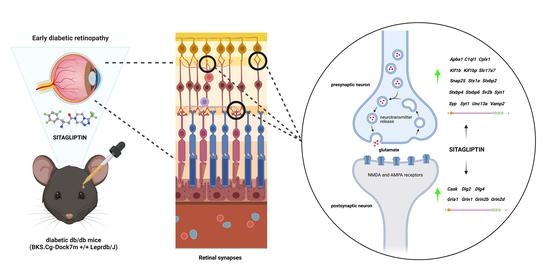
Transcriptomic Analysis Reveals That Retinal Neuromodulation Is a Relevant Mechanism in the Neuroprotective Effect of Sitagliptin in an Experimental Model of Diabetic Retinopathy
Abstract
1. Introduction
2. Results
2.1. Multiple Comparisons between Transcriptomes Revealed a Clear Effect of Both the Diabetic Condition and Sitagliptin Treatment
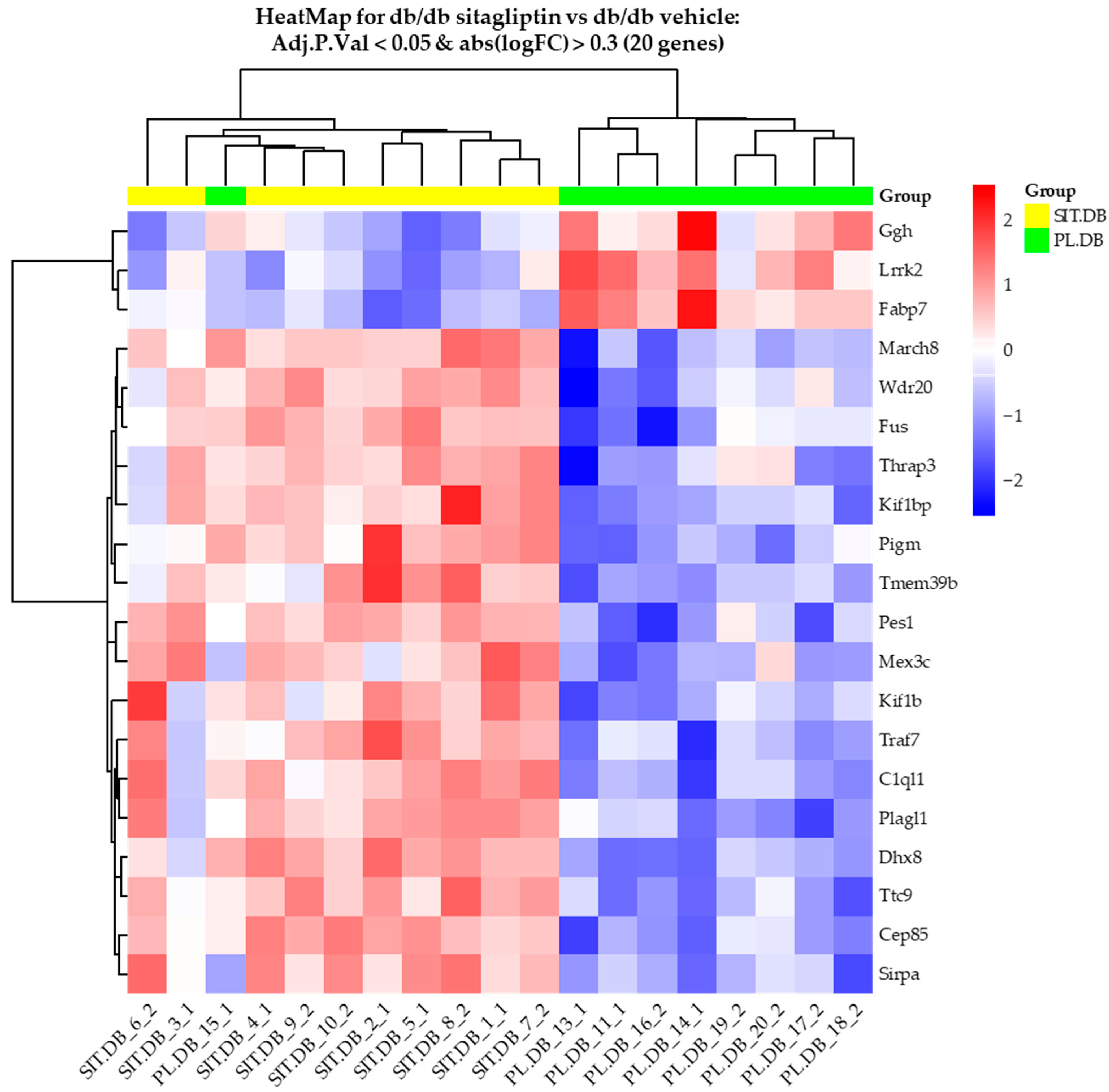
2.2. Db/db Mice Topically Treated with Sitagliptin Showed Different Expression Patterns in the Retina Compared to Those Treated with Vehicle
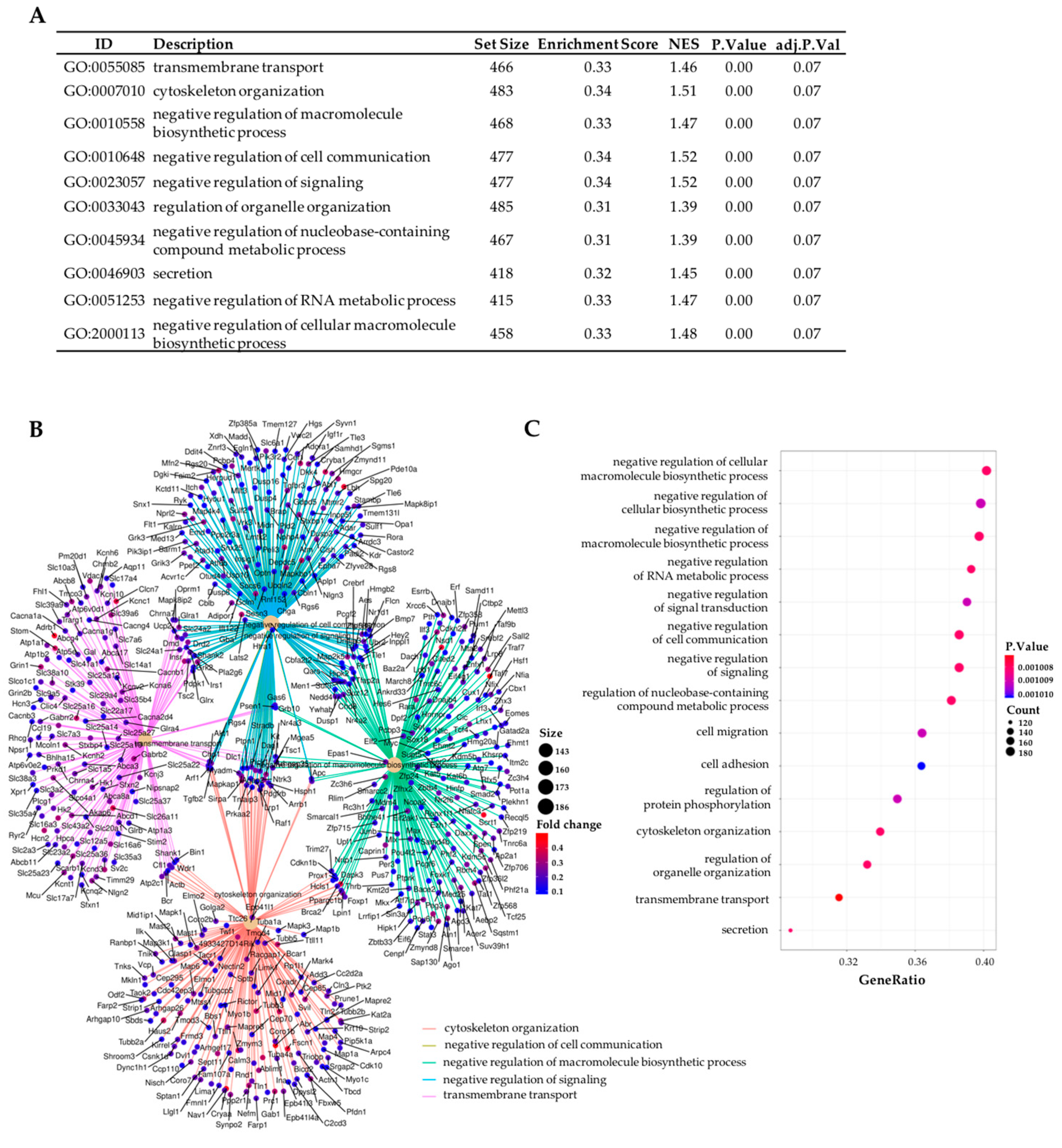
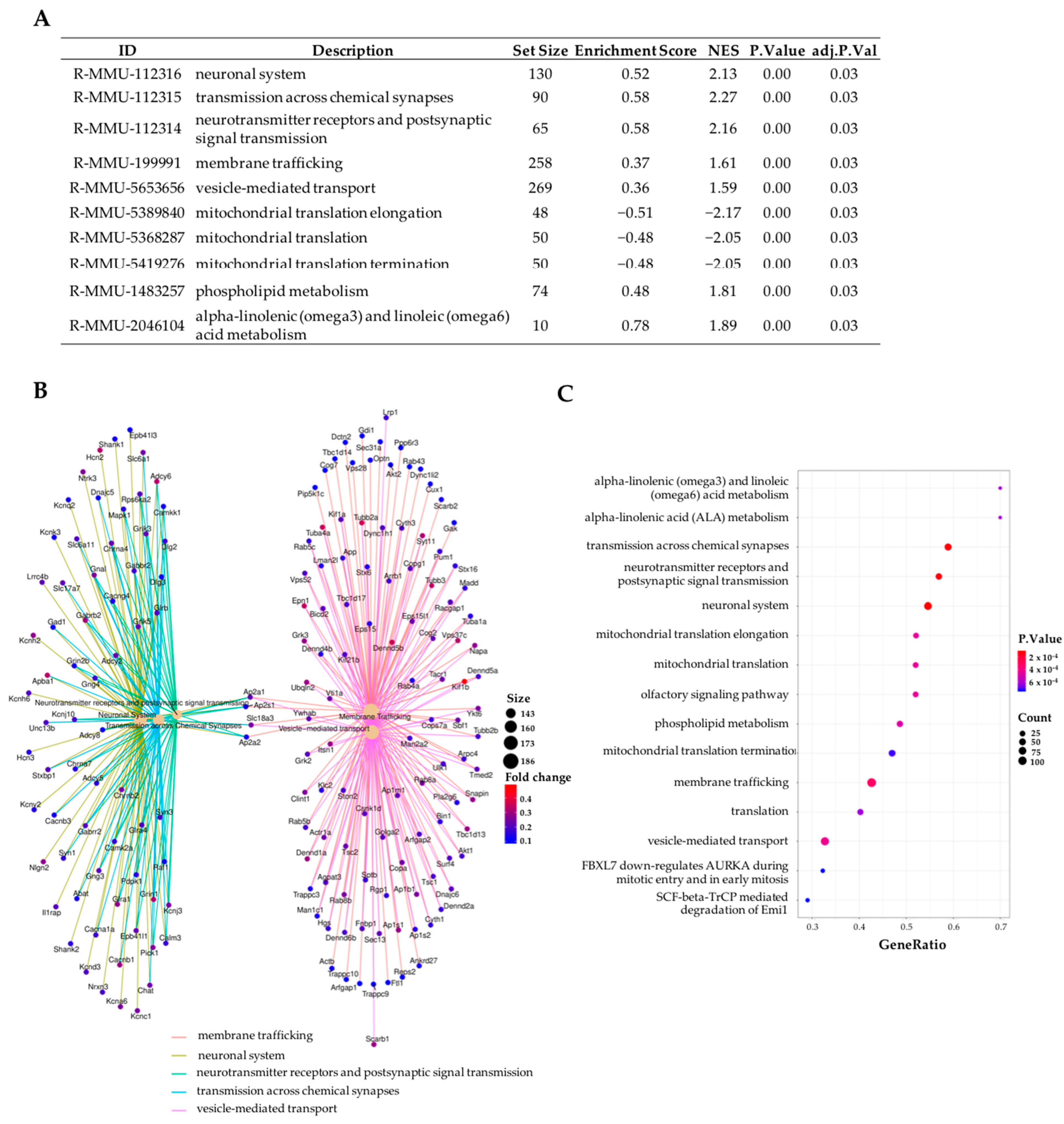
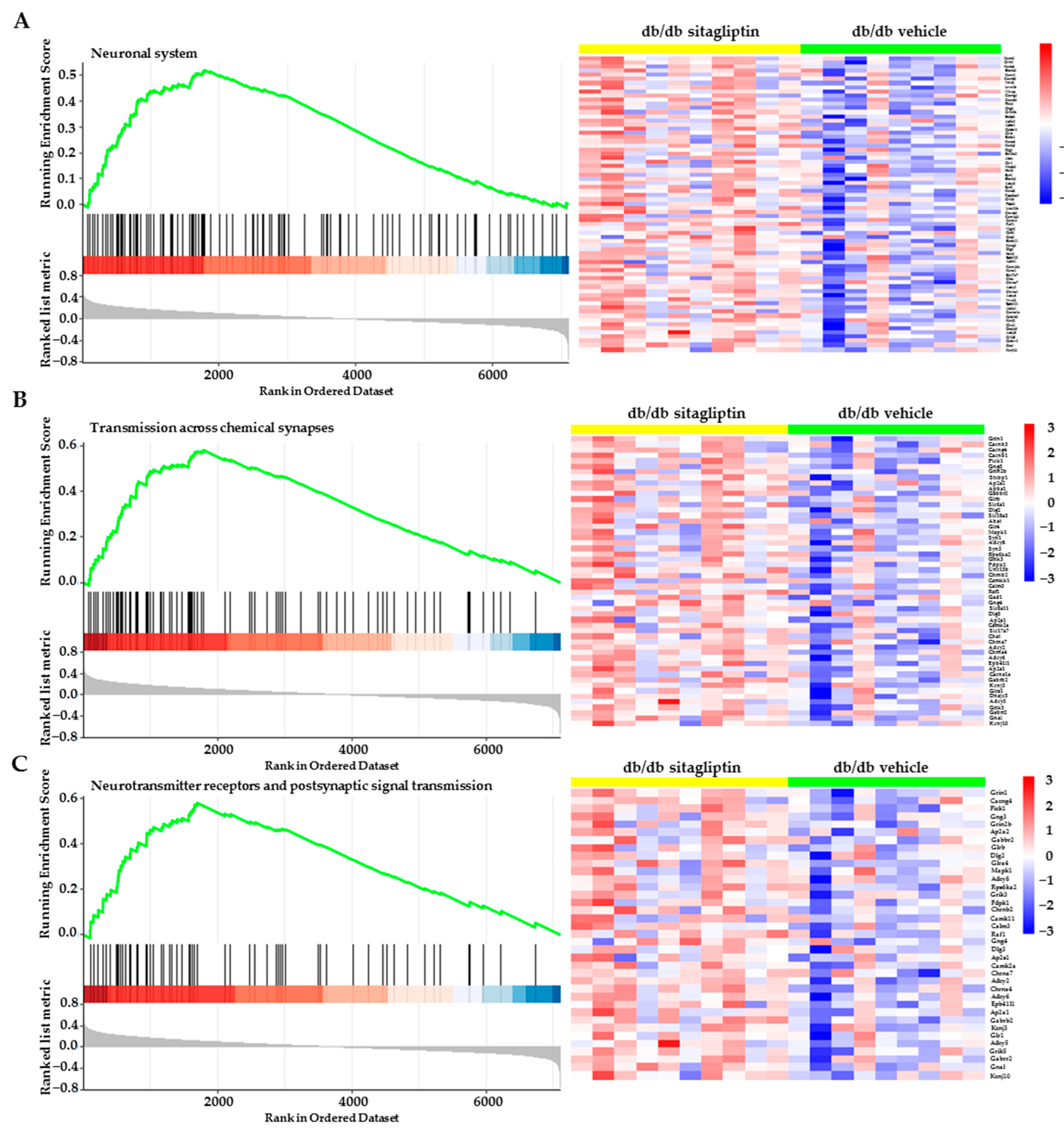
2.3. Gene Set Enrichment Analyses (GSEA) Revealed That the Most Differentiated Biological Process Is Neurotransmission
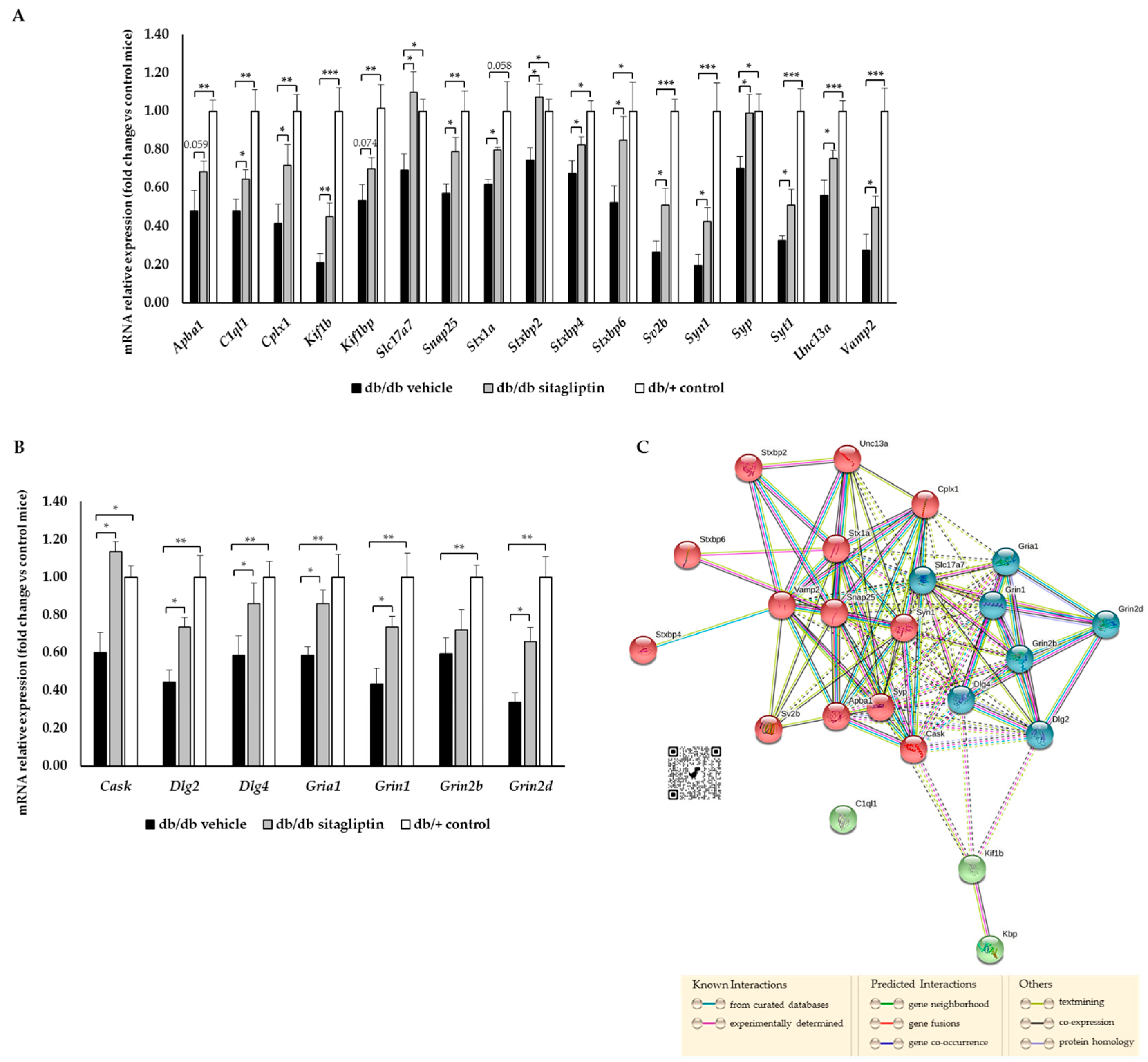
2.4. Topical Administration of Sitagliptin Prevented DR-Induced Down-Regulation of Genes Related to Synapse Formation, Maintenance and Synaptic Transmission
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Interventional Study
4.3. Retinal Tissue Processing
4.4. Transcriptome Analysis
4.5. cDNA Reverse Transcription and Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR) Assay
4.6. Statistical Analysis of RT-PCR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leasher, J.L.; Bourne, R.R.; Flaxman, S.R.; Jonas, J.B.; Keeffe, J.; Naidoo, K.; Pesudovs, K.; Price, H.; White, R.A.; Wong, T.Y.; et al. Vision Loss Expert Group of the Global Burden of Disease Study. Global estimates on the number of people blind or visually impaired by diabetic retinopathy: A meta-analysis from 1990 to 2010. Diabetes Care 2016, 39, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Cheung, C.M.; Larsen, M.; Sharma, S.; Simó, R. Diabetic retinopathy. Nat. Rev. Dis. Primers 2016, 17, 16012. [Google Scholar] [CrossRef] [PubMed]
- Simó, R.; Hernández, C. Novel approaches for treating diabetic retinopathy based on recent pathogenic evidence. Prog. Retin. Eye Res. 2015, 48, 160–180. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.D.; Chew, E.; Duh, E.J.; Sobrin, L.; Sun, J.K.; VanderBeek, B.L.; Wykoff, C.C.; Gardner, T.W. Diabetic Retinopathy: A Position Statement by the American Diabetes Association. Diabetes Care 2017, 40, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Simó, R.; Stitt, A.W.; Gardner, T.W. Neurodegeneration in diabetic retinopathy: Does it really matter? Diabetologia 2018, 61, 1902–1912. [Google Scholar] [CrossRef]
- Simó, R.; Simó-Servat, O.; Bogdanov, P.; Hernández, C. Neurovascular Unit: A New Target for Treating Early Stages of Diabetic Retinopathy. Pharmaceutics 2021, 13, 1320. [Google Scholar] [CrossRef]
- Salcedo, I.; Tweedie, D.; Li, Y.; Greig, N.H. Neuroprotective and neurotrophic actions of glucagon-like peptide-1: An emerging opportunity to treat neurodegenerative and cerebrovascular disorders. Br. J. Pharmacol. 2012, 166, 1586–1599. [Google Scholar] [CrossRef]
- Hernández, C.; Bogdanov, P.; Solà-Adell, C.; Sampedro, J.; Valeri, M.; Genís, X.; Simó-Servat, O.; García-Ramírez, M.; Simó, R. Topical administration of DPP-IV inhibitors prevents retinal neurodegeneration in experimental diabetes. Diabetologia 2017, 60, 2285–2298. [Google Scholar] [CrossRef]
- Hernández, C.; Bogdanov, P.; Corraliza, L.; García-Ramírez, M.; Solà-Adell, C.; Arranz, J.A.; Arroba, A.I.; Valverde, A.M.; Simó, R. Topical administration of GLP-1 receptor agonists prevents retinal neurodegeneration in experimental diabetes. Diabetes 2016, 65, 172–187. [Google Scholar] [CrossRef]
- Röhrborn, D.; Wronkowitz, N.; Eckel, J. DPP4 in Diabetes. Front. Immunol. 2015, 6, 386. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Barber, A.J.; Baccouche, B. Neurodegeneration in diabetic retinopathy: Potential for novel therapies. Vision. Res. 2017, 139, 82–92. [Google Scholar] [CrossRef]
- Kutsyr, O.; Arango-Gonzalez, B.; Fernandez-Sanchez, L.; Maneu, V.; Lax, P.; Ambrosio, A.F.; Ueffing, M.; Cuenca, N. Dipeptidyl deptidase-IV inhibition by sitagliptin slows down retinal neurodegeneration in rd10 mice retinas. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4879. [Google Scholar]
- Gonçalves, A.; Marques, C.; Leal, E.; Ribeiro, C.F.; Reis, F.; Ambrósio, A.F.; Fernandes, R. Dipeptidyl peptidase-IV inhibition prevents blood-retinal barrier breakdown, inflammation and neuronal cell death in the retina of type 1 diabetic rats. Biochim. Biophys. Acta 2014, 1842, 1454–1463. [Google Scholar] [CrossRef]
- Ramos, H.; Bogdanov, P.; Sabater, D.; Huerta, J.; Valeri, M.; Hernández, C.; Simó, R. Neuromodulation Induced by Sitagliptin: A New Strategy for Treating Diabetic Retinopathy. Biomedicines 2021, 9, 1772. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Xu, J.; Sun, Q.; Yu, F.; Cheng, J.; Peng, B.; Liu, W.; Xiao, Z.; Yin, J.; et al. Inhibition of DPP4 enhances inhibitory synaptic transmission through activating the GLP-1/GLP-1R signaling pathway in a rat model of febrile seizures. Biochem. Pharmacol. 2018, 156, 78–85. [Google Scholar] [CrossRef]
- Dietrich, N.; Kolibabka, M.; Busch, S.; Bugert, P.; Kaiser, U.; Lin, J.; Fleming, T.; Morcos, M.; Klein, T.; Schlotterer, A.; et al. The DPP4 inhibitor linagliptin protects from experimental diabetic retinopathy. PLoS ONE 2016, 11, e0167853. [Google Scholar] [CrossRef]
- Chung, Y.R.; Ha, K.H.; Kim, H.C.; Park, S.J.; Lee, K.; Kim, D.J. Dipeptidyl Peptidase-4 Inhibitors versus Other Antidiabetic Drugs Added to Metformin Monotherapy in Diabetic Retinopathy Progression: A Real World-BasedCohort Study. Diabetes Metab. J. 2019, 43, 640–648. [Google Scholar] [CrossRef]
- Tang, H.; Li, G.; Zhao, Y.; Wang, F.; Gower, E.W.; Shi, L.; Wang, T. Comparisons of diabetic retinopathy events associated with glucose-lowering drugs in patients with type 2 diabetes mellitus: A network meta-analysis. Diabetes Obes. Metab. 2018, 20, 1262–1279. [Google Scholar] [CrossRef]
- Taylor, O.M.; Lam, C. The Effect of Dipeptidyl Peptidase-4 Inhibitors on Macrovascular and Microvascular Complications of Diabetes Mellitus: A Systematic Review. Curr. Ther. Res. Clin. Exp. 2020, 25, 100596. [Google Scholar] [CrossRef]
- Fura, A.; Khanna, A.; Vyas, V.; Koplowitz, B.; Chang, S.Y.; Caporuscio, C.; Boulton, D.W.; Christopher, L.J.; Chadwick, K.D.; Hamann, L.G.; et al. Pharmacokinetics of the dipeptidyl peptidase 4 inhibitor saxagliptin in rats, dogs, and monkeys and clinical projections. Drug Metab. Dispos. 2009, 37, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, H.; Binder, R.; Greischel, A. Tissue distribution of the novel DPP-4 inhibitor BI 1356 is dominated by saturable binding to its target in rats. Biopharm. Drug Dispos. 2009, 30, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Sigoillot, S.M.; Iyer, K.; Binda, F.; González-Calvo, I.; Talleur, M.; Vodjdani, G.; Isope, P.; Selimi, F. The Secreted Protein C1QL1 and Its Receptor BAI3 Control the Synaptic Connectivity of Excitatory Inputs Converging on Cerebellar Purkinje Cells. Cell Rep. 2015, 10, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Drerup, C.M.; Lusk, S.; Nechiporuk, A. Kif1B Interacts with KBP to Promote Axon Elongation by Localizing a Microtubule Regulator to Growth Cones. J. Neurosci. 2016, 36, 7014–7026. [Google Scholar] [CrossRef] [PubMed]
- Becherer, U.; Rettig, J. Vesicle pools, docking, priming, and release. Cell Tissue Res. 2006, 326, 393–407. [Google Scholar] [CrossRef]
- Stepien, K.P.; Prinslow, E.A.; Rizo, J. Munc18-1 is crucial to overcome the inhibition of synaptic vesicle fusion by αSNAP. Nat. Commun. 2019, 10, 4326. [Google Scholar] [CrossRef]
- Okamoto, M.; Südhof, T.C. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J. Biol. Chem. 1997, 272, 31459–31464. [Google Scholar] [CrossRef]
- Courtney, N.A.; Bao, H.; Briguglio, J.S.; Chapman, E.R. Synaptotagmin 1 clamps synaptic vesicle fusion in mammalian neurons independent of complexin. Nat. Commun. 2019, 10, 4076. [Google Scholar] [CrossRef]
- Kwon, S.E.; Chapman, E.R. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron 2011, 70, 847–854. [Google Scholar] [CrossRef]
- Bähler, M.; Benfenati, F.; Valtorta, F.; Greengard, P. The synapsins and the regulation of synaptic function. Bioessays 1990, 12, 259–263. [Google Scholar] [CrossRef]
- Bykhovskaia, M. Synapsin regulation of vesicle organization and functional pools. Semin. Cell Dev. Biol. 2011, 22, 387–392. [Google Scholar] [CrossRef]
- Stout, K.A.; Dunn, A.R.; Hoffman, C.; Miller, G.W. The Synaptic Vesicle Glycoprotein 2: Structure, Function, and Disease Relevance. ACS Chem. Neurosci. 2019, 10, 3927–3938. [Google Scholar] [CrossRef]
- Martineau, M.; Guzman, R.E.; Fahlke, C.; Klingauf, J. VGLUT1 functions as a glutamate/proton exchanger with chloride channel activity in hippocampal glutamatergic synapses. Nat. Commun. 2017, 8, 2279. [Google Scholar] [CrossRef]
- Huang, S.; Chen, L.; Bladen, C.; Stys, P.K.; Zamponi, G.W. Differential modulation of NMDA and AMPA receptors by cellular prion protein and copper ions. Mol. Brain 2018, 11, 62. [Google Scholar] [CrossRef]
- Smith, S.B. Diabetic Retinopathy and the NMDA Receptor. Drug News Perspect 2002, 15, 226–232. [Google Scholar] [CrossRef]
- Jeong, J.; Pandey, S.; Li, Y.; Badger, J.D., 2nd; Lu, W.; Roche, K.W. PSD-95 binding dynamically regulates NLGN1 trafficking and function. Proc. Natl. Acad. Sci. USA 2019, 116, 12035–12044. [Google Scholar] [CrossRef]
- Hsueh, Y.P. The role of the MAGUK protein CASK in neural development and synaptic function. Curr. Med. Chem. 2006, 13, 1915–1927. [Google Scholar] [CrossRef]
- Gentleman, R.; Carey, V.; Huber, W.; Irizarry, R.A.; Dudoit, S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet Mol. Biol. 2004, 3. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]


| db/db Sitagliptin vs. db/+ Control | db/db Vehicle vs. db/+ Control | db/db Sitagliptin vs. db/db Vehicle | |
|---|---|---|---|
| UpReg_B | 65 | 15 | 33 |
| DownReg_B | 20 | 13 | 9 |
| UpRegAdj0.01 | 18 | 5 | 0 |
| DownRegAdj0.01 | 6 | 3 | 0 |
| UpRegAdj0.05 | 108 | 10 | 17 |
| DownRegAdj0.05 | 33 | 9 | 3 |
| UpRegAdj0.15 | 369 | 16 | 185 |
| DownRegAdj0.15 | 122 | 14 | 65 |
| UpRegAdj0.25 | 728 | 24 | 494 |
| DownRegAdj0.25 | 291 | 19 | 195 |
| UpRegP0.01 | 363 | 71 | 275 |
| DownRegP0.01 | 116 | 56 | 96 |
| UpRegP0.05 | 852 | 218 | 82 |
| DownRegP0.01 | 376 | 223 | 329 |
| Primers | Nucleotide Sequence | |
|---|---|---|
| Actb | Forward (5′-3′) | 5′-CTAAGGCCAACCGTGAAAG -3′ |
| Reverse (5′-3′) | 5′-CAGTATGTTCGGCTTCCCATTC-3′ | |
| Apba1 | Forward (5′-3′) | 5′-GGTGCTGAGTCATCAAGCATAC-3′ |
| Reverse (5′-3′) | 5′-GAACTTCAACGTAGGTTGGGAA-3′ | |
| B2m | Forward (5′-3′) | 5′-GTATGCTATCCAGAAAACCC-3′ |
| Reverse (5′-3′) | 5′-CTGAAGGACATATCTGACATC-3′ | |
| C1ql1 | Forward (5′-3′) | 5′-GGCACCTACTTTTTCACCTACC-3′ |
| Reverse (5′-3′) | 5′-AGTCGTAGTTCTGGTCTGCAT-3′ | |
| Cask1 | Forward (5′-3′) | 5′-TGAAGAAGTAGTCAAACTGCCAG-3′ |
| Reverse (5′-3′) | 5′-TTTGTCCCGTACATTGCATCC-3′ | |
| Cplx1 | Forward (5′-3′) | 5′-AGTTCGTGATGAAACAAGCCC-3′ |
| Reverse (5′-3′) | 5′-TCTTCCTCCTTCTTAGCAGCA-3′ | |
| Dlg2 | Forward (5′-3′) | 5′-CTGTCACGAGGCAGGAAATAAA-3′ |
| Reverse (5′-3′) | 5′-CGACTTCGTAGTCACGCTTTG-3′ | |
| Dlg4 | Forward (5′-3′) | 5′-TGAGATCAGTCATAGCAGCTACT-3′ |
| Reverse (5′-3′) | 5′-CTTCCTCCCCTAGCAGGTCC-3′ | |
| Gria1 | Forward (5′-3′) | 5′-AAAGGAGTGTACGCCATCTTTG-3′ |
| Reverse (5′-3′) | 5′-TGTCAACGGGAAAACTTGGAG-3′ | |
| Grin1 | Forward (5′-3′) | 5′-ATGCACCTGCTGACATTCG-3′ |
| Reverse (5′-3′) | 5′-TATTGGCCTGGTTTACTGCCT-3′ | |
| Grin2b | Forward (5′-3′) | 5′-GCCATGAACGAGACTGACCC-3′ |
| Reverse (5′-3′) | 5′-GCTTCCTGGTCCGTGTCATC-3′ | |
| Grin2d | Forward (5′-3′) | 5′-GCTGCGAGACTATGGCTTCC-3′ |
| Reverse (5′-3′) | 5′-CCAGTGACGGGTTTACCAGAAA-3′ | |
| Kif1b | Forward (5′-3′) | 5′-GTCAATCGAATGAACGACCTGG-3′ |
| Reverse (5′-3′) | 5′-GCCGATGCAAAAAGTTGAACTG-3′ | |
| Kif1bp | Forward (5′-3′) | 5′-TCTTGACCCGACTGAGCATTT-3′ |
| Reverse (5′-3′) | 5′-ATAATGAGCGGCCTTCTCGAA-3′ | |
| Slc17a7 | Forward (5′-3′) | 5′-GGTGGAGGGGGTCACATAC-3′ |
| Reverse (5′-3′) | 5′-AGATCCCGAAGCTGCCATAGA-3′ | |
| Snap25 | Forward (5′-3′) | 5′-CAACTGGAACGCATTGAGGAA-3′ |
| Reverse (5′-3′) | 5′- GGCCACTACTCCATCCTGATTAT-3′ | |
| Stx1a | Forward (5′-3′) | 5′-CGCTGTCCCGAAAGTTTGTG-3′ |
| Reverse (5′-3′) | 5′-GTGTCTGGTCTCGATCTCACT-3′ | |
| Stxbp2 | Forward (5′-3′) | 5′-AAGGCGGTGGTAGGGGAAA-3′ |
| Reverse (5′-3′) | 5′-CAACAGGATGACAAGATTCGCA-3′ | |
| Stxbp4 | Forward (5′-3′) | 5′-ACAGGTCTAGGTCTGAAGATCC-3′ |
| Reverse (5′-3′) | 5′-CATCCTTGTAACAGTCACCTCC-3′ | |
| Stxbp6 | Forward (5′-3′) | 5′-CTCTTGATGAAAGAATGCTGGGA-3′ |
| Reverse (5′-3′) | 5′-TGACCTTCGTGATAGATGCCT-3′ | |
| Sv2b | Forward (5′-3′) | 5′-AGGTATCGGGACAACTATGAGG-3′ |
| Reverse (5′-3′) | 5′-GCCTTCTGTAACATCGCTCTGT-3′ | |
| Syn1 | Forward (5′-3′) | 5′-AATCACAAAGAGATGCTCAG-3′ |
| Reverse (5′-3′) | 5′-GGACACGCACATCATATTTAG-3′ | |
| Syp | Forward (5′-3′) | 5′-TGCCAACAAGACGGAGAGTG-3′ |
| Reverse (5′-3′) | 5′-TAGTGCCCCCTTTAACGCAG-3′ | |
| Syt1 | Forward (5′-3′) | 5′-ACCCTGGGCTCTGTATCCC-3′ |
| Reverse (5′-3′) | 5′- CCCTGACCACTGAGTGCAAA-3′ | |
| Unc13A | Forward (5′-3′) | 5′-GCTGTGCGTGGGAGTCAAA-3′ |
| Reverse (5′-3′) | 5′-CAGCTATGGTAGTGCTCTTCA-3′ | |
| Vamp2 | Forward (5′-3′) | 5′-ATCATCGTTTACTTCAGCAC-3′ |
| Reverse (5′-3′) | 5′-TGAAAGATATGGCTGAGAGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, H.; Bogdanov, P.; Simó, R.; Deàs-Just, A.; Hernández, C. Transcriptomic Analysis Reveals That Retinal Neuromodulation Is a Relevant Mechanism in the Neuroprotective Effect of Sitagliptin in an Experimental Model of Diabetic Retinopathy. Int. J. Mol. Sci. 2023, 24, 571. https://doi.org/10.3390/ijms24010571
Ramos H, Bogdanov P, Simó R, Deàs-Just A, Hernández C. Transcriptomic Analysis Reveals That Retinal Neuromodulation Is a Relevant Mechanism in the Neuroprotective Effect of Sitagliptin in an Experimental Model of Diabetic Retinopathy. International Journal of Molecular Sciences. 2023; 24(1):571. https://doi.org/10.3390/ijms24010571
Chicago/Turabian StyleRamos, Hugo, Patricia Bogdanov, Rafael Simó, Anna Deàs-Just, and Cristina Hernández. 2023. "Transcriptomic Analysis Reveals That Retinal Neuromodulation Is a Relevant Mechanism in the Neuroprotective Effect of Sitagliptin in an Experimental Model of Diabetic Retinopathy" International Journal of Molecular Sciences 24, no. 1: 571. https://doi.org/10.3390/ijms24010571
APA StyleRamos, H., Bogdanov, P., Simó, R., Deàs-Just, A., & Hernández, C. (2023). Transcriptomic Analysis Reveals That Retinal Neuromodulation Is a Relevant Mechanism in the Neuroprotective Effect of Sitagliptin in an Experimental Model of Diabetic Retinopathy. International Journal of Molecular Sciences, 24(1), 571. https://doi.org/10.3390/ijms24010571