Microbiome and Metabolomics in Liver Cancer: Scientific Technology
Abstract
:1. Introduction
2. Enabling Technologies for Metabolomics Research and Engineering
| Analytical Devices | Scientific Instruments | Key Functions | Metabolic Applications | Ref |
|---|---|---|---|---|
| MS |   | High throughput, high sensitivity/resolution, capable of quickly examining, reduces complexity, improves resolution, specificity, and quantification, allows for isotopic labeling, offers structural information, performed under ambient environmental conditions with the preservation of tissue morphology (DESI-MS). | GC-MS: SCFAs and ketones, carbohydrate metabolites, amino acids. LC-MS: amino acids and their byproducts, bile acids, lipids and fatty acids, sugar metabolites, vitamins, and related compounds. Imaging MS: MALDI/DESI-IMS and nano SIMS. | [31,32,33,34,36,37,38,43,47]. |
| NMR |  | Affords structural evidence, low-sensitivity compared to MS, high-throughput, permits the quantification of isotopic labeling, delivers spatial data (NMR imaging or MRI). | Sugar metabolites, amino acids and amino acid byproducts SCFAs, vitamins, untargeted analysis, and metabolome finger printing. | [22,23,44,45,48] |
| Raman MS |  | 3D info, high-throughput, structural data, non-destructive methods, lower sensitivity versus MS and NMR. | This can be united with fluorescent probes and isotopic labeling for the single-cell-resolved assessment of nutrient assimilation. | [43] |
| UHPLC |  | High-sensitivity detection | Detection and identification of a broad range of metabolites | [49,50] |
| Immunochemistry and enzymatic assays | -- | Low-throughput, high specificity, may provide spatial information (immunohistochemistry or immunofluorescence). | Eicosanoids, uric acid, serotonin, neurotransmitters, lipopolysaccharide, some vitamins, sugar metabolites. | [51,52] |
3. Diagnostics Test of Liver Cancer
- I.
- Physical examination: A general practitioner or gastroenterologist can examine the patient to learn about their health history and identify general risk factors for the development of liver cancer. Examinations include those of the skin, eyes, and areas of the abdomen (signs of jaundice). Additional tests could be necessary to identify the cause of symptoms, depending on the results of the initial physical exam [80,81].
- II.
- III.
- Laparoscopy: For the improved viewing of the liver tissue and adjacent organs, laparoscopic surgeries use a small tube with a camera introduced into the abdomen. Diagnostic laparoscopy is a minimally invasive, low-risk surgical treatment that calls for tiny incisions [84]. An improved understanding of the liver cancer’s current stage, assistance in developing a personalized stem cell treatment strategy, or confirmation of an earlier diagnosis can all be achieved with laparoscopy [85].
- IV.
- Liver biopsy: A surgical procedure called a liver biopsy uses a sample of the patient’s liver tissue to identify the presence of cancer cells [86].
- V.
- VI.
- Genetic screening for cancer: Circulating tumor DNA (ctDNA) analysis is distinct from previously known conventional diagnostic techniques. Cancer biomarker tests such as ctDNA analysis only need small saliva samples or cheek swabs, as opposed to invasive tissue biopsies [89]. Rapid screening is a reliable method of prognostic marker detection. This method can detect potential metastatic disease very early, monitor treatment, and identify genetic and epigenetic changes resulting from primary tumors [90].
4. Microbiome Research and Engineering in HCC Metabolism
5. Microbiome Metabolism for Therapeutic Applications in HCC
6. Liver Transplantation for HCC
7. Systemic Chemotherapy Drugs and Approaches to Improving HCC
8. Conclusions and Challenges for the Future
- Our review indicates a unique liver cancer–metabolomics connection for therapeutic biomarker invention in HCC.
- Liver cancer remains one of the most difficult disease to treat; however, finding the therapeutic biomarker is possible.
- In single-cell studies of liver cancer, the phenomenon of extensive tumor heterogeneity has been noticed, which creates a major barrier for effective cancer interventions.
- Exploiting scientific systems to disrupt these interactions could establish a viable therapeutic strategy for targeting HCC and stopping HCC evolution, thereby improving treatment efficacy.
- We propose that clinical metabolomics may reflect the evolution of therapeutic biomarkers in a successful liver cancer treatment.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2012. CA Cancer J. Clin. 2012, 62, 10–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, E.P.; Ward, E.M.; Siegel, R.; Jemal, A. Cancers with increasing incidence trends in the united states: 1999 through 2008. CA Cancer J. Clin. 2012, 62, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, A.B., 3rd; Abrams, T.A.; Ben-Josef, E.; Bloomston, P.M.; Botha, J.F.; Clary, B.M.; Covey, A.; Curley, S.A.; D’Angelica, M.I.; Davila, R.; et al. Nccn clinical practice guidelines in oncology: Hepatobiliary cancers. J. Natl. Compr. Cancer Netw. JNCCN 2009, 7, 350–391. [Google Scholar] [CrossRef] [PubMed]
- European Association for Study of Liver; European Organisation for Research and Treatment of Cancer. Easl-eortc clinical practice guidelines: Management of hepatocellular carcinoma. Eur. J. Cancer 2012, 48, 599–641. [Google Scholar] [CrossRef] [PubMed]
- MacFie, J.; O’Boyle, C.; Mitchell, C.J.; Buckley, P.M.; Johnstone, D.; Sudworth, P. Gut origin of sepsis: A prospective study investigating associations between bacterial translocation, gastric microflora, and septic morbidity. Gut 1999, 45, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Sano, K.; Ichikawa, T.; Motosugi, U.; Sou, H.; Muhi, A.M.; Matsuda, M.; Nakano, M.; Sakamoto, M.; Nakazawa, T.; Asakawa, M.; et al. Imaging study of early hepatocellular carcinoma: Usefulness of gadoxetic acid-enhanced mr imaging. Radiology 2011, 261, 834–844. [Google Scholar] [CrossRef]
- Kudo, M. Early hepatocellular carcinoma: Definition and diagnosis. Liver Cancer 2013, 2, 69–72. [Google Scholar] [CrossRef]
- Raja, G.; Jung, Y.; Jung, S.H.; Kim, T.-J. 1h-nmr-based metabolomics for cancer targeting and metabolic engineering—A review. Process Biochem. 2020, 99, 112–122. [Google Scholar] [CrossRef]
- Dumas, M.E.; Kinross, J.; Nicholson, J.K. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 2014, 146, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Raja, G.; Gupta, H.; Gebru, Y.A.; Youn, G.S.; Choi, Y.R.; Kim, H.S.; Yoon, S.J.; Kim, D.J.; Kim, T.-J.; Suk, K.T. Recent advances of microbiome-associated metabolomics profiling in liver disease: Principles, mechanisms, and applications. Int. J. Mol. Sci. 2021, 22, 1160. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, L.; Chen, T.; Zhou, K.; Zhang, Z.; Li, J.; Sun, B.; Guo, Y.; Wang, X.; Wang, Y.; et al. A metabolite array technology for precision medicine. Anal. Chem. 2021, 93, 5709–5717. [Google Scholar] [CrossRef]
- Belhaj, M.R.; Lawler, N.G.; Hoffman, N.J. Metabolomics and lipidomics: Expanding the molecular landscape of exercise biology. Metabolites 2021, 11, 151. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for biomarker discovery: Moving to the clinic. BioMed Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef] [Green Version]
- Raja, G.; Cao, S.; Kim, D.-H.; Kim, T.-J. Mechanoregulation of titanium dioxide nanoparticles in cancer therapy. Mater. Sci. Eng. C 2020, 107, 110303. [Google Scholar] [CrossRef]
- Blow, N. Metabolomics: Biochemistry’s new look. Nature 2008, 455, 697–700. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef]
- Raja, G.; Selvaraj, V.; Suk, M.; Suk, K.T.; Kim, T.-J. Metabolic phenotyping analysis of graphene oxide nanosheets exposures in breast cancer cells: Metabolomics profiling techniques. Process Biochem. 2021, 104, 39–45. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30–32. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.; Yde, C.C.; Schmedes, M.S.; Jensen, H.M.; Meier, S.; Bertram, H.C. Strategy for nuclear-magnetic-resonance-based metabolomics of human feces. Anal. Chem. 2015, 87, 5930–5937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, H.A.; Bomhof, M.R.; Vogel, H.J.; Reimer, R.A. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Sci. Rep. 2016, 6, 20683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesan, R.; Vasantha-Srinivasan, P.; Sadhasivam, D.R.; Subramanian, R.; Vimalraj, S.; Suk, K.T. Carbon nanotubes induce metabolomic profile disturbances in zebrafish: Nmr-based metabolomics platform. Front. Mol. Biosci. 2021, 8, 688827. [Google Scholar] [CrossRef] [PubMed]
- Angamuthu, S.; Ramaswamy, C.R.; Thangaswamy, S.; Sadhasivam, D.R.; Nallaswamy, V.D.; Subramanian, R.; Ganesan, R.; Raju, A. Metabolic annotation, interactions and characterization of natural products of mango (Mangifera indica L.): 1h nmr based chemical metabolomics profiling. Process Biochem. 2021, 108, 18–25. [Google Scholar] [CrossRef]
- Raja, G.; Jang, Y.K.; Suh, J.S.; Prabhakaran, V.S.; Kim, T.J. Advanced understanding of genetic risk and metabolite signatures in construction workers via cytogenetics and metabolomics analysis. Process Biochem. 2019, 86, 117–126. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Cavill, R.; Keun, H.C.; Holmes, E.; Lindon, J.C.; Nicholson, J.K.; Ebbels, T.M. Genetic algorithms for simultaneous variable and sample selection in metabonomics. Bioinformatics 2009, 25, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Blanksby, S.J.; Mitchell, T.W. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010, 3, 433–465. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Thompson, C.B. Cellular metabolism and disease: What do metabolic outliers teach us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021, 19, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, C.; Kelly, D.; Cantillon, J.; Cauchi, M.; Yon, M.A.; Bentley, L.; Cox, R.D.; Turner, C. Monitoring type 2 diabetes from volatile faecal metabolome in cushing’s syndrome and single afmid mouse models via a longitudinal study. Sci. Rep. 2019, 9, 18779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.I.; Weir, W.H.; Crowley, J.R.; Hink, T.; Reske, K.A.; Kwon, J.H.; Burnham, C.D.; Dubberke, E.R.; Mucha, P.J.; Henderson, J.P. Metabolomic networks connect host-microbiome processes to human clostridioides difficile infections. J. Clin. Investig. 2019, 129, 3792–3806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-S.; Li, J.; Krautkramer, K.A.; Badri, M.; Battaglia, T.; Borbet, T.C.; Koh, H.; Ng, S.; Sibley, R.A.; Li, Y.; et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. eLife 2018, 7, e37816. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [Green Version]
- Fujisaka, S.; Avila-Pacheco, J.; Soto, M.; Kostic, A.; Dreyfuss, J.M.; Pan, H.; Ussar, S.; Altindis, E.; Li, N.; Bry, L.; et al. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep. 2018, 22, 3072–3086. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via nkt cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, B.K.; Jang, Y.; Lee, E.M.; Jung, D.W.; Moon, J.H.; Lee, H.J.; Lee, D.Y. A systematic approach to metabolic characterization of thyroid-disrupting chemicals and their in vitro biotransformants based on prediction-assisted metabolomic analysis. J. Chromatogr. A 2021, 1649, 462222. [Google Scholar] [CrossRef]
- Raja, G.; Jang, Y.-K.; Suh, J.-S.; Kim, H.-S.; Ahn, S.H.; Kim, T.-J. Microcellular environmental regulation of silver nanoparticles in cancer therapy: A critical review. Cancers 2020, 12, 664. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.A.; Gika, H.G.; Want, E.J.; Wilson, I.D. Liquid chromatography-mass spectrometry based global metabolite profiling: A review. Anal. Chim. Acta 2012, 711, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Loy, A. Stable-isotope probing of human and animal microbiome function. Trends Microbiol. 2018, 26, 999–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röth, D.; Chiang, A.J.; Hu, W.; Gugiu, G.B.; Morra, C.N.; Versalovic, J.; Kalkum, M. Two-carbon folate cycle of commensal lactobacillus reuteri 6475 gives rise to immunomodulatory ethionine, a source for histone ethylation. FASEB J. 2019, 33, 3536–3548. [Google Scholar] [CrossRef]
- Bui, T.P.N.; Ritari, J.; Boeren, S.; de Waard, P.; Plugge, C.M.; de Vos, W.M. Production of butyrate from lysine and the amadori product fructoselysine by a human gut commensal. Nat. Commun. 2015, 6, 10062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagana Gowda, G.A.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Rath, C.M.; Alexandrov, T.; Higginbottom, S.K.; Song, J.; Milla, M.E.; Fischbach, M.A.; Sonnenburg, J.L.; Dorrestein, P.C. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal. Chem. 2012, 84, 9259–9267. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Ma, C.; Liu, C.; Wang, Z.; Yang, J.; Liu, X.; Shen, Z.; Wu, R. Nmr-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget 2016, 7, 29454–29464. [Google Scholar] [CrossRef]
- de Souza, L.P.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef]
- Reher, R.; Aron, A.T.; Fajtová, P.; Stincone, P.; Wagner, B.; Pérez-Lorente, A.I.; Liu, C.; Shalom, I.Y.B.; Bittremieux, W.; Wang, M.; et al. Native metabolomics identifies the rivulariapeptolide family of protease inhibitors. Nat. Commun. 2022, 13, 4619. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, G.; Kim, S.; Yoon, D.; Yoon, C.; Kim, S. H-1 nmr based metabolomics studies of the toxicity of titanium dioxide nanoparticles in zebrafish (danio rerio). Bull. Korean Chem. Soc. 2018, 39, 33–39. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. Metaboanalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. Metaboanalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Eilers, P. Chemometrics. Data analysis for the laboratory and chemical plant. J. Chemom. 2003, 17, 360–361. [Google Scholar] [CrossRef]
- Tistaert, C.; Thierry, L.; Szandrach, A.; Dejaegher, B.; Fan, G.; Frédérich, M.; Vander Heyden, Y. Quality control of citri reticulatae pericarpium: Exploratory analysis and discrimination. Anal. Chim. Acta 2011, 705, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Raja, G.; Kim, S.; Yoon, D.; Yoon, C.; Kim, S. 1h-nmr-based metabolomics studies of the toxicity of mesoporous carbon nanoparticles in zebrafish (danio rerio). Bull. Korean Chem. Soc. 2017, 38, 271–277. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted profiling: Quantitative analysis of 1h nmr metabolomics data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]
- Chang, D.; Banack, C.D.; Shah, S.L. Robust baseline correction algorithm for signal dense nmr spectra. J. Magn. Reson. 2007, 187, 288–292. [Google Scholar] [CrossRef]
- Cottret, L.; Wildridge, D.; Vinson, F.; Barrett, M.P.; Charles, H.; Sagot, M.F.; Jourdan, F. Metexplore: A web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 2010, 38, W132–W137. [Google Scholar] [CrossRef] [PubMed]
- Cottret, L.; Frainay, C.; Chazalviel, M.; Cabanettes, F.; Gloaguen, Y.; Camenen, E.; Merlet, B.; Heux, S.; Portais, J.C.; Poupin, N.; et al. Metexplore: Collaborative edition and exploration of metabolic networks. Nucleic Acids Res. 2018, 46, W495–W502. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. Hmdb: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. Hmdb 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2017, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Yogarajalakshmi, P.; Poonguzhali, T.V.; Ganesan, R.; Karthi, S.; Senthil-Nathan, S.; Krutmuang, P.; Radhakrishnan, N.; Mohammad, F.; Kim, T.-J.; Vasantha-Srinivasan, P. Toxicological screening of marine red algae champia parvula (c. Agardh) against the dengue mosquito vector aedes aegypti (linn.) and its non-toxicity against three beneficial aquatic predators. Aquat. Toxicol. 2020, 222, 105474. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. Kegg: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Bohler, A.; Wu, G.; Kutmon, M.; Pradhana, L.A.; Coort, S.L.; Hanspers, K.; Haw, R.; Pico, A.R.; Evelo, C.T. Reactome from a wikipathways perspective. PLoS Comput. Biol. 2016, 12, e1004941. [Google Scholar] [CrossRef]
- Fabregat, A.; Korninger, F.; Viteri, G.; Sidiropoulos, K.; Marin-Garcia, P.; Ping, P.; Wu, G.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Reactome graph database: Efficient access to complex pathway data. PLoS Comput. Biol. 2018, 14, e1005968. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The metacyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [Green Version]
- Noronha, A.; Danielsdottir, A.D.; Gawron, P.; Johannsson, F.; Jonsdottir, S.; Jarlsson, S.; Gunnarsson, J.P.; Brynjolfsson, S.; Schneider, R.; Thiele, I.; et al. Reconmap: An interactive visualization of human metabolism. Bioinformatics 2017, 33, 605–607. [Google Scholar] [CrossRef]
- Noronha, A.; Modamio, J.; Jarosz, Y.; Guerard, E.; Sompairac, N.; Preciat, G.; Danielsdottir, A.D.; Krecke, M.; Merten, D.; Haraldsdottir, H.S.; et al. The virtual metabolic human database: Integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019, 47, D614–D624. [Google Scholar] [CrossRef] [PubMed]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. Wikipathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef] [PubMed]
- Wanichthanarak, K.; Fan, S.; Grapov, D.; Barupal, D.K.; Fiehn, O. Metabox: A toolbox for metabolomic data analysis, interpretation and integrative exploration. PLoS ONE 2017, 12, e0171046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barupal, D.K.; Fiehn, O. Chemical similarity enrichment analysis (chemrich) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Li, C.; Marcu, A.; Badran, H.; Pon, A.; Budinski, Z.; Patron, J.; Lipton, D.; Cao, X.; Oler, E.; et al. Pathbank: A comprehensive pathway database for model organisms. Nucleic Acids Res. 2020, 48, D470–D478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Xia, J. Omicsnet: A web-based tool for creation and visual analysis of biological networks in 3d space. Nucleic Acids Res. 2018, 46, W514–W522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchweitz, L.F.; Yurkovich, J.T.; Blessing, C.; Kohler, V.; Schwarzkopf, F.; King, Z.A.; Yang, L.; Johannsson, F.; Sigurjonsson, O.E.; Rolfsson, O.; et al. Visualizing metabolic network dynamics through time-series metabolomic data. BMC Bioinform. 2020, 21, 130. [Google Scholar] [CrossRef] [Green Version]
- Nagele, T.; Furtauer, L.; Nagler, M.; Weiszmann, J.; Weckwerth, W. A strategy for functional interpretation of metabolomic time series data in context of metabolic network information. Front. Mol. Biosci. 2016, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Sakaue, M.; Sugimura, K.; Masuzawa, T.; Takeno, A.; Katsuyama, S.; Shinnke, G.; Ikeshima, R.; Kawai, K.; Hiraki, M.; Katsura, Y.; et al. Long-term survival of her2 positive gastric cancer patient with multiple liver metastases who obtained pathological complete response after systemic chemotherapy: A case report. Int. J. Surg. Case Rep. 2022, 94, 107097. [Google Scholar] [CrossRef]
- Xu, M.; Xie, L.-T.; Xiao, Y.-Y.; Liang, P.; Zhao, Q.-Y.; Wang, Z.-M.; Chai, W.-L.; Wei, Y.-T.; Xu, L.-F.; Hu, X.-K.; et al. Chinese clinical practice guidelines for ultrasound-guided irreversible electroporation of liver cancer (version 2022). Hepatobiliary Pancreat. Dis. Int. 2022, 21, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Şahin, E.; Elboğa, U.; Çelen, Y.Z.; Sever, Ö.N.; Çayırlı, Y.B.; Çimen, U. Comparison of 68ga-dota-fapi and 18fdg pet/ct imaging modalities in the detection of liver metastases in patients with gastrointestinal system cancer. Eur. J. Radiol. 2021, 142, 109867. [Google Scholar] [CrossRef] [PubMed]
- Bekki, Y.; Mahamid, A.; Lewis, S.; Ward, S.C.; Simpson, W.; Argiriadi, P.; Kamath, A.; Facciuto, L.; Patel, R.S.; Kim, E.; et al. Radiological and pathological assessment with eob-mri after y90 radiation lobectomy prior to liver resection for hepatocellular carcinoma. HPB 2022, 24, 2185–2192. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzani, C.; Kim, H.J.; Park, E.J.; Turri, G.; Zagolin, G.; Foppa, C.; Baik, S.H.; Spolverato, G.; Spinelli, A.; Choi, G.S. Does laparoscopy increase the risk of peritoneal recurrence after resection for pt4 colon cancer? Results of a propensity score-matched analysis from an international cohort. Eur. J. Surg. Oncol. 2022, 48, 1823–1830. [Google Scholar] [CrossRef]
- Borgstein, A.B.J.; Keywani, K.; Eshuis, W.J.; van Berge Henegouwen, M.I.; Gisbertz, S.S. Staging laparoscopy in patients with advanced gastric cancer: A single center cohort study. Eur. J. Surg. Oncol. 2022, 48, 362–369. [Google Scholar] [CrossRef]
- Hagström, H.; Thiele, M.; Sharma, R.; Simon, T.G.; Roelstraete, B.; Söderling, J.; Ludvigsson, J.F. Risk of cancer in biopsy-proven alcohol-related liver disease: A population-based cohort study of 3410 persons. Clin. Gastroenterol. Hepatol. 2022, 20, 918–929.e8. [Google Scholar] [CrossRef]
- Listopad, S.; Magnan, C.; Asghar, A.; Stolz, A.; Tayek, J.A.; Liu, Z.-X.; Morgan, T.R.; Norden-Krichmar, T.M. Differentiating between liver diseases by applying multiclass machine learning approaches to transcriptomics of liver tissue or blood-based samples. JHEP Rep. 2022, 4, 100560. [Google Scholar] [CrossRef]
- Fujiwara, N.; Kobayashi, M.; Fobar, A.J.; Hoshida, A.; Marquez, C.A.; Koneru, B.; Panda, G.; Taguri, M.; Qian, T.; Raman, I.; et al. A blood-based prognostic liver secretome signature and long-term hepatocellular carcinoma risk in advanced liver fibrosis. Med 2021, 2, 836–850.e10. [Google Scholar] [CrossRef]
- Moy, R.H.; Nguyen, A.; Loo, J.M.; Yamaguchi, N.; Kajba, C.M.; Santhanam, B.; Ostendorf, B.N.; Wu, Y.G.; Tavazoie, S.; Tavazoie, S.F. Functional genetic screen identifies itpr3/calcium/relb axis as a driver of colorectal cancer metastatic liver colonization. Dev. Cell 2022, 57, 1146–1159.e7. [Google Scholar] [CrossRef]
- Calderwood, A.H.; Sawhney, M.S.; Thosani, N.C.; Rebbeck, T.R.; Wani, S.; Canto, M.I.; Fishman, D.S.; Golan, T.; Hidalgo, M.; Kwon, R.S.; et al. American society for gastrointestinal endoscopy guideline on screening for pancreatic cancer in individuals with genetic susceptibility: Methodology and review of evidence. Gastrointest. Endosc. 2022, 95, 827–854.e8. [Google Scholar] [CrossRef]
- McCarville, J.L.; Chen, G.Y.; Cuevas, V.D.; Troha, K.; Ayres, J.S. Microbiota metabolites in health and disease. Annu. Rev. Immunol. 2020, 38, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Beyoğlu, D.; Idle, J.R. Metabolomic insights into the mode of action of natural products in the treatment of liver disease. Biochem. Pharmacol. 2020, 180, 114171. [Google Scholar] [CrossRef]
- Beyoğlu, D.; Idle, J.R. Metabolomic and lipidomic biomarkers for premalignant liver disease diagnosis and therapy. Metabolites 2020, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Beyoğlu, D.; Idle, J.R. Metabolic rewiring and the characterization of oncometabolites. Cancers 2021, 13, 2900. [Google Scholar] [CrossRef] [PubMed]
- Beyoğlu, D.; Idle, J.R. The glycine deportation system and its pharmacological consequences. Pharmacol. Ther. 2012, 135, 151–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyoğlu, D.; Smith, R.L.; Idle, J.R. Dog bites man or man bites dog? The enigma of the amino acid conjugations. Biochem. Pharmacol. 2012, 83, 1331–1339. [Google Scholar] [CrossRef] [Green Version]
- Adamson, R.H.; Bridges, J.W.; Evans, M.E.; Williams, R.T. Species differences in the aromatization of quinic acid in vivo and the role of gut bacteria. Biochem. J. 1970, 116, 437–443. [Google Scholar] [CrossRef] [Green Version]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- James, M.O.; Smith, R.L.; Williams, R.T.; Reidenberg, M. The conjugation of phenylacetic acid in man, sub-human primates and some non-primate species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1972, 182, 25–35. [Google Scholar]
- Mosele, J.I.; Macià, A.; Motilva, M.J. Metabolic and microbial modulation of the large intestine ecosystem by non-absorbed diet phenolic compounds: A review. Molecules 2015, 20, 17429–17468. [Google Scholar] [CrossRef] [Green Version]
- Liebich, H.M.; Först, C. Basic profiles of organic acids in urine. J. Chromatogr. 1990, 525, 1–14. [Google Scholar] [CrossRef] [PubMed]
- McNeil, N.I. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 1984, 39, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar]
- Ganesan, R.; Suk, K.T. Therapeutic potential of human microbiome-based short-chain fatty acids and bile acids in liver disease. Livers 2022, 2, 139–145. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.-Y.; Choo, S.-P.; Trojan, J.; Welling, T.H.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (checkmate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Zhu, A.X.; Finn, R.S.; Edeline, J.; Cattan, S.; Ogasawara, S.; Palmer, D.; Verslype, C.; Zagonel, V.; Fartoux, L.; Vogel, A.; et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (keynote-224): A non-randomised, open-label phase 2 trial. Lancet Oncol. 2018, 19, 940–952. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Kambayashi, Y.; Fujimura, T.; Hidaka, T.; Aiba, S. Biomarkers for predicting efficacies of anti-pd1 antibodies. Front. Med. 2019, 6, 174. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.X.; Yan, H.X.; Liu, Q.; Yang, W.; Wu, H.P.; Dong, W.; Tang, L.; Lin, Y.; He, Y.Q.; Zou, S.S.; et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 2010, 52, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and tlr4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, X.; Liu, P.; Wei, R.; Chen, W.; Rajani, C.; Hernandez, B.Y.; Alegado, R.; Dong, B.; Li, D.; et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016, 7, 19355–19366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut microbiota promotes obesity-associated liver cancer through pge(2)-mediated suppression of antitumor immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.L.; Yu, L.X.; Yang, W.; Tang, L.; Lin, Y.; Wu, H.; Zhai, B.; Tan, Y.X.; Shan, L.; Liu, Q.; et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J. Hepatol. 2012, 57, 803–812. [Google Scholar] [CrossRef]
- Grąt, M.; Wronka, K.M.; Krasnodębski, M.; Masior, Ł.; Lewandowski, Z.; Kosińska, I.; Grąt, K.; Stypułkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Ni, J.; Huang, R.; Zhou, H.; Xu, X.; Li, Y.; Cao, P.; Zhong, K.; Ge, M.; Chen, X.; Hou, B.; et al. Analysis of the relationship between the degree of dysbiosis in gut microbiota and prognosis at different stages of primary hepatocellular carcinoma. Front. Microbiol. 2019, 10, 1458. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef] [Green Version]
- Behary, J.; Amorim, N.; Jiang, X.-T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nature Commun. 2021, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Ferrarini, A.; Di Poto, C.; He, S.; Tu, C.; Varghese, R.S.; Kara Balla, A.; Jayatilake, M.; Li, Z.; Ghaffari, K.; Fan, Z.; et al. Metabolomic Analysis of Liver Tissues for Characterization of Hepatocellular Carcinoma. J. Proteome Res. 2019, 18, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.G.; Dewhirst, F.E.; Tully, J.G.; Paster, B.J.; Yan, L.; Taylor, N.S.; Collins, M.J., Jr.; Gorelick, P.L.; Ward, J.M. Helicobacter hepaticus sp. Nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 1994, 32, 1238–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.M.; Anver, M.R.; Haines, D.C.; Benveniste, R.E. Chronic active hepatitis in mice caused by helicobacter hepaticus. Am. J. Pathol. 1994, 145, 959–968. [Google Scholar] [PubMed]
- Ward, J.M.; Fox, J.G.; Anver, M.R.; Haines, D.C.; George, C.V.; Collins, M.J., Jr.; Gorelick, P.L.; Nagashima, K.; Gonda, M.A.; Gilden, R.V.; et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel helicobacter species. J. Natl. Cancer Inst. 1994, 86, 1222–1227. [Google Scholar] [CrossRef]
- Fox, J.G.; Feng, Y.; Theve, E.J.; Raczynski, A.R.; Fiala, J.L.; Doernte, A.L.; Williams, M.; McFaline, J.L.; Essigmann, J.M.; Schauer, D.B.; et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 2010, 59, 88–97. [Google Scholar] [CrossRef]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996, 334, 693–699. [Google Scholar] [CrossRef]
- Yao, F.Y.; Ferrell, L.; Bass, N.M.; Bacchetti, P.; Ascher, N.L.; Roberts, J.P. Liver transplantation for hepatocellular carcinoma: Comparison of the proposed ucsf criteria with the milan criteria and the pittsburgh modified tnm criteria. Liver Transplant. 2002, 8, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Duffy, J.P.; Vardanian, A.; Benjamin, E.; Watson, M.; Farmer, D.G.; Ghobrial, R.M.; Lipshutz, G.; Yersiz, H.; Lu, D.S.; Lassman, C.; et al. Liver transplantation criteria for hepatocellular carcinoma should be expanded: A 22-year experience with 467 patients at ucla. Ann. Surg. 2007, 246, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Hanje, A.J.; Yao, F.Y. Current approach to down-staging of hepatocellular carcinoma prior to liver transplantation. Curr. Opin. Organ Transplant. 2008, 13, 234–240. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Schwartz, L.; Ricci, S.; Amadori, D.; Santoro, A.; Figer, A.; De Greve, J.; Douillard, J.Y.; Lathia, C.; Schwartz, B.; et al. Phase ii study of sorafenib in patients with advanced hepatocellular carcinoma. J. Clin. Oncol. 2006, 24, 4293–4300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.L.; Kang, Y.K.; Chen, Z.; Tsao, C.J.; Qin, S.; Kim, J.S.; Luo, R.; Feng, J.; Ye, S.; Yang, T.S.; et al. Efficacy and safety of sorafenib in patients in the asia-pacific region with advanced hepatocellular carcinoma: A phase iii randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009, 10, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Reyes, D.K.; Cosgrove, D.; Kamel, I.R.; Bhagat, N.; Geschwind, J.F. Phase ii trial of sorafenib combined with concurrent transarterial chemoembolization with drug-eluting beads for hepatocellular carcinoma. J. Clin. Oncol. 2011, 29, 3960–3967. [Google Scholar] [CrossRef]
- Kudo, M.; Imanaka, K.; Chida, N.; Nakachi, K.; Tak, W.Y.; Takayama, T.; Yoon, J.H.; Hori, T.; Kumada, H.; Hayashi, N.; et al. Phase iii study of sorafenib after transarterial chemoembolisation in japanese and korean patients with unresectable hepatocellular carcinoma. Eur. J. Cancer 2011, 47, 2117–2127. [Google Scholar] [CrossRef]

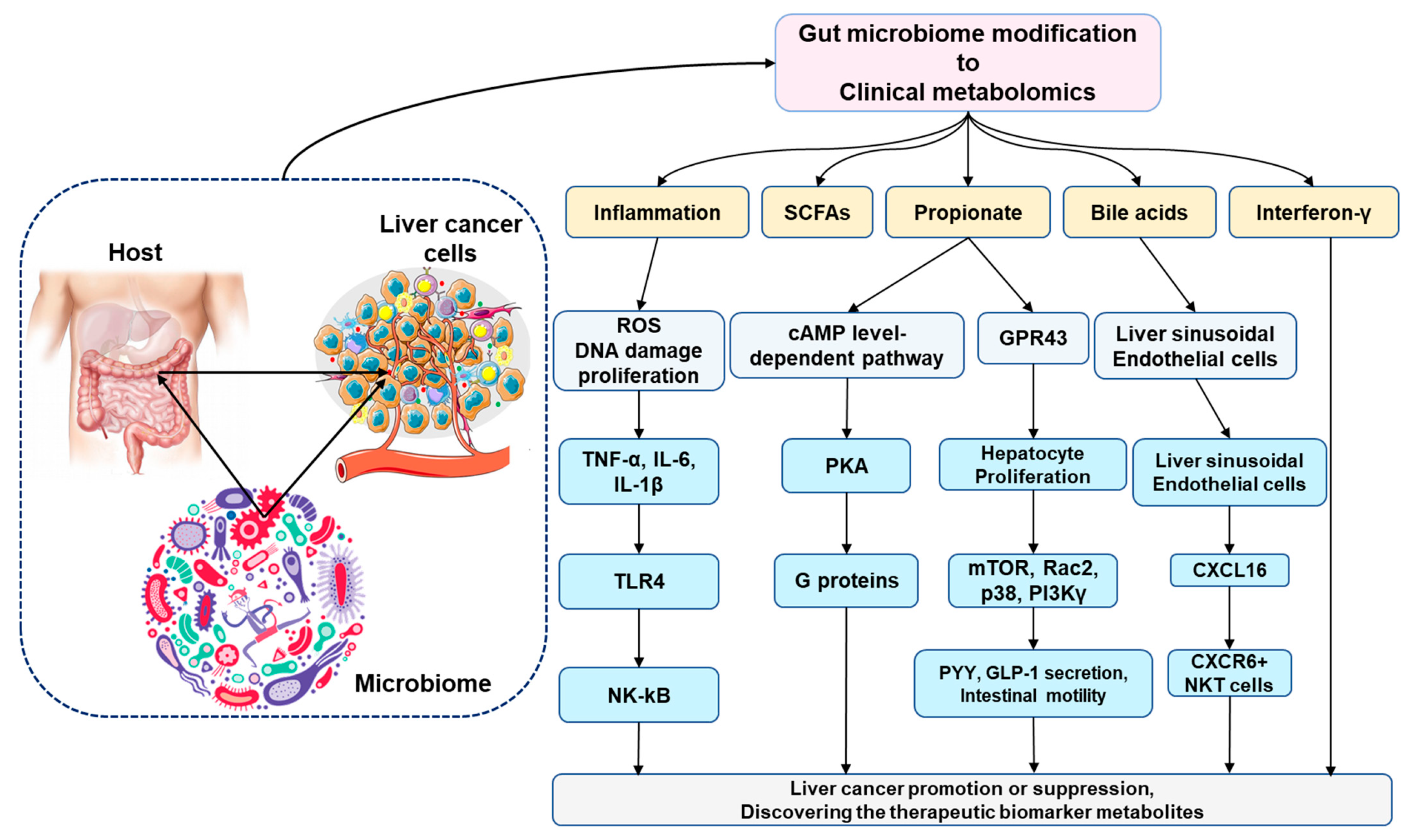
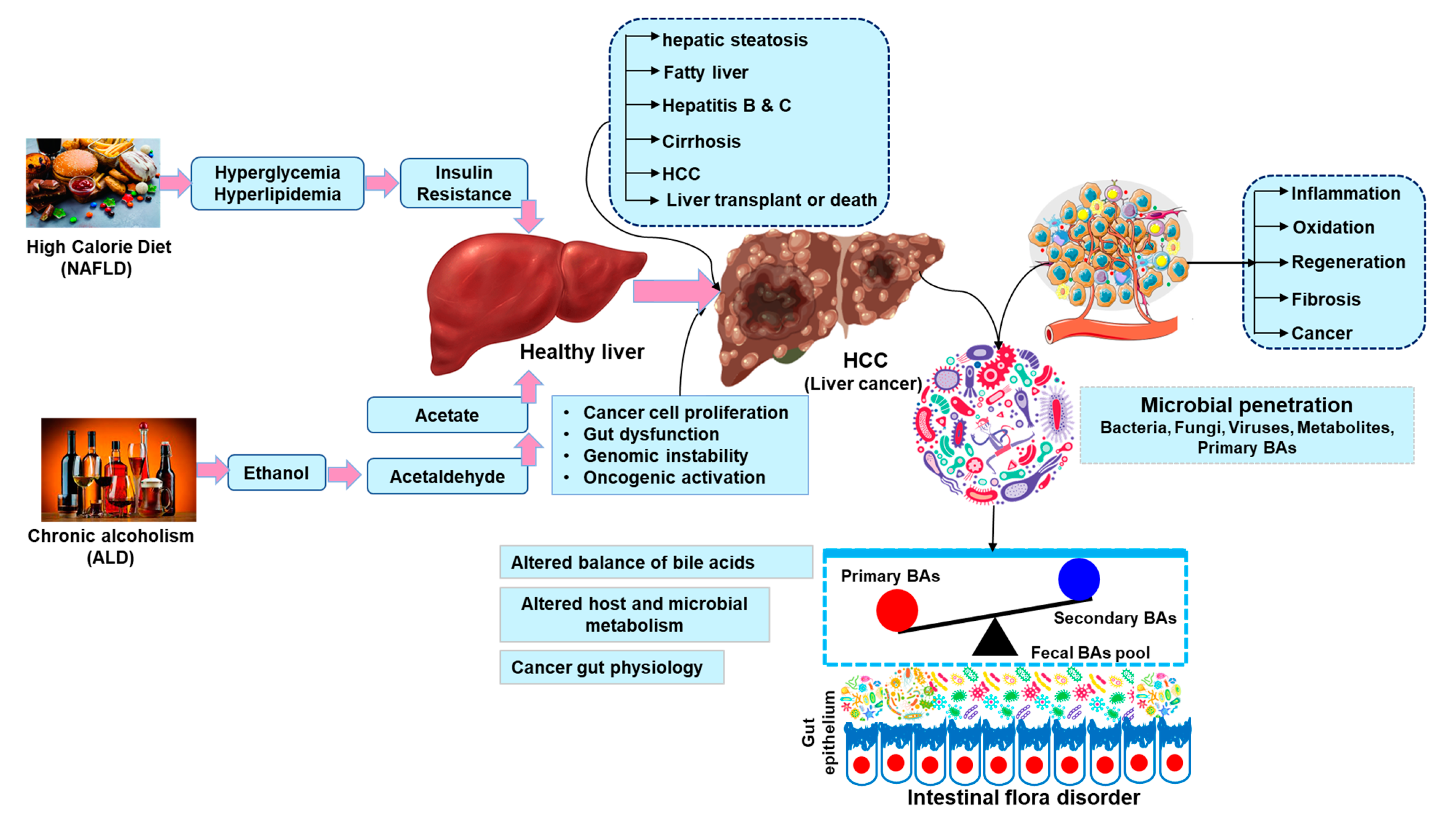
| Platforms | Invention | Ref |
|---|---|---|
| MetaboAnalyst | Web-based analytical pipeline tool, all-in-one metabolomics profiling, data collection, pathway enrichments, data analysis. | [24,54,55] |
| SIMCA-P+ | Pattern recognition of PCA, PLS-DA, OPLS-DA, S-plot, and loading plot, multivariate tool, data mining, interactive graphics. | [11,13,56,57,58] |
| Chenomx Inc., | Correction of the spectral data, metabolite profiling, and quantification. | [58,59,60] |
| MetExplore | Picturing of biological reaction systems and paths, simplifying the analysis of omics data in the biochemical background, and pathways improvement. | [61,62] |
| HMDB | Data bank of NMR, LC-MS, and GC-MS packs, metabolites information, structures, and biological properties. | [63,64,65] |
| KEGG | Databank of genes and genomes; KEGG ortholog for genes and proteins. | [66] |
| Reactome | Information base of biomolecular paths: free/open-source data, curated, and peer-reviewed. | [67,68] |
| Cyc databases | Largest curated collection of metabolic pathways. A wide range of model organisms’ data. | [69] |
| Virtual Metabolic Human | 255 diseases, microbial genes, and human and gut microbiome metabolism database. | [70,71] |
| WikiPathways | Browsable, editable database curated by the research community. | [72] |
| Metabox | Toolbox for integrating proteomics and transcriptomics data for metabolomics data processing and interpretation. | [73] |
| Metscape | Cytoscape plugin, metabolomics correlation networks and KEGG-based metabolic networks integrating gene expression and metabolomics. | [74] |
| ChemRICH | Alternative to biochemical pathway mapping for metabolomic datasets. Not based on biochemistry directly but on structural similarity. The enrichment test is based on the Kolmogorov−Smirnov test (not the hypergeometric test or Fisher’s exact test). | [75] |
| PathBank | Comprehensive, user-friendly resource for metabolic pathways in 10 different model organisms. | [76] |
| OmicsNet | Multi-omics data integration, biological networks (genes, proteins, microRNAs, transcription factors, metabolites). | [77] |
| GEM-Vis | The use of metabolic network maps to visualize time-course metabolomic data. | [78] |
| FEMTO | Combining metabolomic time-series analysis with network data. | [79] |
| Models | Disease | Implicated Microbiota | Ref |
|---|---|---|---|
| Mice | DEN-induced HCC | Changing gut microbiome | [111] |
| DEN-CCL4-induced HCC | Changing gut microbiome | [112] | |
| STZ-HFD-induced NASH-HCC | Atopobium spp.
↑, Bacteroides spp.
↑, Bacteroides vulgatus↑, B. acidifaciens↑, B. uniformis↑, Clostridium cocleatum↑, C. xylanolyticum↑, Desulfovibrio spp. ↑ | [113] | |
| HFHC-induced NAFLD-HCC | Mucispirillum↑, Desulfovibrio↑, Anaerotruncus↑, Desulfovibrionaceae↑, Bifidobacterium↓, Bacteroides↓ | [114] | |
| DMBA-HFD-induced HCC | Changing gut microbiome | [115] | |
| MYC transgenic spontaneous HCC | Gram-positive bacteria ↑, Bacteria mediating primary-to-secondary bile acid conversion ↑, Clostridium scindens ↑ | [38] | |
| DMBA- or DMBA-HFD-induced HCC | Gram-positive bacteria | [116] | |
| Rat | DEN-induced HCC | Lactobacillus species↓, Escherichia coli↑, Atopobium cluster↑, Atopobium↑, Collinsella↑, Coriobacterium↑, Eggerthella↑, Enterococcus species↓, Bifidobacterium species↓, | [117] |
| Human | HCC | Escherichia coli↑ | [118] |
| HCC | Cetobacterium↓, Proteobacteria↑, Desulfococcus↑, Enterobacter↑, Prevotella↑, Veillonella↑, | [119] | |
| HCC | Bifidobacterium↓, Bacteroides↑, Akkermansia↓, | ||
| HCC | Neisseria↑, Enterobacteriaceae↑, Veillonella↑, Limnobacter↑, Enterococcus↓, Phyllobacterium↓, Clostridium↓, Ruminococcus↓, Coprococcus↓ | [120] | |
| HCC | Gut microbial α-diversity↓, Proteobacteria↑, Enterobacteriaceae↑, Bacteroides xylanisolvens↑, B. caecimuris↑, Ruminococcus gnavus↑, Clostridium bolteae↑, Veillonella parvula↑, Oscillospiraceae↓, Erysipelotrichaceae↓ | [121] | |
| HCC | Klebsiella↑, Haemophilus↑, Alistipes↓, Phascolarctobacterium↓, Ruminococcus↓ | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesan, R.; Yoon, S.J.; Suk, K.T. Microbiome and Metabolomics in Liver Cancer: Scientific Technology. Int. J. Mol. Sci. 2023, 24, 537. https://doi.org/10.3390/ijms24010537
Ganesan R, Yoon SJ, Suk KT. Microbiome and Metabolomics in Liver Cancer: Scientific Technology. International Journal of Molecular Sciences. 2023; 24(1):537. https://doi.org/10.3390/ijms24010537
Chicago/Turabian StyleGanesan, Raja, Sang Jun Yoon, and Ki Tae Suk. 2023. "Microbiome and Metabolomics in Liver Cancer: Scientific Technology" International Journal of Molecular Sciences 24, no. 1: 537. https://doi.org/10.3390/ijms24010537
APA StyleGanesan, R., Yoon, S. J., & Suk, K. T. (2023). Microbiome and Metabolomics in Liver Cancer: Scientific Technology. International Journal of Molecular Sciences, 24(1), 537. https://doi.org/10.3390/ijms24010537






