, 2917–2929. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Vannette, R.L.; Fukami, T. Nectar microbes can reduce secondary metabolites in nectar and alter effects on nectar consumption by pollinators. Ecology 2016, 97, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- Vannette, R.L.; Gauthier, M.-P.; Fukami, T. Nectar bacteria, but not yeast, weaken a plant–pollinator mutualism. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122601. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.I.; Lievens, B.; Jacquemyn, H. Impact of Microorganisms on Nectar Chemistry, Pollinator Attraction and Plant Fitness; Nova Science Publishers: Hauppauge, NY, USA, 2015. [Google Scholar]
- Chappell, C.R.; Fukami, T. Nectar yeasts: A natural microcosm for ecology. Yeast 2018, 35, 417–423. [Google Scholar] [CrossRef]
- Ehlers, B.K.; Olesen, J. The fruit-wasp route to toxic nectar in Epipactis orchids? Flora 1997, 192, 223–229. [Google Scholar] [CrossRef]
- Rering, C.C.; Beck, J.J.; Hall, G.W.; McCartney, M.M.; Vannette, R.L. Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. New Phytol. 2017, 220, 750–759. [Google Scholar] [CrossRef]
- Liesivuori, J.; Savolainen, A.H. Methanol and Formic Acid Toxicity: Biochemical Mechanisms. Basic Clin. Pharmacol. Toxicol. 1991, 69, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Darragh, K.; Nelson, D.R.; Ramírez, S.R. The Birth-and-Death Evolution of Cytochrome P450 Genes in Bees. Genome Biol. Evol. 2021, 13, evab261. [Google Scholar] [CrossRef] [PubMed]
- Perdigão, N.; Heinrich, J.; Stolte, C.; Sabir, K.S.; Buckley, M.J.; Tabor, B.; Signal, B.; Gloss, B.S.; Hammang, C.J.; Rost, B.; et al. Unexpected features of the dark proteome. Proc. Natl. Acad. Sci. USA 2015, 112, 15898–15903. [Google Scholar] [CrossRef]
- Perdigão, N.; Rosa, A. Dark Proteome Database: Studies on Dark Proteins. High Throughput 2019, 8, 8. [Google Scholar] [CrossRef]
- Tsybovsky, Y.; Krupenko, S.A. Conserved Catalytic Residues of the ALDH1L1 Aldehyde Dehydrogenase Domain Control Binding and Discharging of the Coenzyme. J. Biol. Chem. 2011, 286, 23357–23367. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Borrelli, K.W.; Greenwood, J.R.; Day, T.; Abel, R.; Farid, R.S.; Harder, E. Docking Covalent Inhibitors: A Parameter Free Approach To Pose Prediction and Scoring. J. Chem. Inf. Model. 2014, 54, 1932–1940. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Bohmer, M.; Sharbati, J.; Zur Bruegge, J.; Einspanier, R.; Sharbati, S. Structural analysis of microRNA-target interaction by sequential seed mutagenesis and stem-loop 3’RACE. PLoS ONE 2013, 8, e81427. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.C. [116] Preparation and properties of 5,10-methenyltetrahydrofolic acid and 10-formultetrahydrofolic acid. Methods Enzymol. 1963, 6, 814–815. [Google Scholar] [CrossRef]

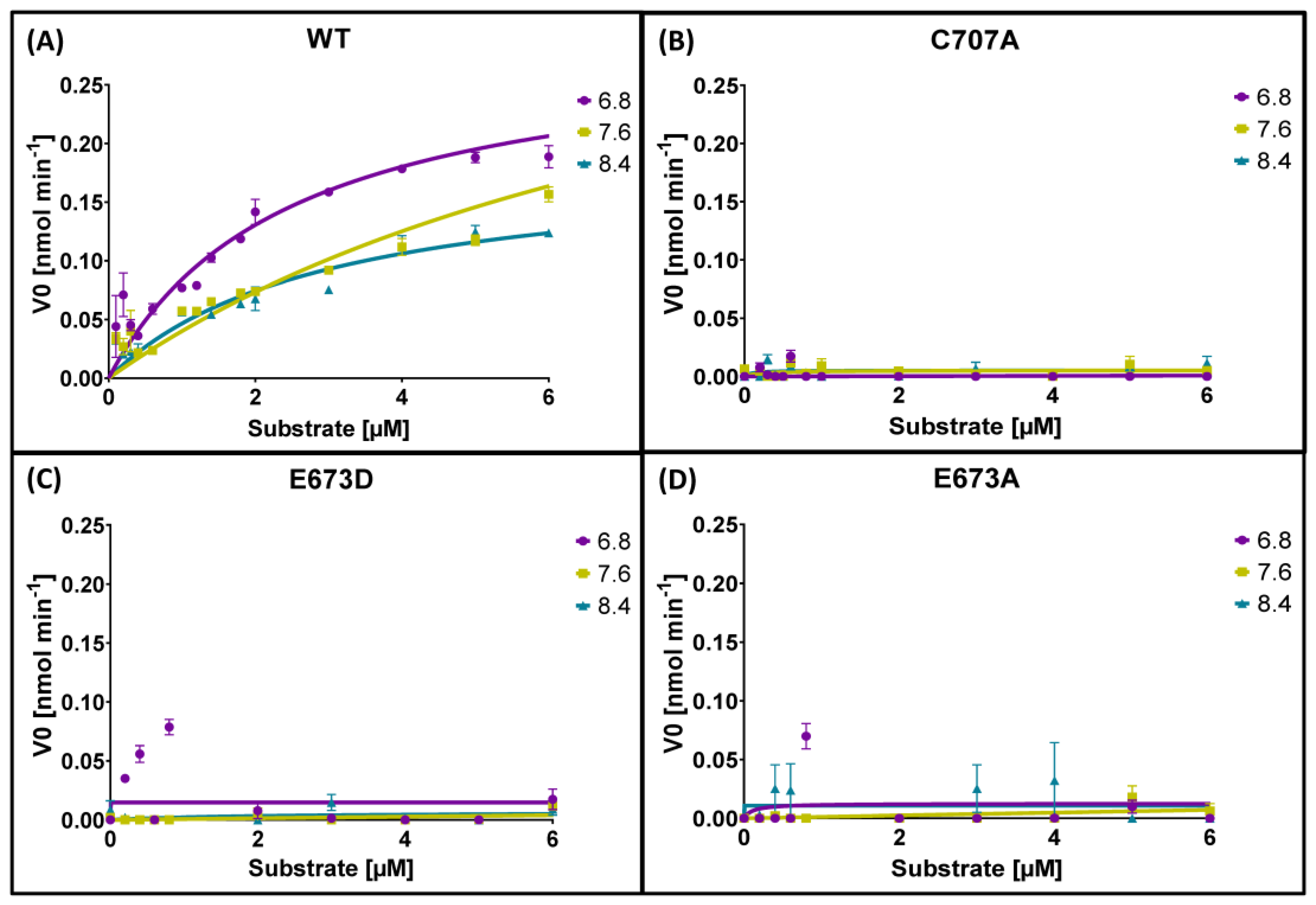
| pH | Enzyme | Vmax | Km | 95% CI Vmax | 95% CI Km | R2 |
|---|---|---|---|---|---|---|
| 6.8 | WT | 0.2907 | 2.455 | 0.2515 to 3.542 | 1.72 to 3.542 | 0.8290 |
| C707A | NA | 8.57 × 1016 | NA | NA | −0.1333 | |
| E673A | 0.01265 | 0.1109 | 0.0026 to 17,623,525 | NA | 0.0207 | |
| E673D | 0.01484 | 8.33 × 10−14 | 0.0078 to 0.02894 | NA to 0.2658 | 0.0262 | |
| 7.6 | WT | 0.4253 | 9.596 | 0.2908 to 83.76 | 5.133 to 23.98 | 0.8858 |
| C707A | 0.00538 | 0.228 | 0.0023 to 0.01327 | NA to 4.951 | −0.0632 | |
| E673A | 0.1299 | 102.8 | 0.0037 to NA | 0.2755 to NA | 0.1713 | |
| E673D | 0.05799 | 81.54 | 0.0021 to NA | NA | 0.1493 | |
| 8.4 | WT | 0.1846 | 2.961 | 0.1520 to 0.2346 | 1.934 to 4.670 | 0.9029 |
| C707A | 0.005269 | 0.09273 | 0.0025 to 26,122,669 | NA | 0.0408 | |
| E673A | 0.0108 | 1.54 × 10−10 | 0.0017 to 9,515,687 | NA | 0.01661 | |
| E673D | 0.006758 | 2.156 | 0.0018 to 1,634,800,901 | NA | 0.0050 |
| Name | 5’–3’ Sequence |
|---|---|
| Amel_FTHFDH_ORF_F | ATGGCGCAACTCAAAGTGGC |
| Amel_ FTHFDH _ORF_R | CTAATATTCTACAGTGATAGTTTTTG |
| Amel_ FTHFDH _BHI_HT_F | TCATACGGATCCATGCACCACCACCACCACCACGCGCAACTCAAAGTGGC |
| Amel_ FTHFDH _NotI_R | TCATACGCGGCCGCCTAATATTCTACAGTGATAGTTTTTG |
| Amel_ FTHFDH _E673A_F | ATCCCTAGCATTAGGTGGAA |
| Amel_ FTHFDH _E673D_F | ATCCCTAGACTTAGGTGGAA |
| Amel_ FTHFDH _E673AD_R | ACTTTCTTCAAATTACTATT |
| Amel_ FTHFDH _C707A_Ass_F | CAAAGGAGAAAACGCAATAG |
| Amel_ FTHFDH _C707A_Ass_R | CTATTGCGTTTTCTCCTTTGTTGAAGAACAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mating, M.; Zou, Y.; Sharbati, S.; Einspanier, R. The Active Site of the Enzyme 10-Formyl-THFDH in the Honey Bee Apis mellifera—A Key Player in Formic Acid Detoxification. Int. J. Mol. Sci. 2023, 24, 354. https://doi.org/10.3390/ijms24010354
Mating M, Zou Y, Sharbati S, Einspanier R. The Active Site of the Enzyme 10-Formyl-THFDH in the Honey Bee Apis mellifera—A Key Player in Formic Acid Detoxification. International Journal of Molecular Sciences. 2023; 24(1):354. https://doi.org/10.3390/ijms24010354
Chicago/Turabian StyleMating, Moritz, Ye Zou, Soroush Sharbati, and Ralf Einspanier. 2023. "The Active Site of the Enzyme 10-Formyl-THFDH in the Honey Bee Apis mellifera—A Key Player in Formic Acid Detoxification" International Journal of Molecular Sciences 24, no. 1: 354. https://doi.org/10.3390/ijms24010354
APA StyleMating, M., Zou, Y., Sharbati, S., & Einspanier, R. (2023). The Active Site of the Enzyme 10-Formyl-THFDH in the Honey Bee Apis mellifera—A Key Player in Formic Acid Detoxification. International Journal of Molecular Sciences, 24(1), 354. https://doi.org/10.3390/ijms24010354






