A Novel Anti-TRPV6 Antibody and Its Application in Cancer Diagnosis In Vitro
Abstract
1. Introduction
2. Results
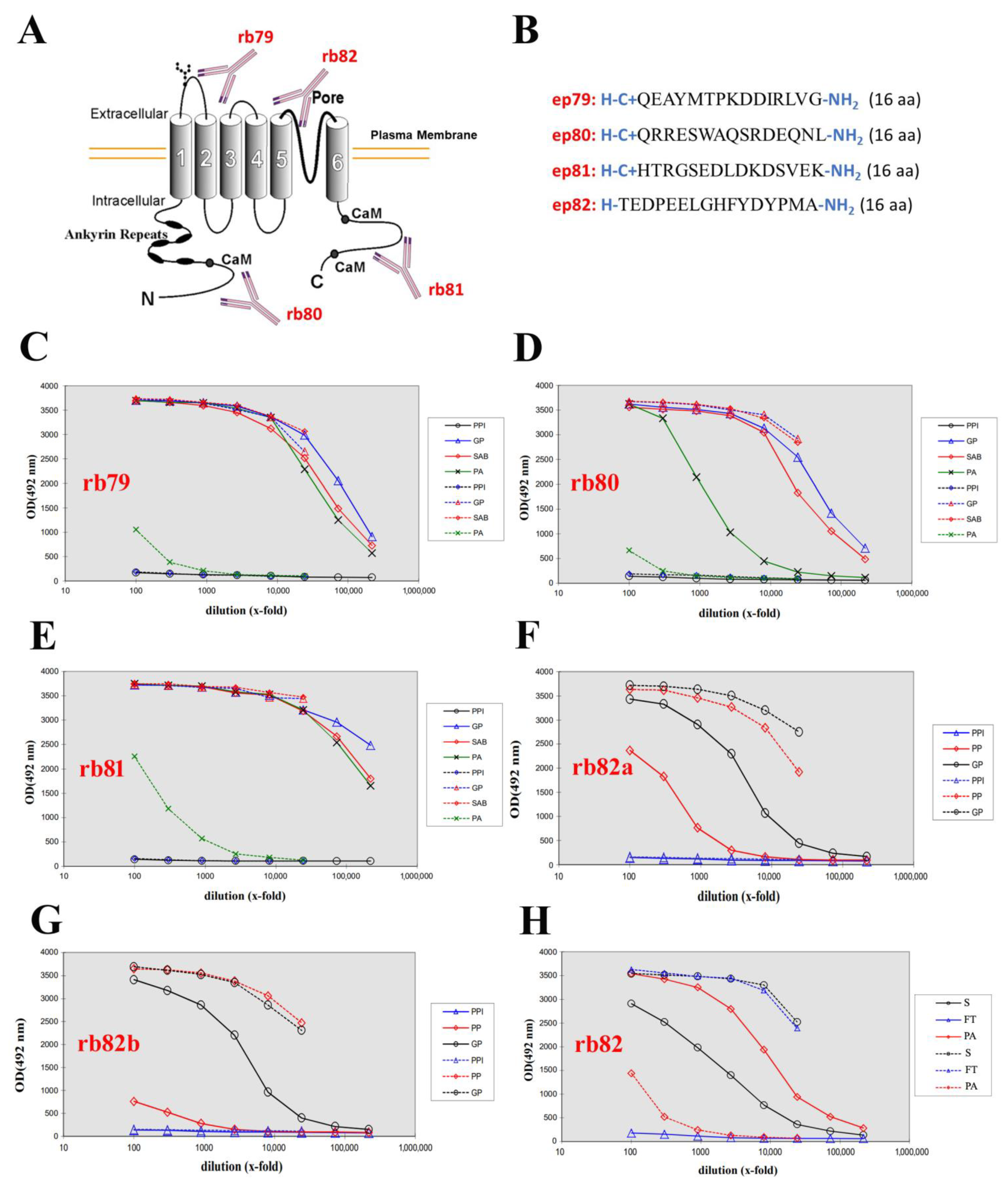
2.1. Generation of Four Rabbit Polyclonal Antibodies
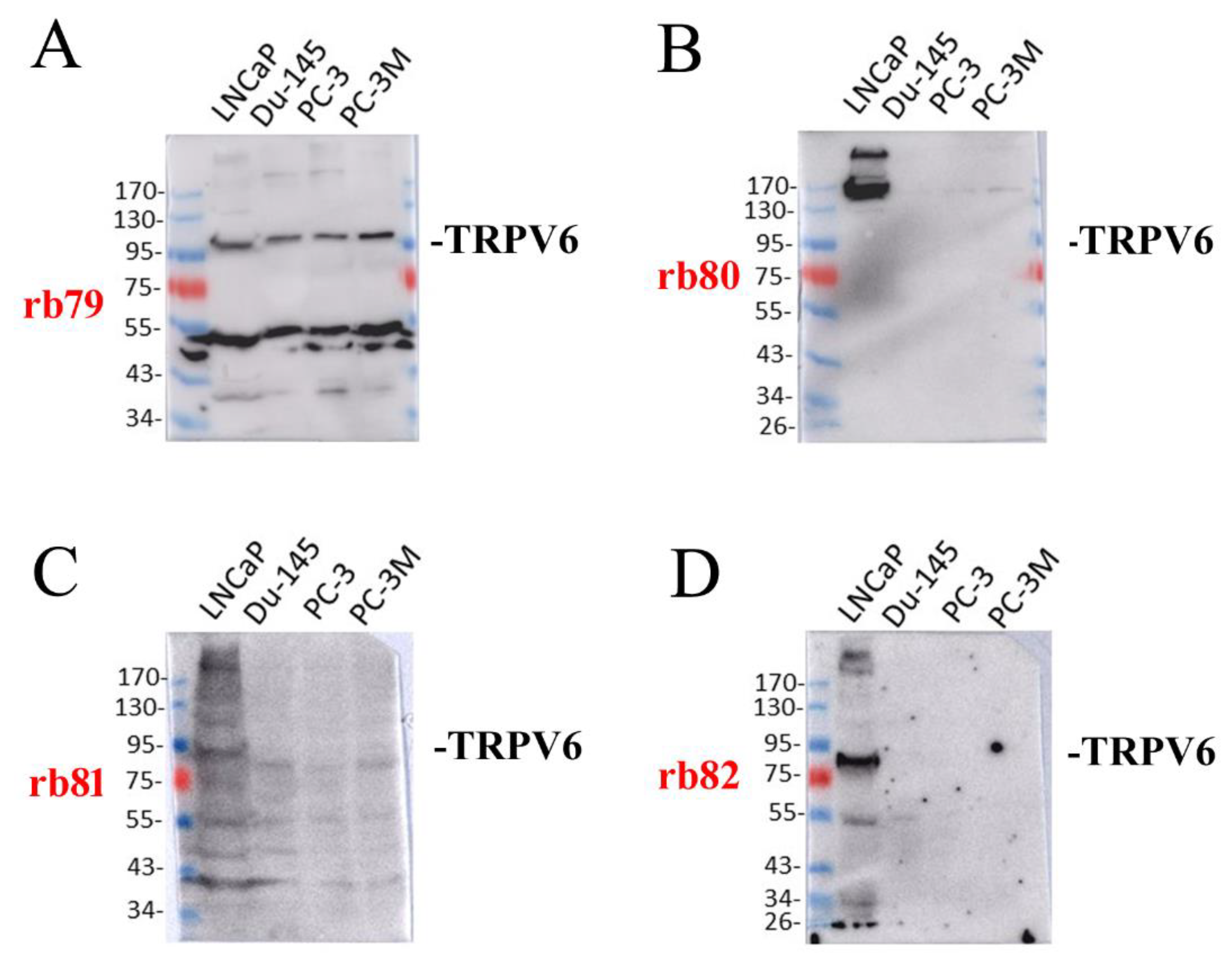
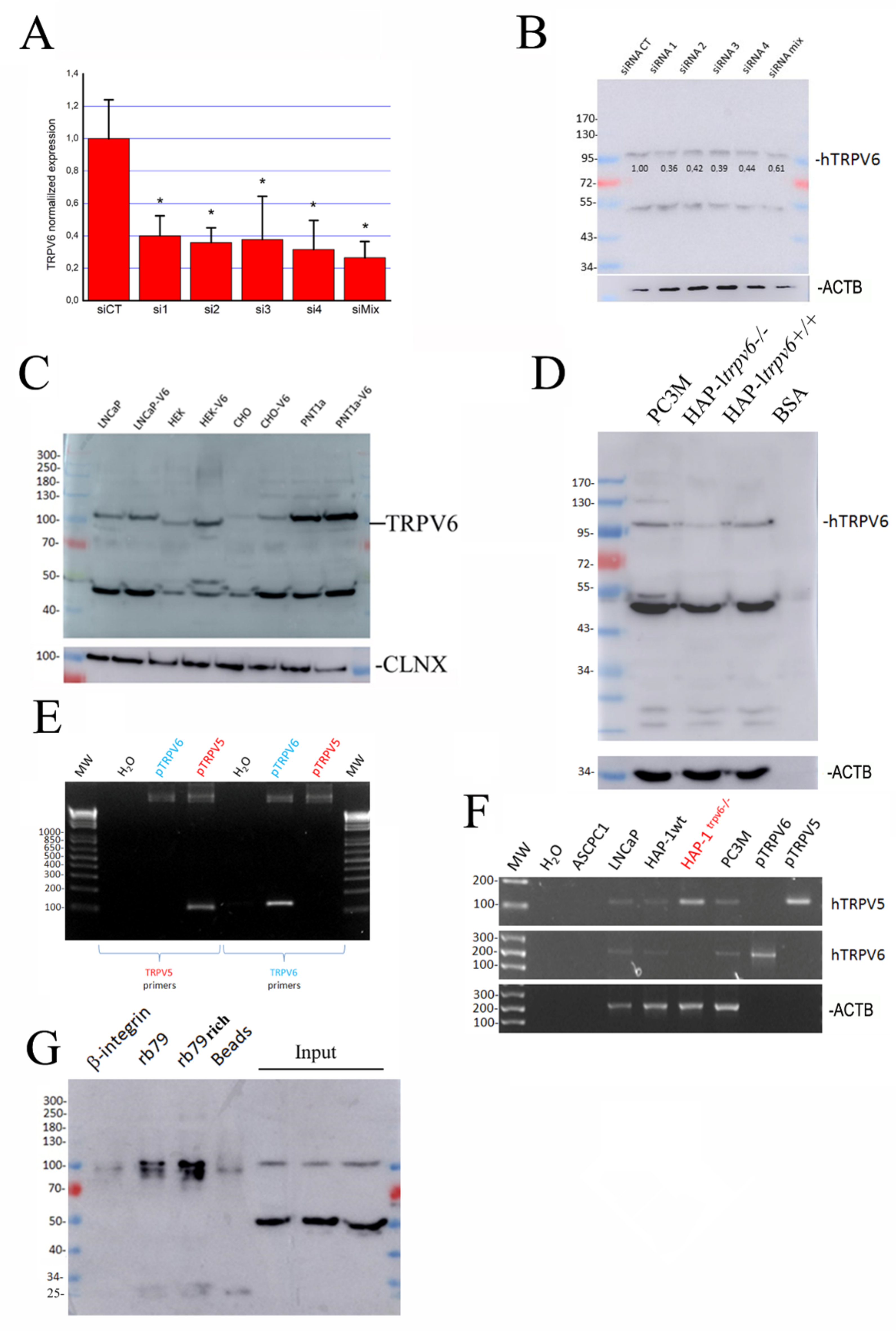
2.2. The Expression of TRPV6 Protein Revealed with Rabbit Polyclonal Antibodies
2.3. Is There Any Splice Variant of the TRPV6 Protein in LNCaP Cells?
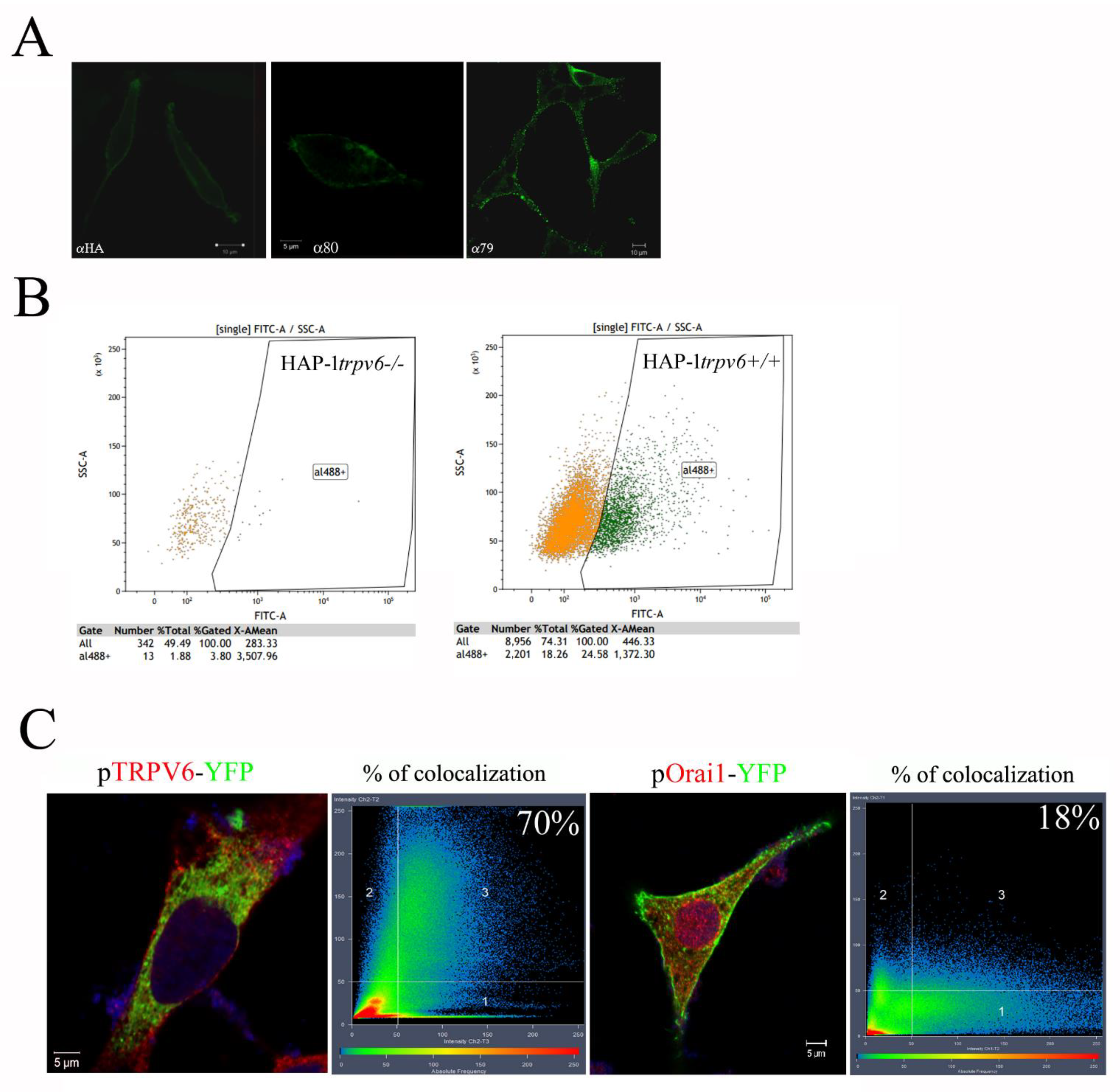
2.4. Use of the Rabbit Antibody 79 in Immunofluorescence Experiences and FACS
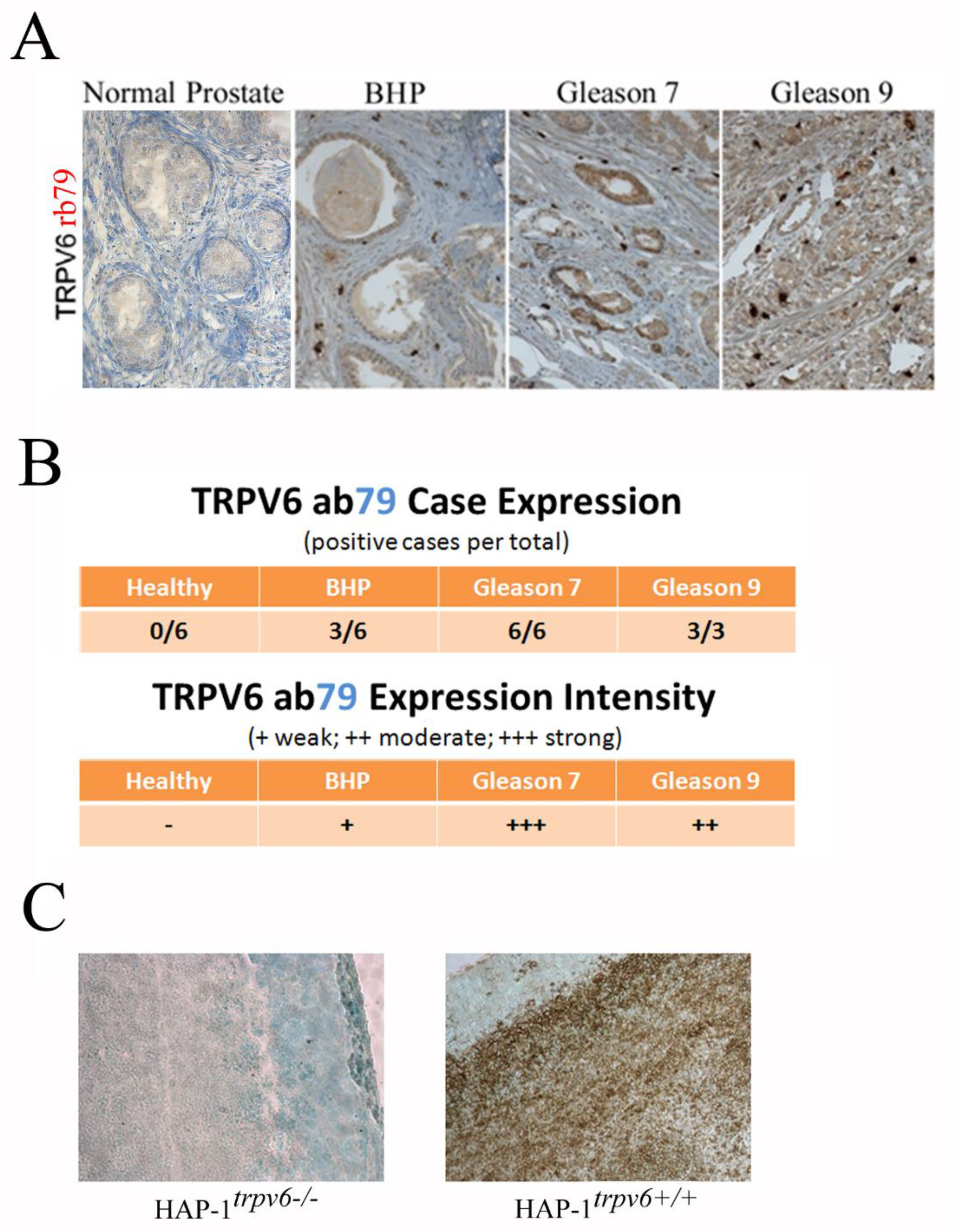
2.5. Use of the Rabbit Antibody 79 in the Diagnosis and Prognostics in Clinics
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. SDS-PAGE and Western Blotting
4.3. RT-PCR
4.4. Quantitative Real-Time PCR
4.5. siRNA Transfection
4.6. Nucleofection
4.7. Confocal Microscopy
4.8. Immunoprecipitation
4.9. FACS
4.10. Immunohistochemistry
4.11. Plasmids
4.12. HAP-1trpv6−/− Model Creation
4.13. Antibody Production
4.14. Reagents
4.15. Data Analysis
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Clapham, D.E.; Runnels, L.W.; Strübing, C. The TRP Ion Channel Family. Nat. Rev. Neurosci. 2001, 2, 387–396. [Google Scholar] [CrossRef]
- Bödding, M.; Flockerzi, V. Ca2+ Dependence of the Ca2+-Selective TRPV6 Channel. J. Biol. Chem. 2004, 279, 36546–36552. [Google Scholar] [CrossRef]
- Hoenderop, J.G.J.; Nilius, B.; Bindels, R.J.M. Epithelial Calcium Channels: From Identification to Function and Regulation. Pflug. Arch. 2003, 446, 304–308. [Google Scholar] [CrossRef]
- Peng, J.B.; Zhuang, L.; Berger, U.V.; Adam, R.M.; Williams, B.J.; Brown, E.M.; Hediger, M.A.; Freeman, M.R. CaT1 Expression Correlates with Tumor Grade in Prostate Cancer. Biochem. Biophys. Res. Commun. 2001, 282, 729–734. [Google Scholar] [CrossRef]
- Nijenhuis, T.; Hoenderop, J.G.J.; Bindels, R.J.M. TRPV5 and TRPV6 in Ca(2+) (Re)Absorption: Regulating Ca(2+) Entry at the Gate. Pflug. Arch. 2005, 451, 181–192. [Google Scholar] [CrossRef]
- Zhuang, L.; Peng, J.-B.; Tou, L.; Takanaga, H.; Adam, R.M.; Hediger, M.A.; Freeman, M.R. Calcium-Selective Ion Channel, CaT1, Is Apically Localized in Gastrointestinal Tract Epithelia and Is Aberrantly Expressed in Human Malignancies. Lab. Investig. 2002, 82, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Bianco, S.D.C.; Peng, J.-B.; Takanaga, H.; Suzuki, Y.; Crescenzi, A.; Kos, C.H.; Zhuang, L.; Freeman, M.R.; Gouveia, C.H.A.; Wu, J.; et al. Marked Disturbance of Calcium Homeostasis in Mice with Targeted Disruption of the Trpv6 Calcium Channel Gene. J. Bone Miner. Res. 2007, 22, 274–285. [Google Scholar] [CrossRef]
- Fecher-Trost, C.; Wissenbach, U.; Beck, A.; Schalkowsky, P.; Stoerger, C.; Doerr, J.; Dembek, A.; Simon-Thomas, M.; Weber, A.; Wollenberg, P.; et al. The in Vivo TRPV6 Protein Starts at a Non-AUG Triplet, Decoded as Methionine, Upstream of Canonical Initiation at AUG. J. Biol. Chem. 2013, 288, 16629–16644. [Google Scholar] [CrossRef]
- Bidaux, G.; Borowiec, A.-S.; Dubois, C.; Delcourt, P.; Schulz, C.; Vanden Abeele, F.; Lepage, G.; Desruelles, E.; Bokhobza, A.; Dewailly, E.; et al. Targeting of Short TRPM8 Isoforms Induces 4TM-TRPM8-Dependent Apoptosis in Prostate Cancer Cells. Oncotarget 2016, 7, 29063–29080. [Google Scholar] [CrossRef] [PubMed]
- Lehen’kyi, V.; Raphaël, M.; Prevarskaya, N. The Role of the TRPV6 Channel in Cancer. J. Physiol. 2012, 590, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Fixemer, T.; Wissenbach, U.; Flockerzi, V.; Bonkhoff, H. Expression of the Ca2+-Selective Cation Channel TRPV6 in Human Prostate Cancer: A Novel Prognostic Marker for Tumor Progression. Oncogene 2003, 22, 7858–7861. [Google Scholar] [CrossRef] [PubMed]
- Raphaël, M.; Lehen’kyi, V.; Vandenberghe, M.; Beck, B.; Khalimonchyk, S.; Vanden Abeele, F.; Farsetti, L.; Germain, E.; Bokhobza, A.; Mihalache, A.; et al. TRPV6 Calcium Channel Translocates to the Plasma Membrane via Orai1-Mediated Mechanism and Controls Cancer Cell Survival. Proc. Natl. Acad. Sci. USA 2014, 111, E3870–E3879. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.-B. TRPV5 and TRPV6 in Transcellular Ca(2+) Transport: Regulation, Gene Duplication, and Polymorphisms in African Populations. Adv. Exp. Med. Biol. 2011, 704, 239–275. [Google Scholar] [CrossRef] [PubMed]
- Wolske, K.; Wyatt, A.; Wissenbach, U. Comments on the Evolution of TRPV6. Ann. Anat. 2021, 238, 151753. [Google Scholar] [CrossRef]
- Popp, M.W.; Maquat, L.E. Leveraging Rules of Nonsense-Mediated MRNA Decay for Genome Engineering and Personalized Medicine. Cell 2016, 165, 1319–1322. [Google Scholar] [CrossRef]
- Wissenbach, U.; Niemeyer, B.; Himmerkus, N.; Fixemer, T.; Bonkhoff, H.; Flockerzi, V. TRPV6 and Prostate Cancer: Cancer Growth beyond the Prostate Correlates with Increased TRPV6 Ca2+ Channel Expression. Biochem. Biophys. Res. Commun. 2004, 322, 1359–1363. [Google Scholar] [CrossRef]
- Wissenbach, U.; Niemeyer, B.A.; Fixemer, T.; Schneidewind, A.; Trost, C.; Cavalie, A.; Reus, K.; Meese, E.; Bonkhoff, H.; Flockerzi, V. Expression of CaT-like, a Novel Calcium-Selective Channel, Correlates with the Malignancy of Prostate Cancer. J. Biol. Chem. 2001, 276, 19461–19468. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion Channels and the Hallmarks of Cancer. Trends Mol. Med. 2010, 16, 107–121. [Google Scholar] [CrossRef]
- Lehen’kyi, V.; Flourakis, M.; Skryma, R.; Prevarskaya, N. TRPV6 Channel Controls Prostate Cancer Cell Proliferation via Ca(2+)/NFAT-Dependent Pathways. Oncogene 2007, 26, 7380–7385. [Google Scholar] [CrossRef]
- Chang, Q.; Hoefs, S.; van der Kemp, A.W.; Topala, C.N.; Bindels, R.J.; Hoenderop, J.G. The Beta-Glucuronidase Klotho Hydrolyzes and Activates the TRPV5 Channel. Science 2005, 310, 490–493. [Google Scholar] [CrossRef]
- Lu, P.; Boros, S.; Chang, Q.; Bindels, R.J.; Hoenderop, J.G. The Beta-Glucuronidase Klotho Exclusively Activates the Epithelial Ca2+ Channels TRPV5 and TRPV6. Nephrol. Dial Transpl. 2008, 23, 3397–3402. [Google Scholar] [CrossRef] [PubMed]
- Essletzbichler, P.; Konopka, T.; Santoro, F.; Chen, D.; Gapp, B.V.; Kralovics, R.; Brummelkamp, T.R.; Nijman, S.M.B.; Bürckstümmer, T. Megabase-Scale Deletion Using CRISPR/Cas9 to Generate a Fully Haploid Human Cell Line. Genome Res. 2014, 24, 2059–2065. [Google Scholar] [CrossRef] [PubMed]

| Accession Number | Forward | Backward | Expected Size (b.p) |
|---|---|---|---|
| TRPV6, NM_018646 | CCCTCAGTGTCTCGAAGTAC | TCAGATCTGATATTCCCAGCTC | 134 |
| TRPV6, NM_018646 | CCCAAGGAGAAAGGGCTAAT | TTGGCAGCTAGAAGGAGAGG | 145 |
| TRPV5, NM_019841 | TCTTCCAACTTCCTCCCTG | CCTCACTAAGGTTCAGTCCAAG | 115 |
| ACTB, NM_001101 | CAGAGCAAGAGAGGCATCCT | GTTGAAGGTCTCAAACATGATC | 209 |
| HPRT, NM_000194 | GGCGTCGTGATTAGTGATGAT | CGAGCAAGACGTTCAGTCCT | 134 |
| TRPV6 siRNA-1 | 5′-CCUGCUGCAGCAGAAGAGG (dTdT)-3′ | ||
| TRPV6 siRNA-2 | 5′-GACUCUCUAUGACCUCACA (dTdT)-3′ | ||
| TRPV6 siRNA-3 | 5′-CGUCAUGUACUUCGCCCGA (dTdT)-3′ | ||
| TRPV6 siRNA-4 | 5′-CCUCCUCAUUGCCAUGAUG (dTdT)-3′ | ||
| siLuciferase, AB_490793 | 5′-CUUACGCCUGAGUACUUCGA (dTdT)-3′ | ||
| Target and Accession Number | Sequence |
|---|---|
| TRPV6, NM_018646 | V6-ex1-F-1: AAGGCAGGAGACAGGAGAC |
| TRPV6, NM_018646 | V6-ex1-F0: GACCTCTACAGGGAGACGG |
| TRPV6, NM_018646 | V6-ex3-B2: CATAGAGCTCAGATGTCATGG |
| TRPV6, NM_018646 | V6-ex4-F3: CAGAACATGAACCTGGTGC |
| TRPV6, NM_018646 | V6-ex5-B3: CGATCTCCTCACTGTTCACA |
| TRPV6, NM_018646 | V6-ex6-B4: CTGTCGTAGGACAGCAACAG |
| TRPV6, NM_018646 | V6-ex7-B5: GGTGGTGATGATAAGTTCCAG |
| TRPV6, NM_018646 | V6-ex8-F4: GGTGCCATATATCTGCTGTAC |
| TRPV6, NM_018646 | V6-ex9-B6: CTACCAGCAGGATGATGATAG |
| TRPV6, NM_018646 | V6-ex10-B7: GGATGGTCTGTCCAAAGAAG |
| TRPV6, NM_018646 | V6-ex11-Fsq: GGCTGGTGCAACGTCATGTAC |
| TRPV6, NM_018646 | V6-ex12-Fex: ATTCTGCTGGCTGATGGC |
| TRPV6, NM_018646 | V6-ex13-Bex: CGATGATGGTAAGGAACAGC |
| TRPV6, NM_018646 | V6-ex14-F5: TTGTGGCCACCACGGTG |
| TRPV6, NM_018646 | V6-ex15-B8: AGGTACTTCGAGACACTGAGG |
| Primers | Expected Size, bp | Targeted Exon |
|---|---|---|
| F-1/B2 | 480 | (5′-UTR)-2 |
| F0/B2 | 460 | (5′-UTR)-2 |
| F1/B1 | 150 | 1-2 |
| F1/B2 | 330 | 1-2-3 |
| F3/B3 | 150 | 4-5 |
| F3/B4 | 290 | 4-5-6 |
| F3/B5 | 500 | 4-5-6-7 |
| F4/B6 | 210 | 8-9 |
| F4/B7 | 260 | 8-9-10 |
| Fsq/Bex | 240 | 11-12 |
| Fex/Bex | 150 | 12-13 |
| F5/B8 | 280 | 14-15 |
| Antibody | Design/Manufacturer | Clone | Epitope | Dilution | IHC Pretreatment and Incubation |
|---|---|---|---|---|---|
| TRPV6 | Design: Dr. LEHEN’KYI, Produced: EUROGENTEC, Ltd., Seraing Belgium | rb79 | QEAYMTPKDDIRLVG | 1/200 IHC | CC2 STD 32 min |
| TRPV6 | Design: Dr. LEHEN’KYI, Produced: EUROGENTEC, Ltd., Seraing, Belgium | rb80 | QRRESWAQSRDEQNL | 1/500 WB | |
| TRPV6 | Design: Dr. LEHEN’KYI, Produced: EUROGENTEC, Ltd., Seraing, Belgium | rb81 | HTRGSEDLDKDSVEKL | 1/500 WB | |
| TRPV6 | Design: Dr. LEHEN’KYI, Produced: EUROGENTEC, Ltd., Seraing, Belgium | rb82 | TEDPEELGHFYDYPMA | 1/500 WB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haustrate, A.; Mihalache, A.; Cordier, C.; Gosset, P.; Prevarskaya, N.; Lehen’kyi, V. A Novel Anti-TRPV6 Antibody and Its Application in Cancer Diagnosis In Vitro. Int. J. Mol. Sci. 2023, 24, 419. https://doi.org/10.3390/ijms24010419
Haustrate A, Mihalache A, Cordier C, Gosset P, Prevarskaya N, Lehen’kyi V. A Novel Anti-TRPV6 Antibody and Its Application in Cancer Diagnosis In Vitro. International Journal of Molecular Sciences. 2023; 24(1):419. https://doi.org/10.3390/ijms24010419
Chicago/Turabian StyleHaustrate, Aurélien, Adriana Mihalache, Clément Cordier, Pierre Gosset, Natalia Prevarskaya, and V’yacheslav Lehen’kyi. 2023. "A Novel Anti-TRPV6 Antibody and Its Application in Cancer Diagnosis In Vitro" International Journal of Molecular Sciences 24, no. 1: 419. https://doi.org/10.3390/ijms24010419
APA StyleHaustrate, A., Mihalache, A., Cordier, C., Gosset, P., Prevarskaya, N., & Lehen’kyi, V. (2023). A Novel Anti-TRPV6 Antibody and Its Application in Cancer Diagnosis In Vitro. International Journal of Molecular Sciences, 24(1), 419. https://doi.org/10.3390/ijms24010419







