Relationships between Diabetes and the Intestinal Microbial Population
Abstract
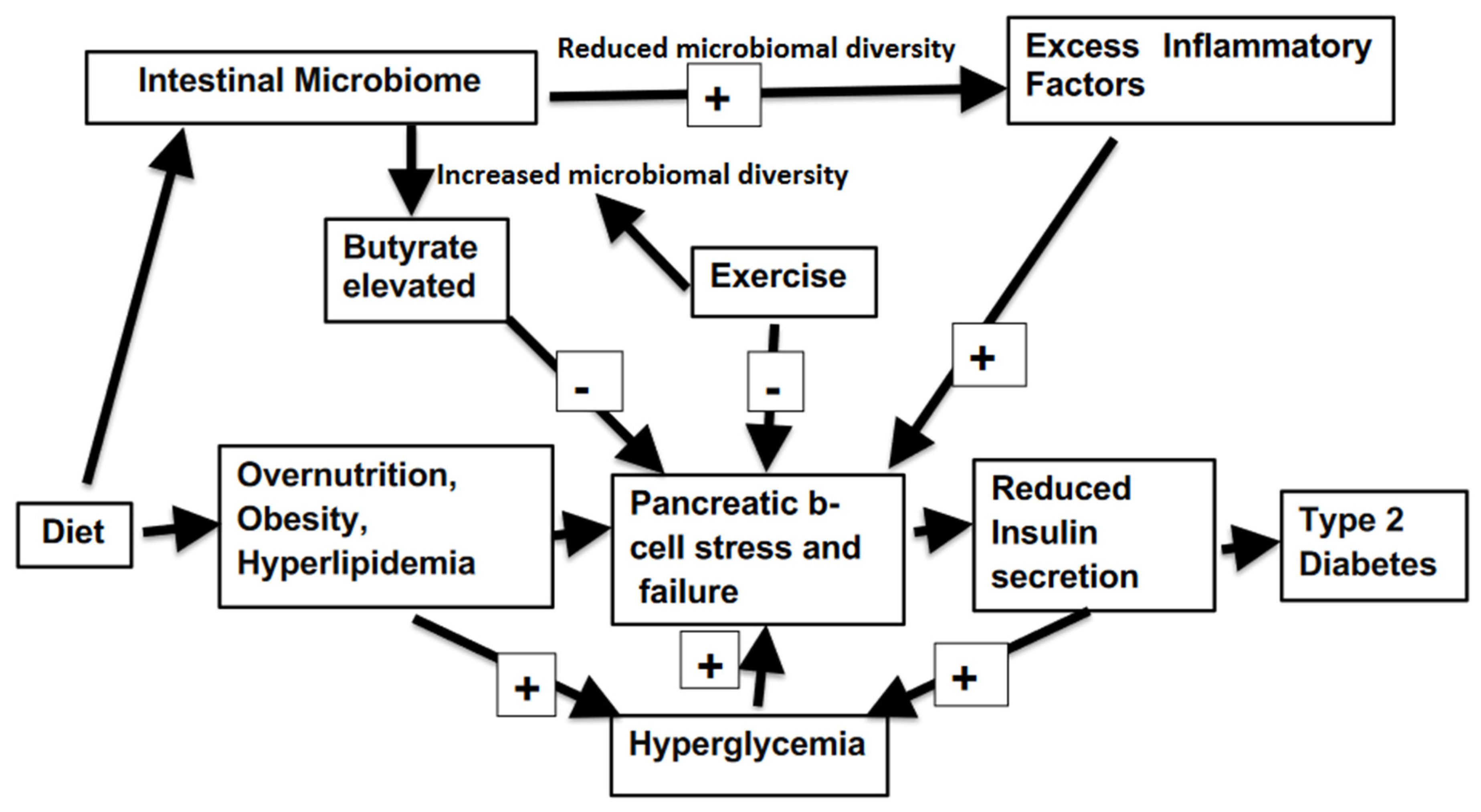
:1. Introduction
2. Type 1 and 2 Diabetes, Similarities and Distinctions
3. Composition of the Human Microbiome and Relevance to Diabetes
3.1. Firmicutes
Relevance to Diabetes
3.2. Bacteriodetes
Relevance to Diabetes
3.3. Actinobacteria
Relevance to Diabetes
3.4. Verrucomicrobiota
Relevance to Diabetes
3.5. Proteobacteria
Relevance to Diabetes
3.6. Bacillota
Relevance to Diabetes
4. Relative Prevalence of Differing Bacterial Classes
5. Bacterial Diversity
6. Bacterial Regulation of Intestinal Permeability
7. Modulation of the Microbiome as a Means of Treating Diabetes
7.1. Diet
7.2. Exercise
7.3. Pharmacological Agents
7.4. Probiotics
7.5. Fecal Transplantation
8. Summary
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, G.; Wei, J.; Liu, P.; Zhang, Q.; Tian, Y.; Hou, G.; Meng, L.; Xin, Y.; Jiang, X. Role of the gut microbiota in type 2 diabetes and related diseases. Metabolism 2021, 117, 154712. [Google Scholar] [CrossRef] [PubMed]
- IDF Diabetes Atlas 2021–10th Edition. Available online: https://idf.org/ (accessed on 12 November 2022).
- Ortega, M.A.; Fraile-Martínez, O.; Naya, I.; García-Honduvilla, N.; Álvarez-Mon, M.; Buján, J.; Asúnsolo; De La Torre, B. Type 2 Diabetes Mellitus Associated with Obesity (Diabesity). The Central Role of Gut Microbiota and Its Translational Applications. Nutrients 2020, 12, 2749. [Google Scholar] [CrossRef] [PubMed]
- Ilonen, J.; Lempainen, J.; Veijola, R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H. Microbiota or short-chain fatty acids: Which regulates diabetes? Cell Mol. Immunol. 2017, 15, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Li, Y.; Stoll, M.L.; Tollefsbol, T.O. The Epigenetic Connection Between the Gut Microbiome in Obesity and Diabetes. Front. Genet. 2020, 10, 1329. [Google Scholar] [CrossRef] [Green Version]
- Triolo, T.M.; Fouts, A.; Pyle, L.; Yu, L.; Gottlieb, P.A.; Steck, A.K.; the Type 1 Diabetes TrialNet Study Group; Greenbaum, C.J.; Atkinson, M.; Baidal, D.; et al. Identical and Nonidentical Twins: Risk and Factors Involved in Development of Islet Autoimmunity and Type 1 Diabetes. Diabetes Care 2018, 42, 192–199. [Google Scholar] [CrossRef] [Green Version]
- Willemsen, G.; Ward, K.J.; Bell, C.; Christensen, K.; Bowden, J.; Dalgård, C.; Harris, J.R.; Kaprio, J.; Lyle, R.; Magnusson, P.; et al. The Concordance and Heritability of Type 2 Diabetes in 34,166 Twin Pairs from International Twin Registers: The Discordant Twin (DISCOTWIN) Consortium. Twin Res. Hum. Genet. 2015, 18, 762–771. [Google Scholar] [CrossRef] [Green Version]
- de Candia, P.; Prattichizzo, F.; Garavelli, S.; De Rosa, V.; Galgani, M.; Di Rella, F.; Spagnuolo, M.I.; Colamatteo, A.; Fusco, C.; Micillo, T.; et al. Type 2 Diabetes: How Much of an Autoimmune Disease? Front. Endocrinol. 2019, 10, 451. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Tian, H.; Liu, Z.; Yin, X.; Xi, B. Preterm birth and risk of type 1 and type 2 diabetes: Systematic review and meta-analysis. Obes. Rev. 2014, 15, 804–811. [Google Scholar] [CrossRef]
- Ikegami, H.; Babaya, N.; Noso, S. β-Cell failure in diabetes: Common susceptibility and mechanisms shared between type 1 and type 2 diabetes. J. Diabetes Investig. 2021, 12, 1526–1539. [Google Scholar] [CrossRef]
- Jones, A.G.; McDonald, T.J.; Shields, B.M.; Hagopian, W.; Hattersley, A.T. Latent Autoimmune Diabetes of Adults (LADA) Is Likely to Represent a Mixed Population of Autoimmune (Type 1) and Nonautoimmune (Type 2) Diabetes. Diabetes Care 2021, 44, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836, PMCID:PMC5433529. [Google Scholar] [CrossRef] [PubMed]
- Pitocco, D.; DI Leo, M.; Tartaglione, L.; De Leva, F.; Petruzziello, C.; Saviano, A.; Pontecorvi, A.; Ojetti, V. The role of gut microbiota in mediating obesity and diabetes mellitus. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1548–1562. [Google Scholar] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, M.M.; Pal, N.; Sharma, P.; Kumawat, M.; Shubham, S.; Verma, V.; Tiwari, R.R.; Singh, B.; Nagpal, R.; Sarma, D.K.; et al. Role of butyrogenic Firmicutes in type-2 diabetes. J. Diabetes Metab. Disord. 2022, 21, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Bamola, V.D.; Ghosh, A.; Kapardar, R.K.; Lal, B.; Cheema, S.; Sarma, P.; Chaudhry, R. Gut microbial diversity in health and disease: Experience of healthy Indian subjects, and colon carcinoma and inflammatory bowel disease patients. Microb. Ecol. Health Dis. 2017, 28, 1322447. [Google Scholar] [CrossRef] [Green Version]
- Tingirikari, J.M.R. Microbiota-accessible pectic poly- and oligosaccharides in gut health. Food Funct. 2018, 9, 5059–5073. [Google Scholar] [CrossRef]
- Cholan, P.M.; Han, A.; Woodie, B.R.; Watchon, M.; Kurz, A.R.; Laird, A.S.; Britton, W.J.; Ye, L.; Holmes, Z.C.; McCann, J.R.; et al. Conserved anti-inflammatory effects and sensing of butyrate in zebrafish. Gut Microbes 2020, 12, 1824563. [Google Scholar] [CrossRef]
- Zheng, P.; Li, Z.; Zhou, Z. Gut microbiome in type 1 diabetes: A comprehensive review. Diabetes/Metab. Res. Rev. 2018, 34, e3043. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Cheng, Y.; Zhu, M.; Xiao, Z.; Ruan, G.; Wei, Y. Gut microbiota: A new target for T2DM prevention and treatment. Front. Endocrinol. 2022, 13, 958218. [Google Scholar] [CrossRef]
- Coppola, S.; Avagliano, C.; Calignano, A.; Canani, R.B. The Protective Role of Butyrate against Obesity and Obesity-Related Diseases. Molecules 2021, 26, 682. [Google Scholar] [CrossRef] [PubMed]
- Halim, M.; Halim, A. The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes). Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1165–1172. [Google Scholar] [CrossRef]
- Riedel, C.U.; Foata, F.; Philippe, D.; Adolfsson, O.; Eikmanns, B.J.; Blum, S. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-κB activation. World J. Gastroenterol. 2006, 12, 3729–3735. [Google Scholar] [CrossRef] [PubMed]
- Halawa, M.R.; El-Salam, M.A.; Mostafa, B.M.; Sallout, S.S. The Gut Microbiome, Lactobacillus acidophilus; Relation with Type 2 Diabetes Mellitus. Curr. Diabetes Rev. 2019, 15, 480–485. [Google Scholar] [CrossRef]
- Kijmanawat, A.; Panburana, P.; Reutrakul, S.; Tangshewinsirikul, C. Effects of probiotic supplements on insulin resistance in gestational diabetes mellitus: A double-blind randomized controlled trial. J. Diabetes Investig. 2018, 10, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, V.F.; Elias-Oliveira, J.; Pereira, Í.S.; Pereira, J.A.; Barbosa, S.C.; Machado, M.S.G.; Carlos, D. Akkermansia muciniphila and Gut Immune System: A Good Friendship That Attenuates Inflammatory Bowel Disease, Obesity, and Diabetes. Front. Immunol. 2022, 13, 934695. [Google Scholar] [CrossRef]
- Zhang, J.; Ni, Y.; Qian, L.; Fang, Q.; Zheng, T.; Zhang, M.; Gao, Q.; Ni, J.; Hou, X.; Bao, Y.; et al. Decreased Abundance of Akkermansia muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv. Sci. 2021, 8, e2100536. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; Van Raalte, D.H.; Herrema, H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731. [Google Scholar] [CrossRef]
- Soyucen, E.; Gulcan, A.; Aktuglu-Zeybek, A.C.; Onal, H.; KIYKIM, E.; Aydin, A. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr. Int. 2014, 56, 336–343. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef] [PubMed]
- Bojarczuk, A.; Skąpska, S.; Khaneghah, A.M.; Marszałek, K. Health benefits of resistant starch: A review of the literature. J. Funct. Foods 2022, 93, 105094. [Google Scholar] [CrossRef]
- Cheng, S.H.; Yang, T.Y.; Hsu, C.C.; Wei, Y.H.; Wu, C.C.; Tsai, Y.C. Lactobacillus paragasseri BBM171 Ameliorates Allergic Airway Inflammation Induced by Ovalbumin in Mice via Modulating the Th1/Th2 Balance. Microorganisms 2022, 10, 2041. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Bueno, A.A.; de Souza, R.G.M.; Mota, J.F. Gut microbiota, probiotics and diabetes. Nutr. J. 2014, 13, 60. [Google Scholar] [CrossRef] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Hung, W.-C.; Tsai, H.-J.; Chang, C.-C.; Chiu, Y.-W.; Hwang, S.-J.; Kuo, M.-C.; Chen, S.-C.; Dai, C.-Y.; Tsai, Y.-C. The Association of Targeted Gut Microbiota with Body Composition in Type 2 Diabetes Mellitus. Int. J. Med Sci. 2021, 18, 511–519. [Google Scholar] [CrossRef]
- Lontchi-Yimagou, E.; Dasgupta, R.; Anoop, S.; Kehlenbrink, S.; Koppaka, S.; Goyal, A.; Venkatesan, P.; Livingstone, R.; Ye, K.; Chapla, A.; et al. An Atypical Form of Diabetes Among Individuals with Low BMI. Diabetes Care 2022, 45, 1428–1437. [Google Scholar] [CrossRef]
- George, A.M.; Jacob, A.G.; Fogelfeld, L. Lean diabetes mellitus: An emerging entity in the era of obesity. World J. Diabetes 2015, 6, 613–620. [Google Scholar] [CrossRef]
- Koh, H.-C.E.; Cao, C.; Mittendorfer, B. Insulin Clearance in Obesity and Type 2 Diabetes. Int. J. Mol. Sci. 2022, 23, 596. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, P.; Metos, J.; Babu, P.V.A. Impact of type 1 diabetes on the composition and functional potential of gut microbiome in children and adolescents: Possible mechanisms, current knowledge, and challenges. Gut Microbes 2021, 13, 1926841. [Google Scholar] [CrossRef] [PubMed]
- Knip, M.; Honkanen, J. Modulation of Type 1 Diabetes Risk by the Intestinal Microbiome. Curr. Diabetes Rep. 2017, 17, 105. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Zhao, X.; Sun, L.; Liu, Y.; Lv, Y.; Gang, X.; Wang, G. Gut Microbiota Profile in Patients with Type 1 Diabetes Based on 16S rRNA Gene Sequencing: A Systematic Review. Dis. Markers 2020, 2020, 3936247. [Google Scholar] [CrossRef]
- Siljander, H.; Honkanen, J.; Knip, M. Microbiome and type 1 diabetes. Ebiomedicine 2019, 46, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Moffa, S.; Mezza, T.; Cefalo, C.M.A.; Cinti, F.; Impronta, F.; Sorice, G.P.; Santoro, A.; Di Giuseppe, G.; Pontecorvi, A.; Giaccari, A. The Interplay between Immune System and Microbiota in Diabetes. Mediat. Inflamm. 2019, 2019, 9367404. [Google Scholar] [CrossRef]
- Bessac, A.; Cani, P.D.; Meunier, E.; Dietrich, G.; Knauf, C. Inflammation and Gut-Brain Axis During Type 2 Diabetes: Focus on the Crosstalk Between Intestinal Immune Cells and Enteric Nervous System. Front. Neurosci. 2018, 12, 725. [Google Scholar] [CrossRef]
- Liu, Y.; Lou, X. Type 2 diabetes mellitus-related environmental factors and the gut microbiota: Emerging evidence and challenges. Clinics 2020, 75, e1277. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef]
- Ho, J.; Nicolucci, A.C.; Virtanen, H.; Schick, A.; Meddings, J.; Reimer, R.A.; Huang, C. Effect of Prebiotic on Microbiota, Intestinal Permeability, and Glycemic Control in Children with Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2019, 104, 4427–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaiss, C.A.; Levy, M.; Grosheva, I.; Zheng, D.; Soffer, E.; Blacher, E.; Braverman, S.; Tengeler, A.C.; Barak, O.; Elazar, M.; et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 2018, 359, 1376–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiarelli, F.; Giannini, C.; Primavera, M. Prediction and prevention of type 1 diabetes in children. Clin. Pediatr. Endocrinol. 2019, 28, 43–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huda, M.N.; Kim, M.; Bennett, B.J. Modulating the Microbiota as a Therapeutic Intervention for Type 2 Diabetes. Front. Endocrinol. 2021, 12, 632335. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 2021, 12, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, S.P.; Denu, J.M. Short-chain fatty acids activate acetyltransferase p300. eLife 2021, 10, e72171. [Google Scholar] [CrossRef]
- Pedersen, S.S.; Prause, M.; Williams, K.; Barrès, R.; Billestrup, N. Butyrate inhibits IL-1β-induced inflammatory gene expression by suppression of NF-κB activity in pancreatic beta cells. J. Biol. Chem. 2022, 298, 102312. [Google Scholar] [CrossRef]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; Van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Harbison, J.E.; Roth-Schulze, A.J.; Giles, L.; Tran, C.D.; Ngui, K.M.; Penno, M.; Thomson, R.L.; Wentworth, J.M.; Colman, P.G.; Craig, M.E.; et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: A prospective cohort study. Pediatr. Diabetes 2019, 20, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, S.-X.; Cai, Y.; Xie, K.-L.; Zhang, W.-L.; Zheng, F. Effects of combined aerobic and resistance training on the glycolipid metabolism and inflammation levels in type 2 diabetes mellitus. J. Phys. Ther. Sci. 2015, 27, 2365–2371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasini, E.; Corsetti, G.; Assanelli, D.; Testa, C.; Romano, C.; Dioguardi, F.S.; Aquilani, R. Effects of chronic exercise on gut microbiota and intestinal barrier in human with type 2 diabetes. Minerva Med. 2019, 110, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxidative Med. Cell. Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef] [PubMed]
- Yun, E.-J.; Imdad, S.; Jang, J.; Park, J.; So, B.; Kim, J.-H.; Kang, C. Diet Is a Stronger Covariate than Exercise in Determining Gut Microbial Richness and Diversity. Nutrients 2022, 14, 2507. [Google Scholar] [CrossRef] [PubMed]
- LaMoia, T.E.; Shulman, G.I. Cellular and Molecular Mechanisms of Metformin Action. Endocr. Rev. 2020, 42, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.G.; Stratigou, T.; Tsagarakis, S. Metformin and gut microbiota: Their interactions and their impact on diabetes. Hormones (Athens) 2019, 18, 141–144. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Krogh Pedersen, H.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Chae, S.; Jo, S.; Jerng, U.; Bae, S. The Relationship between the Gut Microbiome and Metformin as a Key for Treating Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 3566. [Google Scholar] [CrossRef]
- Craciun, C.I.; Neag, M.A.; Catinean, A.; Mitre, A.O.; Rusu, A.; Bala, C.; Roman, G.; Buzoianu, A.D.; Muntean, D.M.; Craciun, A.E. The Relationships between Gut Microbiota and Diabetes Mellitus, and Treatments for Diabetes Mellitus. Biomedicines 2022, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Salles, B.I.M.; Cioffi, D.; Ferreira, S.R.G. Probiotics supplementation and insulin resistance: A systematic review. Diabetol. Metab. Syndr. 2020, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Y.; Yan, Y.; Tian, S.; Zheng, D.; Leng, D.; Wang, C.; Jiao, J.; Wang, Z.; Bai, Y. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front. Cell. Infect. Microbiol. 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D.; De Vos, W.M. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef] [Green Version]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2020, 70, 1174–1182. [Google Scholar] [CrossRef]
- Michael, H.; Amimo, J.O.; Rajashekara, G.; Saif, L.J.; Vlasova, A.N. Mechanisms of Kwashiorkor-Associated Immune Suppression: Insights from Human, Mouse, and Pig Studies. Front. Immunol. 2022, 13, 826268. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bondy, S.C. Relationships between Diabetes and the Intestinal Microbial Population. Int. J. Mol. Sci. 2023, 24, 566. https://doi.org/10.3390/ijms24010566
Bondy SC. Relationships between Diabetes and the Intestinal Microbial Population. International Journal of Molecular Sciences. 2023; 24(1):566. https://doi.org/10.3390/ijms24010566
Chicago/Turabian StyleBondy, Stephen C. 2023. "Relationships between Diabetes and the Intestinal Microbial Population" International Journal of Molecular Sciences 24, no. 1: 566. https://doi.org/10.3390/ijms24010566





