Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets
Abstract
1. Introduction
2. Glycosaminoglycans: Structure, Biosynthesis, and Catabolism
2.1. Glycosaminoglycans Biosynthesis
2.1.1. Keratan Sulfate
2.1.2. Heparan Sulfate (HS)
2.1.3. Chondroitin (CS) and Dermatan Sulfate (DS)
2.2. Glycosaminoglycans Degradation
3. Intracellular Organelle Impairment
3.1. Lysosomal Impairment
3.2. Autophagy Impairment

3.3. Mitochondrial Impairment

3.4. Endoplasmic Reticulum Stress
3.5. Apoptosis Activation
3.6. Immune Response Activation
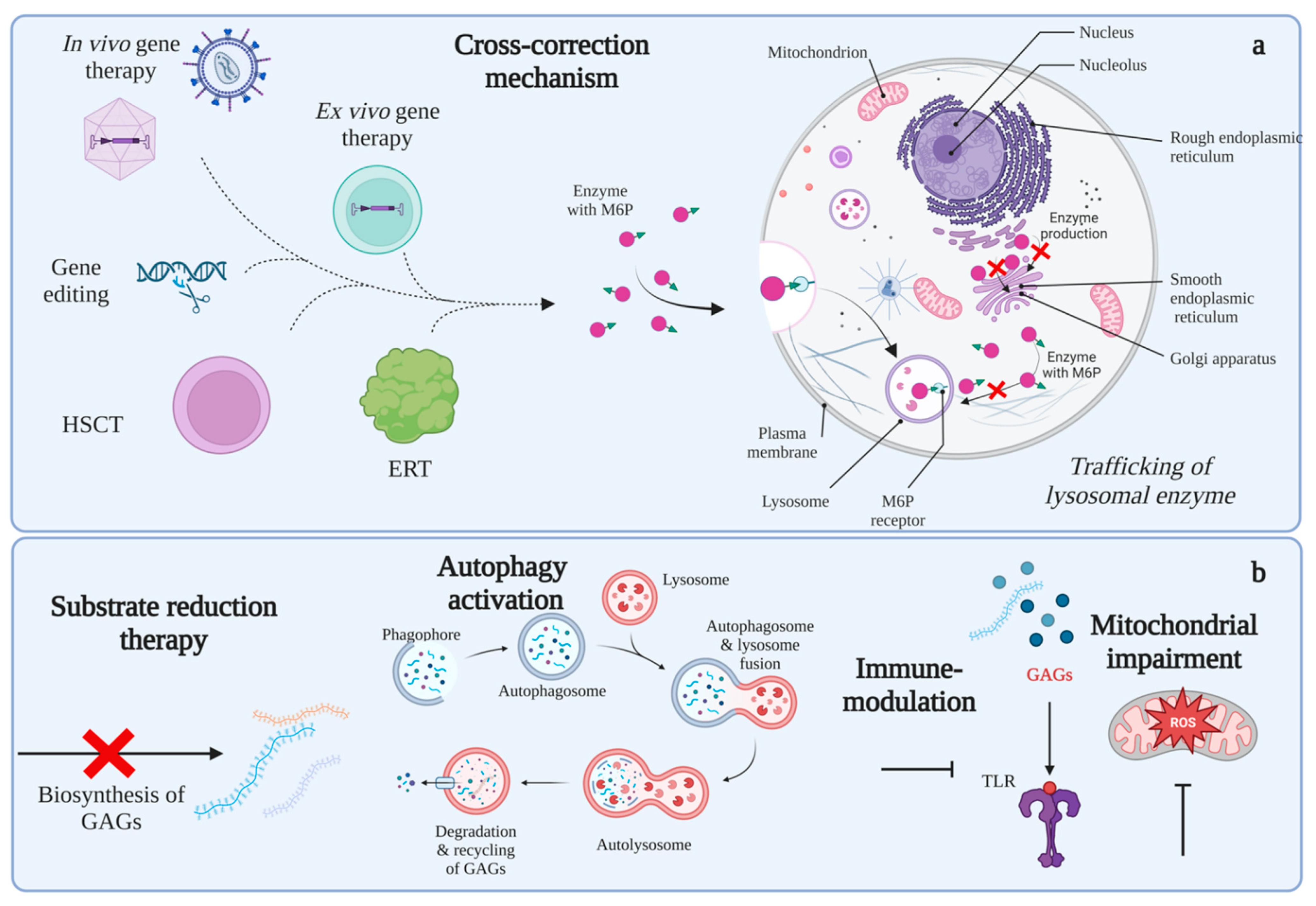
4. Modulation of Altered Intracellular Pathways as a Potential Therapeutic Approach
4.1. Substrate Reduction Therapy (SRT)
4.2. Autophagy Recovery
4.3. Mitochondria and Oxidative Stress
4.4. Immune Response Modulation
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, S.A.; Mason, R.W.; Kobayashi, H.; Yamaguchi, S.; Tomatsu, S. Advances in glycosaminoglycan detection. Mol. Genet. Metab. 2020, 130, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Fecarotta, S.; Tarallo, A.; Damiano, C.; Minopoli, N.; Parenti, G. Pathogenesis of Mucopolysaccharidoses, an Update. Int. J. Mol. Sci. 2020, 21, 2515. [Google Scholar] [CrossRef] [PubMed]
- Salazar, D.A.; Rodríguez-López, A.; Herreño, A.; Barbosa, H.; Herrera, J.; Ardila, A.; Barreto, G.E.; González, J.; Alméciga-Díaz, C.J. Systems biology study of mucopolysaccharidosis using a human metabolic reconstruction network. Mol. Genet. Metab. 2016, 117, 129–139. [Google Scholar] [CrossRef]
- De Pasquale, V.; Pavone, L.M. Heparan sulfate proteoglycans: The sweet side of development turns sour in mucopolysaccharidoses. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 165539. [Google Scholar] [CrossRef]
- Stepien, K.M.; Roncaroli, F.; Turton, N.; Hendriksz, C.J.; Roberts, M.; Heaton, R.A.; Hargreaves, I. Mechanisms of Mitochondrial Dysfunction in Lysosomal Storage Disorders: A Review. J. Clin. Med. 2020, 9, 2596. [Google Scholar] [CrossRef] [PubMed]
- Pierzynowska, K.; Gaffke, L.; Cyske, Z.; Węgrzyn, G.; Buttari, B.; Profumo, E.; Saso, L. Oxidative Stress in Mucopolysaccharidoses: Pharmacological Implications. Molecules 2021, 26, 5616. [Google Scholar] [CrossRef] [PubMed]
- Sawamoto, K.; Stapleton, M.; Alméciga-Díaz, C.J.; Espejo-Mojica, A.J.; Losada, J.C.; Suarez, D.A.; Tomatsu, S. Therapeutic Options for Mucopolysaccharidoses: Current and Emerging Treatments. Drugs 2019, 79, 1103–1134. [Google Scholar] [CrossRef]
- Poletto, E.; Baldo, G.; Gomez-Ospina, N. Genome Editing for Mucopolysaccharidoses. Int. J. Mol. Sci. 2020, 21, 500. [Google Scholar] [CrossRef]
- Leal, A.F.; Espejo-Mojica, A.J.; Sánchez, O.F.; Ramírez, C.M.; Reyes, L.H.; Cruz, J.C.; Alméciga-Díaz, C.J. Lysosomal storage diseases: Current therapies and future alternatives. J. Mol. Med. 2020, 98, 931–946. [Google Scholar] [CrossRef]
- Leal, A.F.; Alméciga-Díaz, C.J. Efficient CRISPR/Cas9 nickase-mediated genome editing in an in vitro model of mucopolysaccharidosis IVA. Gene Ther. 2022, 1–8. [Google Scholar] [CrossRef]
- Alméciga-Diaz, C.; Guevara, J.; Espejo, A.; Rodriguez, A.; Echeverri, O.; Barrera, L. Therapies for Mucopolysaccharidoses: An Overview. In Mucopolysaccharidoses Update (2 Volume Set); Tomatsu, S., Lavery, C., Giugliani, R., Harmatz, P., Scarpa, M., Węgrzyn, G., Orii, T., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2018. [Google Scholar]
- Sawamoto, K.; Chen, H.-H.; Alméciga-Díaz, C.; Mason, R.W.; Tomatsu, S. Gene therapy for Mucopolysaccharidoses. Mol. Genet. Metab. 2017, 123, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Díaz, J.C.L.; del Castillo, J.C.; Rodriguez-López, E.A.; Alméciga-Díaz, C.J. Advances in the Development of Pharmacological Chaperones for the Mucopolysaccharidoses. Int. J. Mol. Sci. 2020, 21, 232. [Google Scholar] [CrossRef] [PubMed]
- Wiesinger, A.-M.; Bigger, B.; Giugliani, R.; Scarpa, M.; Moser, T.; Lampe, C.; Kampmann, C.; Lagler, F.B. The Inflammation in the Cytopathology of Patients With Mucopolysaccharidoses-Immunomodulatory Drugs as an Approach to Therapy. Front. Pharmacol. 2022, 13, 863667. [Google Scholar] [CrossRef] [PubMed]
- Frohbergh, M.; Ge, Y.; Meng, F.; Karabul, N.; Solyom, A.; Lai, A.; Iatridis, J.; Schuchman, E.H.; Simonaro, C.M. Dose Responsive Effects of Subcutaneous Pentosan Polysulfate Injection in Mucopolysaccharidosis Type VI Rats and Comparison to Oral Treatment. PLoS ONE 2014, 9, e100882. [Google Scholar] [CrossRef]
- Capuozzo, A.; Montefusco, S.; Cacace, V.; Sofia, M.; Esposito, A.; Napolitano, G.; Nusco, E.; Polishchuk, E.; Pizzo, M.T.; De Risi, M.; et al. Fluoxetine ameliorates mucopolysaccharidosis type IIIA. Mol. Ther. 2022, 30, 1432–1450. [Google Scholar] [CrossRef]
- Argüello, G.; Balboa, E.; Tapia, P.; Castro, J.; Yañez, M.; Mattar, P.; Pulgar, R.; Zanlungo, S. Genistein Activates Transcription Factor EB and Corrects Niemann–Pick C Phenotype. Int. J. Mol. Sci. 2021, 22, 4220. [Google Scholar] [CrossRef]
- Pierzynowska, K.; Gaffke, L.; Cyske, Z.; Puchalski, M.; Rintz, E.; Bartkowski, M.; Osiadły, M.; Pierzynowski, M.; Mantej, J.; Piotrowska, E.; et al. Autophagy stimulation as a promising approach in treatment of neurodegenerative diseases. Metab. Brain Dis. 2018, 33, 989–1008. [Google Scholar] [CrossRef]
- Rintz, E.; Pierzynowska, K.; Podlacha, M.; Węgrzyn, G. Has resveratrol a potential for mucopolysaccharidosis treatment? Eur. J. Pharm. 2020, 888, 173534. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Mancera, R.L. The Structure of Glycosaminoglycans and their Interactions with Proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. [Google Scholar] [CrossRef]
- Solakyildirim, K. Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques. Anal. Bioanal. Chem. 2019, 411, 3731–3741. [Google Scholar] [CrossRef]
- Nikitovic, D.; Pérez, S. Preface for the Special Issue on the Exploration of the Multifaceted Roles of Glycosaminoglycans: GAGs. Biomolecules 2021, 11, 1630. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H.; Mulloy, B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022. [Google Scholar]
- Haouari, W.; Dubail, J.; Poüs, C.; Cormier-Daire, V.; Bruneel, A. Inherited Proteoglycan Biosynthesis Defects—Current Laboratory Tools and Bikunin as a Promising Blood Biomarker. Genes 2021, 12, 1654. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, N.B.; Domowicz, M.S. Chemistry and Function of Glycosaminoglycans in the Nervous System. Adv. Neurobiol. 2014, 9, 89–115. [Google Scholar] [CrossRef] [PubMed]
- Prydz, K.; Dalen, K.T. Synthesis and sorting of proteoglycans. J. Cell Sci. 2000, 113 (Pt 2), 193–205. [Google Scholar] [CrossRef]
- Funderburgh, J.L. MINI REVIEW Keratan sulfate: Structure, biosynthesis, and function. Glycobiology 2000, 10, 951–958. [Google Scholar] [CrossRef]
- Caterson, B.; Melrose, J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 2018, 28, 182–206. [Google Scholar] [CrossRef]
- Smith, P.D.; Coulson-Thomas, V.J.; Foscarin, S.; Kwok, J.C.; Fawcett, J.W. “GAG-ing with the neuron”: The role of glycosaminoglycan patterning in the central nervous system. Exp. Neurol. 2015, 274, 100–114. [Google Scholar] [CrossRef]
- Mochizuki, H.; Yoshida, K.; Shibata, Y.; Kimata, K. Tetrasulfated disaccharide unit in heparan sulfate: Enzymatic formation and tissue distribution. J. Biol. Chem. 2008, 283, 31237–31245. [Google Scholar] [CrossRef]
- Farrugia, B.L.; Lord, M.S.; Melrose, J.; Whitelock, J.M. The Role of Heparan Sulfate in Inflammation, and the Development of Biomimetics as Anti-Inflammatory Strategies. J. Histochem. Cytochem. 2018, 66, 321–336. [Google Scholar] [CrossRef]
- Wilusz, R.E.; Sanchez-Adams, J.; Guilak, F. The structure and function of the pericellular matrix of articular cartilage. Matrix Biol. 2014, 39, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.J.; Farrugia, B.L.; Biose, I.J.; Bix, G.J.; Melrose, J. Perlecan, A Multi-Functional, Cell-Instructive, Matrix-Stabilizing Proteoglycan With Roles in Tissue Development Has Relevance to Connective Tissue Repair and Regeneration. Front. Cell Dev. Biol. 2022, 10, 856261. [Google Scholar] [CrossRef] [PubMed]
- Pichert, A.; Schlorke, D.; Franz, S.; Arnhold, J. Functional aspects of the interaction between interleukin-8 and sulfated glycosaminoglycans. Biomatter 2012, 2, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Crijns, H.; Vanheule, V.; Proost, P. Targeting Chemokine—Glycosaminoglycan Interactions to Inhibit Inflammation. Front. Immunol. 2020, 11, 483. [Google Scholar] [CrossRef]
- Bosch, M.E.; Kielian, T. Neuroinflammatory paradigms in lysosomal storage diseases. Front. Neurosci. 2015, 9, 417. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.S.; Routhe, L.J.; Moos, T. The vascular basement membrane in the healthy and pathological brain. J. Cereb. Blood Flow Metab. 2017, 37, 3300–3317. [Google Scholar] [CrossRef] [PubMed]
- Amendum, P.C.; Khan, S.; Yamaguchi, S.; Kobayashi, H.; Ago, Y.; Suzuki, Y.; Celik, B.; Rintz, E.; Hossain, J.; Xiao, W.; et al. Glycosaminoglycans as Biomarkers for Mucopolysaccharidoses and Other Disorders. Diagnostics 2021, 11, 1563. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.A.; Sun, M.; Barnum, C.E.; Weiss, S.N.; Huegel, J.; Shetye, S.S.; Lin, L.; Saez, D.; Adams, S.M.; Iozzo, R.V.; et al. Decorin and biglycan are necessary for maintaining collagen fibril structure, fiber realignment, and mechanical properties of mature tendons. Matrix Biol. 2017, 64, 81–93. [Google Scholar] [CrossRef]
- Koosha, E.; Eames, B.F. Two Modulators of Skeletal Development: BMPs and Proteoglycans. J. Dev. Biol. 2022, 10, 15. [Google Scholar] [CrossRef]
- Filocamo, M.; Tomanin, R.; Bertola, F.; Morrone, A. Biochemical and molecular analysis in mucopolysaccharidoses: What a paediatrician must know. Ital. J. Pediatr. 2018, 44, 35–45. [Google Scholar] [CrossRef]
- Celik, B.; Tomatsu, S.C.; Tomatsu, S.; Khan, S.A. Epidemiology of Mucopolysaccharidoses Update. Diagnostics 2021, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Peracha, H.; Ballhausen, D.; Wiesbauer, A.; Rohrbach, M.; Gautschi, M.; Mason, R.W.; Giugliani, R.; Suzuki, Y.; Orii, K.E.; et al. Epidemiology of mucopolysaccharidoses. Mol. Genet. Metab. 2017, 121, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, S.; Blatterer, J.; Speicher, M.R.; Bhavani, G.S.; Boons, G.-J.; Ilse, M.-B.; Andrae, D.; Sproß, J.; Vaz, F.M.; Kircher, S.G.; et al. Novel subtype of mucopolysaccharidosis caused by arylsulfatase K (ARSK) deficiency. J. Med. Genet. 2021, 59, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Giugliani, R.; Tomatsu, S.; Lavery, C.; Harmatz, P.; Scarpa, M.; Węgrzyn, G.; Orii, T. Overview of Mucopolysaccharidoses: Diagnosis, Natural History, Clinical pictures. In Mucopolysaccharidoses Update (2 Volume Set); Tomatsu, S., Lavery, C., Giugliani, R., Harmatz, P., Scarpa, M., Węgrzyn, G., Orii, T., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2018; pp. 17–19. [Google Scholar]
- Parini, R.; Biondi, A. The new frame for Mucopolysaccharidoses. Ital. J. Pediatr. 2018, 44, 117. [Google Scholar] [CrossRef]
- Lawrence, R.; Brown, J.R.; Lorey, F.; Dickson, P.I.; Crawford, B.E.; Esko, J.D. Glycan-based biomarkers for mucopolysaccharidoses. Mol. Genet. Metab. 2014, 111, 73–83. [Google Scholar] [CrossRef]
- Saftig, P.; Puertollano, R. How Lysosomes Sense, Integrate, and Cope with Stress. Trends Biochem. Sci. 2021, 46, 97–112. [Google Scholar] [CrossRef]
- Gaffke, L.; Pierzynowska, K.; Podlacha, M.; Brokowska, J.; Węgrzyn, G. Changes in cellular processes occurring in mucopolysaccharidoses as underestimated pathomechanisms of these diseases. Cell Biol. Int. 2021, 45, 498–506. [Google Scholar] [CrossRef]
- Lakpa, K.L.; Khan, N.; Afghah, Z.; Chen, X.; Geiger, J.D. Lysosomal Stress Response (LSR): Physiological Importance and Pathological Relevance. J. Neuroimmune Pharmacol. 2021, 16, 219–237. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142. [Google Scholar] [CrossRef]
- Simonaro, C.M. Lysosomes, Lysosomal Storage Diseases, and Inflammation. J. Inborn Errors Metab. Screen. 2019, 4, 1–8. [Google Scholar] [CrossRef]
- Moskot, M.; Montefusco, S.; Jakóbkiewicz-Banecka, J.; Mozolewski, P.; Węgrzyn, A.; Di Bernardo, D.; Węgrzyn, G.; Medina, D.L.; Ballabio, A.; Gabig-Cimińska, M. The Phytoestrogen Genistein Modulates Lysosomal Metabolism and Transcription Factor EB (TFEB) Activation. J. Biol. Chem. 2014, 289, 17054–17069. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Martina, J.A.; Puertollano, R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J. Biol. Chem. 2018, 293, 12525–12534. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J. Cell Sci. 2018, 131, jcs208884. [Google Scholar] [CrossRef]
- Chauhan, S.; Kumar, S.; Jain, A.; Ponpuak, M.; Mudd, M.H.; Kimura, T.; Choi, S.W.; Peters, R.; Mandell, M.; Bruun, J.-A.; et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell 2016, 39, 13–27. [Google Scholar] [CrossRef]
- Jia, J.; Bissa, B.; Brecht, L.; Allers, L.; Choi, S.W.; Gu, Y.; Zbinden, M.; Burge, M.R.; Timmins, G.; Hallows, K.; et al. AMPK, a Regulator of Metabolism and Autophagy, Is Activated by Lysosomal Damage via a Novel Galectin-Directed Ubiquitin Signal Transduction System. Mol. Cell 2020, 77, 951–969.e9. [Google Scholar] [CrossRef]
- Li, F.-Y.; Wang, S.-F.; Bernardes, E.S.; Liu, F.-T. Galectins in Host Defense Against Microbial Infections. Adv. Exp. Med. Biol. 2020, 1204, 141–167. [Google Scholar] [CrossRef]
- Patel, S.; Homaei, A.; El-Seedi, H.R.; Akhtar, N. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed. Pharmacother. 2018, 105, 526–532. [Google Scholar] [CrossRef]
- Qi, X.; Man, S.M.; Malireddi, R.S.; Karki, R.; Lupfer, C.; Gurung, P.; Neale, G.; Guy, C.S.; Lamkanfi, M.; Kanneganti, T.-D. Cathepsin B modulates lysosomal biogenesis and host defense against Francisella novicida infection. J. Exp. Med. 2016, 213, 2081–2097. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Seranova, E.; Connolly, K.J.; Zatyka, M.; Rosenstock, T.; Barrett, T.; Tuxworth, R.I.; Sarkar, S. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem. 2017, 61, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Monaco, A.; Fraldi, A. Protein aggregation and autophagy dysfunction: New lessons from mucopolysaccharidoses. Autophagy 2021, 17, 3875–3876. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Fraldi, A.; Jahreiss, L.; Spampanato, C.; Venturi, C.; Medina, D.L.; De Pablo, R.; Tacchetti, C.; Rubinsztein, D.C.; Ballabio, A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008, 17, 119–129. [Google Scholar] [CrossRef]
- Monaco, A.; Maffia, V.; Sorrentino, N.C.; Sambri, I.; Ezhova, Y.; Giuliano, T.; Cacace, V.; Nusco, E.; De Risi, M.; De Leonibus, E.; et al. The Amyloid Inhibitor CLR01 Relieves Autophagy and Ameliorates Neuropathology in a Severe Lysosomal Storage Disease. Mol. Ther. 2020, 28, 1167–1176. [Google Scholar] [CrossRef]
- Pshezhetsky, A.V. Lysosomal storage of heparan sulfate causes mitochondrial defects, altered autophagy, and neuronal death in the mouse model of mucopolysaccharidosis III type C. Autophagy 2016, 12, 1059–1060. [Google Scholar] [CrossRef]
- Maeda, M.; Seto, T.; Kadono, C.; Morimoto, H.; Kida, S.; Suga, M.; Nakamura, M.; Kataoka, Y.; Hamazaki, T.; Shintaku, H. Autophagy in the Central Nervous System and Effects of Chloroquine in Mucopolysaccharidosis Type II Mice. Int. J. Mol. Sci. 2019, 20, 5829. [Google Scholar] [CrossRef]
- Fedele, A.O.; Proud, C.G. Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci. Rep. 2020, 40, BSR20200905. [Google Scholar] [CrossRef]
- Bartolomeo, R.; Cinque, L.; De Leonibus, C.; Forrester, A.; Salzano, A.C.; Monfregola, J.; De Gennaro, E.; Nusco, E.; Azario, I.; Lanzara, C.; et al. mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J. Clin. Investig. 2017, 127, 3717–3729. [Google Scholar] [CrossRef]
- Puentes-Tellez, M.A.; Sánchez, O.F.; Rojas-Rodriguez, F.; Benincore-Flórez, E.; Barbosa, H.; Díaz, C.J.A. Evaluation of HIV–1 derived lentiviral vectors as transductors of Mucopolysaccharidosis type IV a fibroblasts. Gene 2021, 780, 145527. [Google Scholar] [CrossRef]
- Alméciga-Diaz, C.J.; Hidalgo, O.A.; Olarte-Avellaneda, S.; Rodríguez-López, A.; Guzman, E.; Garzón, R.; Pimentel-Vera, L.N.; Puentes-Tellez, M.A.; Rojas-Rodriguez, A.F.; Gorshkov, K.; et al. Identification of Ezetimibe and Pranlukast as Pharmacological Chaperones for the Treatment of the Rare Disease Mucopolysaccharidosis Type IVA. J. Med. Chem. 2019, 62, 6175–6189. [Google Scholar] [CrossRef] [PubMed]
- Vitry, S.; Bruyère, J.; Hocquemiller, M.; Bigou, S.; Ausseil, J.; Colle, M.-A.; Prévost, M.-C.; Heard, J.M. Storage Vesicles in Neurons Are Related to Golgi Complex Alterations in Mucopolysaccharidosis IIIB. Am. J. Pathol. 2010, 177, 2984–2999. [Google Scholar] [CrossRef] [PubMed]
- Viana, G.M.; Nascimento, C.C.D.; Paredes-Gamero, E.J.; D’Almeida, V. Altered Cellular Homeostasis in Murine MPS I Fibroblasts: Evidence of Cell-Specific Physiopathology. JIMD Rep. 2017, 36, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Hampe, C.; Yund, B.; Orchard, P.; Lund, T.; Wesley, J.; McIvor, R. Differences in MPS I and MPS II Disease Manifestations. Int. J. Mol. Sci. 2021, 22, 7888. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.; Bigger, B.W. The role of innate immunity in mucopolysaccharide diseases. J. Neurochem. 2019, 148, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.B.; Brunn, G.J.; Kodaira, Y.; Platt, J.L. Receptor-Mediated Monitoring of Tissue Well-Being Via Detection of Soluble Heparan Sulfate by Toll-Like Receptor 4. J. Immunol. 2002, 168, 5233–5239. [Google Scholar] [CrossRef]
- Balderas, P.M.D.O. Mitochondria–plasma membrane interactions and communication. J. Biol. Chem. 2021, 297, 101164. [Google Scholar] [CrossRef]
- Peoples, J.N.; Saraf, A.; Ghazal, N.; Pham, T.T.; Kwong, J.Q. Mitochondrial dysfunction and oxidative stress in heart disease. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Juan, C.; de la Lastra, J.P.; Plou, F.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Runwal, G.; Stamatakou, E.; Siddiqi, F.H.; Puri, C.; Zhu, Y.; Rubinsztein, D.C. LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019, 9, 10147. [Google Scholar] [CrossRef] [PubMed]
- Beese, C.J.; Brynjólfsdóttir, S.H.; Frankel, L.B. Selective Autophagy of the Protein Homeostasis Machinery: Ribophagy, Proteaphagy and ER-Phagy. Front. Cell Dev. Biol. 2020, 7, 373. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Hou, C.; Lai, Y.; Long, J.; Liu, J.; Zhong, Q.; Diao, J. SNARE-mediated membrane fusion in autophagy. Semin. Cell Dev. Biol. 2016, 60, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Filippon, L.; Vanzin, C.S.; Biancini, G.B.; Pereira, I.N.; Manfredini, V.; Sitta, A.; Peralba, M.d.C.R.; Schwartz, I.V.D.; Giugliani, R.; Vargas, C.R. Oxidative stress in patients with mucopolysaccharidosis type II before and during enzyme replacement therapy. Mol. Genet. Metab. 2011, 103, 121–127. [Google Scholar] [CrossRef]
- Arfi, A.; Richard, M.; Gandolphe, C.; Bonnefont-Rousselot, D.; Thérond, P.; Scherman, D. Neuroinflammatory and oxidative stress phenomena in MPS IIIA mouse model: The positive effect of long-term aspirin treatment. Mol. Genet. Metab. 2011, 103, 18–25. [Google Scholar] [CrossRef]
- Jeyakumar, M.; Smith, D.A.; Williams, I.M.; Borja, M.C.; Neville, D.C.A.; Butters, T.D.; Dwek, R.A.; Platt, F. NSAIDs increase survival in the Sandhoff disease mouse: Synergy withN-butyldeoxynojirimycin. Ann. Neurol. 2004, 56, 642–649. [Google Scholar] [CrossRef]
- Donida, B.; Marchetti, D.P.; Biancini, G.B.; Deon, M.; Manini, P.R.; da Rosa, H.T.; Moura, D.J.; Saffi, J.; Bender, F.; Burin, M.G.; et al. Oxidative stress and inflammation in mucopolysaccharidosis type IVA patients treated with enzyme replacement therapy. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2015, 1852, 1012–1019. [Google Scholar] [CrossRef]
- Donida, B.; Marchetti, D.P.; Jacques, C.E.D.; Ribas, G.; Deon, M.; Manini, P.; da Rosa, H.T.; Moura, D.J.; Saffi, J.; Giugliani, R.; et al. Oxidative profile exhibited by Mucopolysaccharidosis type IVA patients at diagnosis: Increased keratan urinary levels. Mol. Genet. Metab. Rep. 2017, 11, 46–53. [Google Scholar] [CrossRef]
- Orhan, H.; Coolen, S.; Meerman, J.H. Quantification of urinary o,o′-dityrosine, a biomarker for oxidative damage to proteins, by high performance liquid chromatography with triple quadrupole tandem mass spectrometry: A comparison with ion-trap tandem mass spectrometry. J. Chromatogr. B 2005, 827, 104–108. [Google Scholar] [CrossRef]
- Graille, M.; Wild, P.; Sauvain, J.-J.; Hemmendinger, M.; Canu, I.G.; Hopf, N. Urinary 8-isoprostane as a biomarker for oxidative stress. A systematic review and meta-analysis. Toxicol. Lett. 2020, 328, 19–27. [Google Scholar] [CrossRef]
- Gaffke, L.; Pierzynowska, K.; Krzelowska, K.; Piotrowska, E.; Węgrzyn, G. Changes in expressions of genes involved in the regulation of cellular processes in mucopolysaccharidoses as assessed by fibroblast culture-based transcriptomic analyses. Metab. Brain Dis. 2020, 35, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.I.; Abreu, D.; McGill, J.B.; Urano, F. Monogenic and syndromic diabetes due to endoplasmic reticulum stress. J. Diabetes Complicat. 2020, 35, 107618. [Google Scholar] [CrossRef] [PubMed]
- Quijada-Fraile, P.; Canales, E.A.; Martín-Hernández, E.; Ballesta-Martínez, M.J.; Guillén-Navarro, E.; Pintos-Morell, G.; Moltó-Abad, M.; Moreno-Martínez, D.; Morillo, S.G.; Blasco-Alonso, J.; et al. Clinical features and health-related quality of life in adult patients with mucopolysaccharidosis IVA: The Spanish experience. Orphanet J. Rare Dis. 2021, 16, 464. [Google Scholar] [CrossRef] [PubMed]
- Sawamoto, K.; González, J.V.; Piechnik, M.; Otero, F.J.; Couce, M.L.; Suzuki, Y.; Tomatsu, S. Mucopolysaccharidosis IVA: Diagnosis, Treatment, and Management. Int. J. Mol. Sci. 2020, 21, 1517. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ren, D. IER3IP1 deficiency leads to increased β-cell death and decreased β-cell proliferation. Oncotarget 2017, 8, 56768–56779. [Google Scholar] [CrossRef] [PubMed]
- Osaki, Y.; Matsuhisa, K.; Che, W.; Kaneko, M.; Asada, R.; Masaki, T.; Imaizumi, K.; Saito, A. Calnexin promotes the folding of mutant iduronate 2-sulfatase related to mucopolysaccharidosis type II. Biochem. Biophys. Res. Commun. 2019, 514, 217–223. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, D. Endoplasmic reticulum-associated degradation and beyond: The multitasking roles for HRD1 in immune regulation and autoimmunity. J. Autoimmun. 2020, 109, 102423. [Google Scholar] [CrossRef]
- Osaki, Y.; Saito, A.; Kanemoto, S.; Kaneko, M.; Matsuhisa, K.; Asada, R.; Masaki, T.; Orii, K.; Fukao, T.; Tomatsu, S.; et al. Shutdown of ER-associated degradation pathway rescues functions of mutant iduronate 2-sulfatase linked to mucopolysaccharidosis type II. Cell Death Dis. 2018, 9, 808. [Google Scholar] [CrossRef]
- Chen, K.C.; Qu, S.; Chowdhury, S.; Noxon, I.C.; Schonhoft, J.D.; Plate, L.; Powers, E.T.; Kelly, J.W.; Lander, G.C.; Wiseman, R.L. The endoplasmic reticulum HSP40 co-chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. EMBO J. 2017, 36, 2296–2309. [Google Scholar] [CrossRef]
- Matsuhisa, K.; Imaizumi, K. Loss of Function of Mutant IDS Due to Endoplasmic Reticulum-Associated Degradation: New Therapeutic Opportunities for Mucopolysaccharidosis Type II. Int. J. Mol. Sci. 2021, 22, 12227. [Google Scholar] [CrossRef]
- Álvarez, V.J.; Bravo, S.B.; Chantada-Vazquez, M.P.; Colón, C.; De Castro, M.J.; Morales, M.; Vitoria, I.; Tomatsu, S.; Otero-Espinar, F.J.; Couce, M.L. Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA. Int. J. Mol. Sci. 2020, 22, 226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huo, C.; Liu, Y.; Su, R.; Zhao, Y.; Li, Y. Mechanism and Disease Association With a Ubiquitin Conjugating E2 Enzyme: UBE2L3. Front. Immunol. 2022, 13, 793610. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, D. Friend or foe: Endoplasmic reticulum protein 29 (ERp29) in epithelial cancer. FEBS Openbio 2015, 5, 91–98. [Google Scholar] [CrossRef][Green Version]
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell. Mol. Immunol. 2021, 18, 1106–1121. [Google Scholar] [CrossRef]
- Obeng, E. Apoptosis (programmed cell death) and its signals—A review. Braz. J. Biol. 2021, 81, 1133–1143. [Google Scholar] [CrossRef]
- Patergnani, S.; Danese, A.; Bouhamida, E.; Aguiari, G.; Previati, M.; Pinton, P.; Giorgi, C. Various Aspects of Calcium Signaling in the Regulation of Apoptosis, Autophagy, Cell Proliferation, and Cancer. Int. J. Mol. Sci. 2020, 21, 8323. [Google Scholar] [CrossRef]
- Pereira, V.G.; Gazarini, M.L.; Rodrigues, L.C.; da Silva, F.H.; Han, S.W.; Martins, A.M.; Tersariol, I.L.; D’Almeida, V. Evidence of lysosomal membrane permeabilization in mucopolysaccharidosis type I: Rupture of calcium and proton homeostasis. J. Cell. Physiol. 2010, 223, 335–342. [Google Scholar] [CrossRef]
- Kobolák, J.; Molnar, K.; Varga, E.; Bock, I.; Jezsó, B.; Téglási, A.; Zhou, S.; Giudice, M.L.; Hoogeveen-Westerveld, M.; Pijnappel, W.P.; et al. Modelling the neuropathology of lysosomal storage disorders through disease-specific human induced pluripotent stem cells. Exp. Cell Res. 2019, 380, 216–233. [Google Scholar] [CrossRef]
- Wu, Y.-P.; Proia, R.L. Deletion of macrophage-inflammatory protein 1α retards neurodegeneration in Sandhoff disease mice. Proc. Natl. Acad. Sci. USA 2004, 101, 8425–8430. [Google Scholar] [CrossRef]
- Villani, G.R.D.; Gargiulo, N.; Faraonio, R.; Castaldo, S.; Gonzalez y Reyero, E.; Di Natale, P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J. Neurosci. Res. 2007, 85, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Martell, L.; Lau, K.; Mei, M.; Burnett, V.; Decker, C.; Foehr, E.D. Biomarker analysis of Morquio syndrome: Identification of disease state and drug responsive markers. Orphanet J. Rare Dis. 2011, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Simonaro, C.M.; Haskins, M.E.; Schuchman, E.H. Articular Chondrocytes from Animals with a Dermatan Sulfate Storage Disease Undergo a High Rate of Apoptosis and Release Nitric Oxide and Inflammatory Cytokines: A Possible Mechanism Underlying Degenerative Joint Disease in the Mucopolysaccharidoses. Lab. Investig. 2001, 81, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Tessitore, A.; Pirozzi, M.; Auricchio, A. Abnormal autophagy, ubiquitination, inflammation and apoptosis are dependent upon lysosomal storage and are useful biomarkers of mucopolysaccharidosis VI. Pathogenetics 2009, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Eliyahu, E.; Wolfson, T.; Ge, Y.; Jepsen, K.J.; Schuchman, E.H.; Simonaro, C.M. Anti-TNF-Alpha Therapy Enhances the Effects of Enzyme Replacement Therapy in Rats with Mucopolysaccharidosis Type VI. PLoS ONE 2011, 6, e22447. [Google Scholar] [CrossRef]
- Mandolfo, O.; Parker, H.; Bigger, B. Innate Immunity in Mucopolysaccharide Diseases. Int. J. Mol. Sci. 2022, 23, 1999. [Google Scholar] [CrossRef]
- Simonaro, C.M.; Ge, Y.; Eliyahu, E.; He, X.; Jepsen, K.J.; Schuchman, E.H. Involvement of the Toll-like receptor 4 pathway and use of TNF-alpha antagonists for treatment of the mucopolysaccharidoses. Proc. Natl. Acad. Sci. USA 2010, 107, 222–227. [Google Scholar] [CrossRef]
- Parker, H.; Ellison, S.M.; Holley, R.J.; O’Leary, C.; Liao, A.; Asadi, J.; Glover, E.; Ghosh, A.; Jones, S.; Wilkinson, F.L.; et al. Haematopoietic stem cell gene therapy with IL -1Ra rescues cognitive loss in mucopolysaccharidosis IIIA. EMBO Mol. Med. 2020, 12, e11185. [Google Scholar] [CrossRef]
- Jeyakumar, M.; Thomas, R.; Elliot-Smith, E.; Smith, D.A.; Van Der Spoel, A.C.; D’Azzo, A.; Perry, V.H.; Butters, T.D.; Dwek, R.A.; Platt, F. Central nervous system inflammation is a hallmark of pathogenesis in mouse models of GM1 and GM2 gangliosidosis. Brain 2003, 126 (Pt 4), 974–987. [Google Scholar] [CrossRef]
- Vitner, E.B.; Platt, F.M.; Futerman, A.H. Common and Uncommon Pathogenic Cascades in Lysosomal Storage Diseases. J. Biol. Chem. 2010, 285, 20423–20427. [Google Scholar] [CrossRef]
- Watson, H.A.; Holley, R.J.; Langford-Smith, K.J.; Wilkinson, F.L.; van Kuppevelt, T.H.; Wynn, R.F.; Wraith, J.E.; Merry, C.L.R.; Bigger, B.W. Heparan Sulfate Inhibits Hematopoietic Stem and Progenitor Cell Migration and Engraftment in Mucopolysaccharidosis I. J. Biol. Chem. 2014, 289, 36194–36203. [Google Scholar] [CrossRef]
- Wilkinson, F.L.; Holley, R.J.; Langford-Smith, K.J.; Badrinath, S.; Liao, A.; Langford-Smith, A.; Cooper, J.D.; Jones, S.A.; Wraith, J.E.; Wynn, R.F.; et al. Neuropathology in Mouse Models of Mucopolysaccharidosis Type I, IIIA and IIIB. PLoS ONE 2012, 7, e35787. [Google Scholar] [CrossRef]
- Amara, A.; Lorthioir, O.; Valenzuela, A.; Magerus, A.; Thelen, M.; Montes, M.; Virelizier, J.-L.; Delepierre, M.; Baleux, F.; Lortat-Jacob, H.; et al. Stromal cell-derived factor-1alpha associates with heparan sulfates through the first beta-strand of the chemokine. J. Biol. Chem. 1999, 274, 23916–23925. [Google Scholar] [CrossRef]
- Spillmann, D.; Witt, D.; Lindahl, U. Defining the Interleukin-8-binding Domain of Heparan Sulfate. J. Biol. Chem. 1998, 273, 15487–15493. [Google Scholar] [CrossRef]
- O’Callaghan, P.; Zhang, X.; Li, J.-P. Heparan Sulfate Proteoglycans as Relays of Neuroinflammation. J. Histochem. Cytochem. 2018, 66, 305–319. [Google Scholar] [CrossRef]
- Proudfoot, A.E.I.; Handel, T.M.; Johnson, Z.; Lau, E.K.; LiWang, P.; Clark-Lewis, I.; Borlat, F.; Wells, T.N.C.; Kosco-Vilbois, M.H. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA 2003, 100, 1885–1890. [Google Scholar] [CrossRef]
- Brennan, T.V.; Lin, L.; Huang, X.; Cardona, D.; Li, Z.; Dredge, K.; Chao, N.J.; Yang, Y. Heparan sulfate, an endogenous TLR4 agonist, promotes acute GVHD after allogeneic stem cell transplantation. Blood 2012, 120, 2899–2908. [Google Scholar] [CrossRef]
- Goodall, K.; Poon, I.; Phipps, S.; Hulett, M.D. Soluble Heparan Sulfate Fragments Generated by Heparanase Trigger the Release of Pro-Inflammatory Cytokines through TLR-4. PLoS ONE 2014, 9, e109596. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. The heparanome and regulation of cell function: Structures, functions and challenges. Front. Biosci. 2008, 13, 4309–4338. [Google Scholar] [CrossRef]
- Lau, E.K.; Paavola, C.D.; Johnson, Z.; Gaudry, J.-P.; Geretti, E.; Borlat, F.; Kungl, A.J.; Proudfoot, A.E.; Handel, T.M. Identification of the Glycosaminoglycan Binding Site of the CC Chemokine, MCP-1. J. Biol. Chem. 2004, 279, 22294–22305. [Google Scholar] [CrossRef]
- Dwyer, C.A.; Scudder, S.L.; Lin, Y.; Dozier, L.; Phan, D.; Allen, N.J.; Patrick, G.N.; Esko, J.D. Neurodevelopmental Changes in Excitatory Synaptic Structure and Function in the Cerebral Cortex of Sanfilippo Syndrome IIIA Mice. Sci. Rep. 2017, 7, 46576. [Google Scholar] [CrossRef]
- Holley, R.J.; Deligny, A.; Wei, W.; Watson, H.A.; Niñonuevo, M.R.; Dagälv, A.; Leary, J.A.; Bigger, B.W.; Kjellén, L.; Merry, C.L.; et al. Mucopolysaccharidosis type I, unique structure of accumulated heparan sulfate and increased N-sulfotransferase activity in mice lacking α-l-iduronidase. J. Biol. Chem. 2011, 286, 37515–37524. [Google Scholar] [CrossRef]
- Daly, T.M.; Lorenz, R.G.; Sands, M.S. Abnormal Immune Function In Vivo in a Murine Model of Lysosomal Storage Disease. Pediatr. Res. 2000, 47, 757–762. [Google Scholar] [CrossRef][Green Version]
- Blum, J.S.; Wearsch, P.A.; Cresswell, P. Pathways of Antigen Processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef]
- DiRosario, J.; Divers, E.; Wang, C.; Etter, J.; Charrier, A.; Jukkola, P.; Auer, H.; Best, V.; Newsom, D.L.; McCarty, D.M.; et al. Innate and adaptive immune activation in the brain of MPS IIIB mouse model. J. Neurosci. Res. 2009, 87, 978–990. [Google Scholar] [CrossRef]
- Chen, H.H.; Sawamoto, K.; Mason, R.W.; Kobayashi, H.; Yamaguchi, S.; Suzuki, Y.; Orii, K.; Orii, T.; Tomatsu, S. Enzyme replacement therapy for mucopolysaccharidoses; past, present, and future. J. Hum. Genet. 2019, 64, 1153–1171. [Google Scholar] [CrossRef]
- Concolino, D.; Deodato, F.; Parini, R. Enzyme replacement therapy: Efficacy and limitations. Ital. J. Pediatr. 2018, 44 (Suppl. S2), 117–126. [Google Scholar] [CrossRef]
- Taylor, M.; Khan, S.; Stapleton, M.; Wang, J.; Chen, J.; Wynn, R.; Yabe, H.; Chinen, Y.; Boelens, J.J.; Mason, R.W.; et al. Hematopoietic Stem Cell Transplantation for Mucopolysaccharidoses: Past, Present, and Future. Biol. Blood Marrow Transplant. 2019, 25, e226–e246. [Google Scholar] [CrossRef]
- Yabe, H.; Tanaka, A.; Chinen, Y.; Kato, S.; Sawamoto, K.; Yasuda, E.; Shintaku, H.; Suzuki, Y.; Orii, T.; Tomatsu, S. Hematopoietic stem cell transplantation for Morquio A syndrome. Mol. Genet. Metab. 2016, 117, 84–94. [Google Scholar] [CrossRef]
- Rintz, E.; Higuchi, T.; Kobayashi, H.; Galileo, D.S.; Wegrzyn, G.; Tomatsu, S. Promoter considerations in the design of lentiviral vectors for use in treating lysosomal storage diseases. Mol. Ther.-Methods Clin. Dev. 2022, 24, 71–87. [Google Scholar] [CrossRef]
- Leal, A.F.; Fnu, N.; Benincore-Flórez, E.; Pachón, A.M.H.; Echeverri-Peña, O.Y.; Alméciga-Díaz, C.J.; Tomatsu, S. The landscape of CRISPR/Cas9 for inborn errors of metabolism. Mol. Genet. Metab. 2022, 138, 106968. [Google Scholar] [CrossRef]
- Gambello, M.J.; Li, H. Current strategies for the treatment of inborn errors of metabolism. J. Genet. Genom. 2018, 45, 61–70. [Google Scholar] [CrossRef]
- Friso, A.; Tomanin, R.; Salvalaio, M.; Scarpa, M. Genistein reduces glycosaminoglycan levels in a mouse model of mucopolysaccharidosis type II. Br. J. Pharmacol. 2010, 159, 1082–1091. [Google Scholar] [CrossRef]
- Malinowska, M.; Wilkinson, F.L.; Langford-Smith, K.J.; Langford-Smith, A.; Brown, J.R.; Crawford, B.E.; Vanier, M.T.; Grynkiewicz, G.; Wynn, R.F.; Wraith, J.E.; et al. Genistein Improves Neuropathology and Corrects Behaviour in a Mouse Model of Neurodegenerative Metabolic Disease. PLoS ONE 2010, 5, e14192. [Google Scholar] [CrossRef]
- Jakóbkiewicz-Banecka, J.; Piotrowska, E.; Narajczyk, M.; Barańska, S.; Węgrzyn, G. Genistein-mediated inhibition of glycosaminoglycan synthesis, which corrects storage in cells of patients suffering from mucopolysaccharidoses, acts by influencing an epidermal growth factor-dependent pathway. J. Biomed. Sci. 2009, 16, 26. [Google Scholar] [CrossRef]
- Piotrowska, E.; Jakóbkiewicz-Banecka, J.; Baranska, S.; Tylki-Szymanska, A.; Czartoryska, B.; Wegrzyn, A.; Wegrzyn, G. Genistein-mediated inhibition of glycosaminoglycan synthesis as a basis for gene expression-targeted isoflavone therapy for mucopolysaccharidoses. Eur. J. Hum. Genet. 2006, 14, 846–852. [Google Scholar] [CrossRef]
- Marucha, J.; Tylki-Szymańska, A.; Jakóbkiewicz-Banecka, J.; Piotrowska, E.; Kloska, A.; Czartoryska, B.; Węgrzyn, G. Improvement in the range of joint motion in seven patients with mucopolysaccharidosis type II during experimental gene expression-targeted isoflavone therapy (GET IT). Am. J. Med. Genet. Part A 2011, 155, 2257–2262. [Google Scholar] [CrossRef]
- Ghosh, A.; Rust, S.; Langford-Smith, K.; Weisberg, D.; Canal, M.; Breen, C.; Hepburn, M.; Tylee, K.; Vaz, F.M.; Vail, A.; et al. High dose genistein in Sanfilippo syndrome: A randomised controlled trial. J. Inherit. Metab. Dis. 2021, 44, 1248–1262. [Google Scholar] [CrossRef]
- Roberts, A.L.K.; Thomas, B.J.; Wilkinson, A.S.; Fletcher, J.M.; Byers, S. Inhibition of Glycosaminoglycan Synthesis Using Rhodamine B in a Mouse Model of Mucopolysaccharidosis Type IIIA. Pediatr. Res. 2006, 60, 309–314. [Google Scholar] [CrossRef]
- Derrick-Roberts, A.L.K.; Jackson, M.R.; Pyragius, C.E.; Byers, S. Substrate Deprivation Therapy to Reduce Glycosaminoglycan Synthesis Improves Aspects of Neurological and Skeletal Pathology in MPS I Mice. Diseases 2017, 5, 5. [Google Scholar] [CrossRef]
- Chua, J.S.; Kuberan, B. Synthetic Xylosides: Probing the Glycosaminoglycan Biosynthetic Machinery for Biomedical Applications. Accounts Chem. Res. 2017, 50, 2693–2705. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.; He, Q.Q.; Singh, A.A.; Ferro, V. Mucopolysaccharidosis type II (Hunter syndrome): Clinical and biochemical aspects of the disease and approaches to its diagnosis and treatment. Adv. Carbohydr. Chem. Biochem. 2020, 77, 71–117. [Google Scholar] [CrossRef] [PubMed]
- Guffon, N.; Chowdary, P.; Teles, E.L.; Hughes, D.; Hennermann, J.B.; Huot-Marchand, P.; Faudot-Vernier, E.; Lacombe, O.; Fiquet, A.; Richard, M.; et al. Oral treatment for mucopolysaccharidosis VI: Outcomes of the first phase IIa study with odiparcil. J. Inherit. Metab. Dis. 2022, 45, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Stepien, K.M.; Cufflin, N.; Donald, A.; Jones, S.; Church, H.; Hargreaves, I.P. Secondary Mitochondrial Dysfunction as a Cause of Neurodegenerative Dysfunction in Lysosomal Storage Diseases and an Overview of Potential Therapies. Int. J. Mol. Sci. 2022, 23, 10573. [Google Scholar] [CrossRef]
- Yubero, D.; Montero, R.; O’Callaghan, M.; Pineda, M.; Meavilla, S.; Delgadillo, V.; Sierra, C.; Altimira, L.; Navas, P.; Pope, S.; et al. Coenzyme Q10 and Pyridoxal Phosphate Deficiency Is a Common Feature in Mucopolysaccharidosis Type III. JIMD Rep. 2016, 25, 1–7. [Google Scholar]
- Osellame, L.D.; Rahim, A.A.; Hargreaves, I.P.; Gegg, M.E.; Richard-Londt, A.; Brandner, S.; Waddington, S.N.; Schapira, A.H.; Duchen, M.R. Mitochondria and Quality Control Defects in a Mouse Model of Gaucher Disease—Links to Parkinson’s Disease. Cell Metab. 2013, 17, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Montero, R.; Yubero, D.; Salgado, M.C.; González, M.J.; Campistol, J.; O’Callaghan, M.D.M.; Pineda, M.; Delgadillo, V.; Maynou, J.; Fernandez, G.; et al. Plasma coenzyme Q10 status is impaired in selected genetic conditions. Sci. Rep. 2019, 9, 793. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Li, Y.; Yu, S.; Zhao, Y. SIRT1/PGC-1α Signaling Promotes Mitochondrial Functional Recovery and Reduces Apoptosis after Intracerebral Hemorrhage in Rats. Front. Mol. Neurosci. 2017, 10, 443. [Google Scholar] [CrossRef]
- Orii, K.; Lim, A.; Tomatsu, S.; Stapleton, M.; Suzuki, Y.; Simonaro, C.M.; Schuchman, E.H.; Fukao, T.; Matsumoto, T. Safety Study of Sodium Pentosan Polysulfate for Adult Patients with Mucopolysaccharidosis Type II. Diagnostics 2019, 9, 226. [Google Scholar] [CrossRef]
- Polgreen, L.E.; Kunin-Batson, A.; Rudser, K.; Vehe, R.K.; Utz, J.J.; Whitley, C.B.; Dickson, P. Pilot study of the safety and effect of adalimumab on pain, physical function, and musculoskeletal disease in mucopolysaccharidosis types I and II. Mol. Genet. Metab. Rep. 2017, 10, 75–80. [Google Scholar] [CrossRef]
- Moser, T.; Akgün, K.; Proschmann, U.; Sellner, J.; Ziemssen, T. The role of TH17 cells in multiple sclerosis: Therapeutic implications. Autoimmun. Rev. 2020, 19, 102647. [Google Scholar] [CrossRef] [PubMed]
- Lugt, M.T.V.; Chen, X.; Escolar, M.L.; Carella, B.A.; Barnum, J.L.; Windreich, R.M.; Hill, M.J.; Poe, M.; Marsh, R.A.; Stanczak, H.; et al. Reduced-intensity single-unit unrelated cord blood transplant with optional immune boost for nonmalignant disorders. Blood Adv. 2020, 4, 3041–3052. [Google Scholar] [CrossRef]
- Guo, N.; DeAngelis, V.; Zhu, C.; Schuchman, E.H.; Simonaro, C.M. Pentosan Polysulfate Treatment of Mucopolysaccharidosis Type IIIA Mice. JIMD Rep. 2019, 43, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Simonaro, C.M.; Tomatsu, S.; Sikora, T.; Kubaski, F.; Frohbergh, M.; Guevara, J.M.; Wang, R.Y.; Vera, M.; Kang, J.L.; Smith, L.J.; et al. Pentosan Polysulfate: Oral Versus Subcutaneous Injection in Mucopolysaccharidosis Type I Dogs. PLoS ONE 2016, 11, e0153136. [Google Scholar] [CrossRef] [PubMed]
- Hennermann, J.B.; Gökce, S.; Solyom, A.; Mengel, E.; Schuchman, E.H.; Simonaro, C.M. Treatment with pentosan polysulphate in patients with MPS I: Results from an open label, randomized, monocentric phase II study. J. Inherit. Metab. Dis. 2016, 39, 831–837. [Google Scholar] [CrossRef]
- Sjöström, E.O.; Culot, M.; Leickt, L.; Åstrand, M.; Nordling, E.; Gosselet, F.; Kaiser, C. Transport study of interleukin-1 inhibitors using a human in vitro model of the blood-brain barrier. Brain Behav. Immun.-Health 2021, 16, 100307. [Google Scholar] [CrossRef]
| GAG | Monosaccharides Constituents | Chemical Structure |
|---|---|---|
| Keratan sulfate | Galactose N-acetylglucosamine | 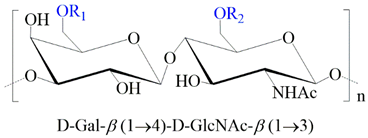 |
| Heparan sulfate | Glucuronic acid N-acetylglucosamine |  |
| Chondroitin sulfate | Glucuronic acid N-acetylgalactosamine | 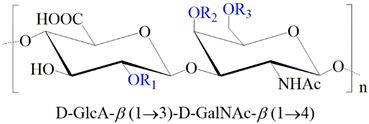 |
| Dermatan sulfate | Iduronic acid N-acetylgalactosamine | 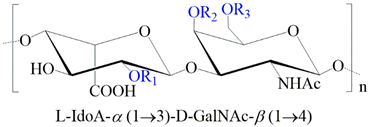 |
| Hyaluronan | Glucuronic acid N-acetylglucosamine | 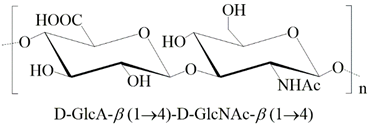 |
| Type (Eponym) | OMIM | Affected Gene | Clinical Characteristics | GAGs |
|---|---|---|---|---|
| MPS I (Hurler) | 607014 (Hurler) | IDUA | Corneal clouding, multiple dysostoses, organomegaly, cardiac disease, CNS impairment | DS and HS |
| 607015 (Scheie) | Corneal clouding, joint stiffness, normal CNS | DS and HS | ||
| 607016 (Hurler-Scheie) | Intermediate phenotype between Hurler and Scheie phenotypes | DS and HS | ||
| MPS II (Hunter) | 309900 | IDS | Multiple dysostoses, organomegaly, CNS impairment | DS and HS |
| MPS IIIA (Sanfilippo A) | 252900 | SGSH | CNS impairment, hyperactivity, mild somatic manifestations | HS |
| MPS IIIB (Sanfilippo B) | 252920 | NAGLU | Like MPS IIIA | HS |
| MPS III C (Sanfilippo C) | 252930 | HGSNAT | Like MPS IIIA | HS |
| MPS IIID (Sanfilippo D) | 252940 | GNS | Like MPS IIIA | HS |
| MPS IVA (Morquio A) | 253000 | GALNS | Short stature, skeletal dysplasia, corneal | KS and CS |
| MPS IVB (Morquio B) | 253010 | GLB1 | Like MPS IVA | KS |
| MPS VI (Maroteaux-Lamy) | 253200 | ARSB | Multiple dysostoses, corneal clouding, cardiac disease | DS |
| MPS VII (Sly) | 253220 | GUSB | Multiple dysostoses, hepatosplenomegaly, mild to severe central nervous system involvement | HS, DS, and CS |
| MPS IX (Natowicz) | 601492 | HYAL1 | Short stature, soft-tissue masses | HA |
| MPS X | 619698 | ARSK | Short stature, coarse facial features, dysostosis multiplex, cardiac and ophthalmological abnormalities. | DS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leal, A.F.; Benincore-Flórez, E.; Rintz, E.; Herreño-Pachón, A.M.; Celik, B.; Ago, Y.; Alméciga-Díaz, C.J.; Tomatsu, S. Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets. Int. J. Mol. Sci. 2023, 24, 477. https://doi.org/10.3390/ijms24010477
Leal AF, Benincore-Flórez E, Rintz E, Herreño-Pachón AM, Celik B, Ago Y, Alméciga-Díaz CJ, Tomatsu S. Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets. International Journal of Molecular Sciences. 2023; 24(1):477. https://doi.org/10.3390/ijms24010477
Chicago/Turabian StyleLeal, Andrés Felipe, Eliana Benincore-Flórez, Estera Rintz, Angélica María Herreño-Pachón, Betul Celik, Yasuhiko Ago, Carlos Javier Alméciga-Díaz, and Shunji Tomatsu. 2023. "Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets" International Journal of Molecular Sciences 24, no. 1: 477. https://doi.org/10.3390/ijms24010477
APA StyleLeal, A. F., Benincore-Flórez, E., Rintz, E., Herreño-Pachón, A. M., Celik, B., Ago, Y., Alméciga-Díaz, C. J., & Tomatsu, S. (2023). Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets. International Journal of Molecular Sciences, 24(1), 477. https://doi.org/10.3390/ijms24010477










