MicroRNAs as Biomarkers for Coronary Artery Disease Related to Type 2 Diabetes Mellitus—From Pathogenesis to Potential Clinical Application
Abstract
:1. Introduction
2. Biology of MicroRNAs
3. Type 2 Diabetes Mellitus (T2DM), Atherosclerosis and Coronary Artery Disease (CAD)—The Vicious Circle Paradigm
3.1. The Role of MicroRNAs in the Initiation of T2DM-Associated Atherosclerosis
3.1.1. MicroRNAs in Chronic Hyperglycemia-Induced Endothelial Dysfunction
3.1.2. MicroRNAs in Diabetes-Associated Endothelial to Mesenchymal Transition
3.1.3. MicroRNAs in Monocyte Differentiation/Macrophage Activation under Diabetic Condition
3.2. The Role of MicroRNAs in the Progression of T2DM-Associated Atherosclerosis
3.2.1. MicroRNAs in Vascular Smooth Muscle Cell Proliferation and Migration under Diabetic Condition
3.2.2. MicroRNAs in Platelet Hyperactivity under Diabetic Condition
3.2.3. MicroRNAs in Diabetes-Associated Calcification
4. Clinical Research on Circulating MicroRNAs in T2DM and CAD
4.1. MicroRNAs as Potential Biomarkers for T2DM
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples | Ethnicity | Age [Years] | Gender (Male/Female, n) | BMI [kg/m2] | Duration of T2DM [Years] | HbA1c [%] | Value of Biomarker (AUC; 95% CI; SV [%]; SP [%]) | Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| let-7b-5p | Up | Serum | RT-qPCR | T2DM (29) HC (25) | Emirati | 55.6 ± 9.0 42.8 ± 12.7 | 13/16 9/16 | 31.5 ± 6.0 28.3 ± 6.5 | Newly diagnosed | 7.6 ± 1.6 5.2 ± 0.4 | N/A | Aljaibeji et al., 2022 [185] |
| miR-766-3p | Down | Serum | qPCR | T2DM (108) HC (68) | Chinese | 46.80 ± 18.43 46.56 ± 16.85 | 62/46 40/28 | 25.61 ± 6.59 24.14 ± 4.54 | Newly diagnosed | 9.60 ± 2.48 5.57 ± 0.81 | 0.880 88.9; 75.0 | Cao et al., 2022 [221] |
| miR-33a, miR-122 | Up | Whole blood | RT-qPCR | T2DM (50) HC (50) | Iranian | 55.9 ± 8.9 47.4 ± 9.2 | Only male | 27.0 ± 3.4 25.0 ± 3.3 | Diagnosed | N/A | N/A | Masoudi et al., 2022 [193] |
| miR-499 | Down | Serum | RT-qPCR | T2DM (60) HC (60) | Egyptian | Age-matched | Sex-matched | BMI-matched | N/A | N/D | 0.970 90.0; 96.6 | Oraby et al., 2022 [226] |
| miR-145-5p | Down | Plasma | RT-qPCR | T2DM (20) HC (20) | Iranian | 57.05 ± 1.99 51.07 ± 2.29 | N/A | 28.25 ± 0.95 26.94 ± 0.08 | Diagnosed | 8.15 ± 0.4 5.29 ± 0.06 | 0.77 (0.60–0.93) | Shahrokhi et al., 2022 [192] |
| miR-107 | Up | Serum | RT-qPCR | T2DM (53) HC (54) | Lithuanian | 65 (44–83) 62 (48–80) | 24/29 25/29 | 34.14 ± 5.92 28.07 ± 5.25 | 15 (5–30) – | 8.23 ± 2.14 5.46 ± 0.49 | N/A | Šimonienė et al., 2022 [242] |
| miR-21 | Up | Plasma | RT-qPCR | T2DM (24) HC (29) | Iranian | 54.42 ± 7.76 50.42 ± 6.14 | 15/9 19/10 | N/A | Newly diagnosed | 7.16 ± 0.16 5.15 ± 0.54 | 0.78 (0.64–0.92) 79.17; 81.48 | Yazdanpanah et al., 2022 [227] |
| miR-720 | Up | Whole blood | RT-qPCR | T2DM (50) HC (50) | Chinese | 57 ± 8.2 55 ± 7.8 | 24/26 27/23 | 26.2 ± 4.1 23.1 ± 3.8 | Newly diagnosed | 9.89 ± 2.74 3.21 ± 1.27 | N/A | Lu et al., 2021 [191] |
| miR-135a | Up | Saliva | RT-qPCR | T2DM (40) HC (40) | Iranian | 47 ± 1.6 46 ± 1.4 | 26/54 1 | 27.6 ± 1.3 26.4 ± 1.9 | Diagnosed | 7.6 ± 0.3 4.0 ± 0.2 | 0.007 95.0; 95.0 | Monfared et al., 2021 [200] |
| miR-126 | Down | 1 100.0; 100.0 | ||||||||||
| miR-33a-5p | Up | Plasma | RT-qPCR | T2DM (20) HC (20) | Iranian | 57.05 ± 1.99 51.07 ± 2.29 | 10/10 10/10 | 28.25 ± 0.95 26.94 ± 0.08 | Diagnosed | 8.15 ± 0.4 5.29 ± 0.06 | 0.71 (0.542–0.889) | Saeidi et al., 2021 [230] |
| miR-7-1-5p | Down | NS | ||||||||||
| miR-770-5p | Up | Serum | RT-qPCR | T2DM (20) HC (20) | Chinese | 32–61 29–64 | 14/8 14/8 | N/A | Newly diagnosed | N/A | N/A | Wang et al., 2021 [190] |
| miR-30a-5p, miR-126-3p, miR-182-5p, miR-1299 | Up | Whole blood | RT-qPCR | T2DM (92) HC (974) | South African | 58.15 ± 10.62 45.22 ± 15.3 | 19/73 286/688 | 31.5 ± 8.0 27.4 ± 7.8 | Newly diagnosed | 7.3 ± 1.9 5.6 ± 0.5 | N/A | Weale et al., 2021 [232] |
| miR-30a-5p, miR-30e-3p, miR-126-3p, miR-182-5p, miR-1299 | T2DM (188) HC (974) | 57.88 ± 11.97 45.22 ± 15.3 | 37/151 286/688 | 30.7 ± 6.4 27.4 ± 7.8 | Diagnosed | 8.9 ± 2.4 5.6 ± 0.5 | ||||||
| miR-126-3p | Up | Whole blood | RT-qPCR | T2DM (94) HC (972) | South African | 58.4 ± 10.6 45.2 ± 15.3 | 19/75 284/688 | 31.3 ± 8.0 27.4 ± 7.9 | Newly diagnosed | 7.4 5.6 | 0.646 (0.576–0.717) 55.6; 70.8 | Weale et al., 2021 [199] |
| miR-122 | Up | Whole blood | RT-qPCR | T2DM (30) HC (30) | Iranian | 53.03 ± 9.66 55.37 ± 8.47 | 15/15 15/15 | 30.27 ± 3.11 29.80 ± 2.89 | Diagnosed | 7.29 ± 1.22 4.54 ± 0.20 | N/A | Zeinali et al., 2021 [194] |
| miR-126-3p, miR-146a | Down | |||||||||||
| miR-29, miR-155 | Up | Serum | qPCR | T2DM (59) HC (72) | Xinjiang Uygurian | 48.45 ± 7.36 44.56 ± 3.58 | 27/32 36/36 | 28.50 ± 4.69 21.94 ± 1.33 | Diagnosed | N/A | N/A | Zhu et al., 2021 [189] |
| miR-330 | Up | Serum | RT-qPCR | T2DM (100) HC (100) | Indian | > 50 (40.0%) > 50 (50.0%) | 57/43 55/45 | > 25 (34.0%) > 25 (15.0%) | Newly diagnosed | N/A | N/A | Ali Beg et al., 2020 [188] |
| let-7f-5p, miR-24-3p, miR-214-3p | Down | Whole blood | miSript miRNA PCR array, RT-qPCR | T2DM (40) HC (16) | Greek | 59 (35–75) 45 (19–52) | 19/21 7/9 | 29.3 (21.5–46.5) 24 (21.3–24.0) | 5 (0–26) – | 6.7 (5.2–12.1) – | N/A | Avgeris et al., 2020 [175] |
| miR-34a | Up | Plasma | RT-qPCR | T2DM (30) HC (30) | Indian | 38.9 ± 5.8 40.6 ± 5.95 | 19/11 17/13 | 27.8 ± 6.29 23.33 ± 3.57 | 4.55 ± 4.3 – | 7.51 ± 1.22 4.89 ± 0.29 | N/A | Banerjee et al., 2020 [235] |
| miR-126-5p, miR-181b | Down | Whole blood | RT-qPCR | T2DM (30) HC (30) | Iranian | 55.4 ± 5.3 53.5 ± 7.2 | 14/16 16/14 | N/A | Newly diagnosed | 8.62 ± 1.74 5.1 ± 0.24 | N/A | Dehghani et al., 2020 [207] |
| miR-103a | Up | Plasma | RT-qPCR | T2DM (48) HC (50) | Han Chinese | 52.6 ± 9.13 45.62 ± 8.58 | 26/22 26/24 | 25.52 ± 2.89 24.56 ± 3.40 | Newly diagnosed | 9.16 ± 1.05 5.15 ± 0.32 | 0.998 (0.993–1.0) 97.9; 98.0 | Luo et al., 2020 [225] |
| miR-103b | Down | 0.964 (0.920–1.0) 98.0; 91.7 | ||||||||||
| miR-135 | Up | Plasma | RT-qPCR | T2DM (40) HC (40) | Iranian | 53.69 ± 5.69 33.59 ± 7.58 | N/A | 30.11 ± 1.01 25.23 ± 2.43 | Newly diagnosed | 7.63 ± 0.41 4.70 ± 2.30 | N/A 50.5; 91.2 | Monfared et al., 2020 [229] |
| miR-222 | Up | Plasma | RT-qPCR | T2DM (30) HC (30) | Iranian | 52.42 ± 8.77 51.44 ± 6.04 | 20/10 21/9 | 28.17 ± 5.46 27.60 ± 3.9 | Newly diagnosed | 7.34 ± 1.08 5.76 ± 0.41 | N/A | Sadeghzadeh et al., 2020 [239] |
| miR-15a | Down | |||||||||||
| miR-19a, miR-130a, miR-148b, miR-223 | Up | Serum | RT-qPCR | T2DM (102) HC (68) | Mongolian (Chinese) | N/A | N/A | N/A | Newly diagnosed | N/A | N/A | Yan et al., 2020 [238] |
| let-7b-5p, miR-1-3p, miR-24-3p, miR-34a-5p, miR-98-5p, 133a-3p | Down | Whole blood | qPCR | T2DM (40) HC (37) | Greek | 59 (35–75) 49 (19–69) | 19/21 19/18 | 29.3 (21.5–46.5) 26.9 (21.3–36.3) | 5 (0–26) – | 6.7 (5.2–12.1) 5.6 (5.0–6.1) | N/A | Kokkinopoulou et al., 2019 [234] |
| miR-21 2 | Up | Plasma | RT-qPCR | T2DM (27) HC (44) | Italian | 61.69 ± 7.59 59.3 ± 9.82 | 10/17 15/29 | 29.26 ± 5.83 25.11 ± 3.32 | Newly diagnosed | 6.64 ± 0.6 5.80 ± 0.38 | 0.699 93.0; 35.0 | La Sala et al., 2019 [228] |
| miR-30c | Down | Plasma | qPCR | T2DM (47) HC (32) | Han Chinese | 60.5 ± 11.1 58.6 ± 8.1 | 23/24 17/15 | 24.76 ± 3.29 24.49 ± 2.30 | Newly diagnosed | 9.15 ± 1.02 5.36 ± 0.35 | 0.916 (0.853–0.980) 87.9; 87.2 | Luo et al., 2019 [224] |
| miR-486-3p | Up | Plasma | RT-qPCR | T2DM (29) HC (30) | Israeli Arab/Jewish | 64 ± 10 31 ± 11 | 18/11 15/15 | 30 ± 5 25 ± 4 | Newly diagnosed | 7.7 ± 1.9 5.1 ± 0.3 | N/A | Meerson et al., 2019 [240] |
| miR-423 | Down | |||||||||||
| miR-342 | Up | Serum | RT-qPCR | T2DM (50) HC (50) | Egyptian | 62.06 ± 1.26 62.22 ± 0.69 | Only female | 27.58 ± 0.28 23.82 ± 0.14 | 12.06 ± 0.30 – | 10.75 ± 0.17 4.10 ± 0.68 | N/A | Seleem et al., 2019 [233] |
| miR-450 | Down | |||||||||||
| miR-3666 | Up | Serum | qPCR | T2DM (60) HC (30) | Chinese | 45.81 ± 5.92 N/A | 36/24 N/A | 25.12 ± 0.31 N/A | Diagnosed | N/A | N/A | Tan et al., 2019 [214] |
| miR-146a | Down | Plasma, PBMC | RT-qPCR | T2DM (30) HC (30) | Iranian | 57 (48–61) 50.5 (45.75–61) | 11/19 9/21 | 27.13 ± 4.15 27.22 ± 3.26 | 8.53 ± 1.29 – | 7.4 (6.7–8) 5.1 (5–5.4) | N/A | Alipoor et al., 2018 [206] |
| miR-9, miR-375 | Up | Whole blood | RT-qPCR | T2DM (30) HC (30) | Bahrainis | 60 ± 12 56 ± 5.1 | 12/18 14/16 | 25.7 ± 5.2 24.2 ± 4.6 | 15 ± 4.4 – | 8.68 ± 2.6 5.03 ± 0.7 | 0.783 (0.665–0.902) 3 | Al-Muhtaresh et al., 2018 [231] |
| miR-210 | Up | Plasma | RT-qPCR | T2DM (54) HC (20) | Egyptian | 56.5 ± 7.7 58.1 ± 1.1 | 29/25 11/9 | 30.7 ± 5.3 23.2 ± 0.2 | 10.8 ± 7.8 – | 8.3 ± 1.1 4.8 ± 0.4 | 0.950 87.0; 100.0 | Amr et al., 2018 [223] |
| miR-126 | Down | 0.960 96.3; 95.0 | ||||||||||
| let-7b 3, miR-29a, miR-144 3 | Up | Plasma | Microarray, RT-qPCR | T2DM (112) HC (94) | Han Chinese | 54.75 ± 7.53 52.84 ± 8.85 | 69/43 53/41 | 27.11 ± 3.17 23.86 ± 3.27 | Newly diagnosed | 7.58 ± 1.54 5.16 ± 0.39 | 0.871 (0.822–0.919) 3 79.5; 81.9 | Liang et al., 2018 [173] |
| miR-142 3 | Down | |||||||||||
| let-7e-5p, let-7f-5p, miR-15b-5p, miR-99b-5p, miR-103a-3p | Up | Whole blood | sRNA-Seq, RT-qPCR | T2DM (12) HC (12) | South African | 54.8 ± 7.5 52.1 ± 7.8 | Only female | 33.5 ± 8.9 27.3 ± 5.8 | Newly diagnosed | N/A | N/A | Matsha et al., 2018 [177] |
| miR-30d | Up | Plasma | RT-qPCR | T2DM (30) HC (30) | Asian Indian | 50.5 ± 6.3 42.1 ± 7.8 | 21/9 21/9 | 27.3 ± 4.6 27.3 ± 4.7 | 3.10 ± 0.99 – | 8.4 ± 2.0 5.5 ± 0.4 | N/A | Sucharita et al., 2018 [187] |
| miR-126 | Down | Whole blood | RT-qPCR | T2DM (45) HC (45) | Bahrainis | 61 ± 12 53 ± 8.6 | 23/22 21/24 | 25.4 ± 4.8 24 ± 4.5 | 16 ± 6 – | 7.4 ± 8.3 3.64 ± 1.1 | 0.932 (0.858–1.000) | Al-Kafaji et al., 2017 [198] |
| miR-148a-3p | Up | Plasma | RT-qPCR | T2DM (9) HC (9) | Italian | 60.2 ± 8.0 57.9 ± 8.9 | 2/7 4/5 | 29.6 ± 7.8 23.7 ± 3.3 | Newly diagnosed | 6.4 ± 2.7 5.5 ± 2.4 | N/A | de Candia et al., 2017 [220] |
| miR-222-3p, miR-342-3p | Down | |||||||||||
| miR-26b, miR-126, miR-140, miR-223 | Down | Plasma, Platelet | RT-qPCR | T2DM (28) HC (23) | Hungarian | 53 (50–59) 53 (34–60) | 15/13 12/11 | 32.9 (30.3–40.2) 24 (22.1–25.9) | 10 (8.0–14.5) – | 7.5 (7.0–8.8) – | N/A | Fejes et al., 2017 [147] |
| miR-126-3p | Down | Plasma (MPs) | RT-qPCR | T2DM (68) HC (53) | Italian | 60 ± 1 57 ± 1 | 42/26 30/23 | 30 ± 1.6 25 ± 0.4 | >5 – | N/A | N/A | Giannella et al., 2017 [205] |
| miR-223-3p | Down | PBMC | RT-qPCR | T2DM (16) HC (18) | Han Chinese | 57 ± 9 53 ± 11 | 8/8 12/6 | N/A | Newly diagnosed | N/A | N/A | Long et al., 2017 [237] |
| miR-217 | Up | Serum | qPCR | T2DM (186) HC (195) | Chinese | 54.87 ± 11.65 54.12 ± 9.45 | 95/91 99/96 | 25.30 ± 3.11 25.10 ± 3.27 | 6.39 ± 6.31 – | 8.10 ± 2.09 5.36 ± 0.32 | N/A | Shao et al., 2017 [219] |
| miR-34a, miR-125b | Up | PBMC | RT-qPCR | T2DM (73) HC (52) | Chinese | 56.81 ± 11.85 Age-matched | 38/35 Sex-matched | N/A | 4.54 ± 5.41 – | 8.50 ± 2.09 5.82 ± 1.07 | N/A | Shen et al., 2017 [236] |
| miR-7 | Up | Serum | RT-qPCR | T2DM (76) HC (74) | Chinese | 48.5 ± 14.5 48.8 ± 15.2 | 50/26 41/33 | 25.2 ± 3.7 23.0 ± 2.7 | 1.8 ± 2.6 – | 9.9 ± 2.9 5.3 ± 0.4 | 0.76 (0.68–0.83) | Wan et al., 2017 [218] |
| Serum (exosome-free) | 0.75 (0.67–0.83) | |||||||||||
| miR-18a | Up | PBMC | RT-qPCR | T2DM (117) HC (105) | Chinese | 51.68 ± 8.77 49.26 ± 9.09 | 68/49 58/47 | 27.44 ± 3.08 24.18 ± 2.86 | Newly diagnosed | 7.51 ± 1.42 5.19 ± 0.42 | 0.851 (0.800–0.902) 3 78.6; 80.0 | Wang et al., 2017 [212] |
| miR-34c | Down | |||||||||||
| miR-96-5p, miR-144-3p, miR-454-3p, miR-455-5p | Up | Serum | miRNA qPCR array, RT-qPCR | T2DM (10) HC (5) | Chinese | 58.2 ± 7.7 56.4 ± 3.7 | 4/6 2/3 | N/A | Newly diagnosed | N/A | N/A | Yang et al., 2017 [176] |
| miR-409-3p, miR-665, miR-766-3p | Down | |||||||||||
| miR-574-3p | Down | Serum | RT-qPCR | T2DM (64) HC (44) | Ecuadorian | 61 (37–85) 53 (32–87) | 24/40 13/31 | 29.5 (22–49) 28.7 (23–42) | Diagnosed | 7.0 (3.2–12.5) 5.6 (3.9–6.9) | N/A | Baldeón et al., 2016 [211] |
| miR-451a, miR-4534 | Up | Serum | Microarray, RT-qPCR | T2DM (154) HC (69) | Chinese | 61.1 ± 12.4 54.2 ± 10.7 | 70/84 30/39 | N/A | Newly diagnosed | N/A | N/A | Ding et al., 2016 [178] |
| miR-320d, miR-572, miR-3960 | Down | |||||||||||
| miR-221, miR-222 | Up | Serum | RT-qPCR | T2DM (30) HC (20) | Chinese | 60.79 ± 11.11 59.78 ± 11.23 | Only female | 28.88 ± 1.18 20.12 ± 1.69 | Newly diagnosed | 7.60 ± 0.33 4.56 ± 0.45 | N/A | Li et al., 2016 [217] |
| miR-30c | Down | Plasma, Platelet | RT-qPCR | T2DM (40) HC (50) | Han Chinese | 58.6 ± 6.2 52.2 ± 5.5 | 21/29 31/19 | 27.3 ± 4.2 23.6 ± 2.8 | Diagnosed | 7.3 ± 0.5 5.3 ± 0.2 | N/A | Luo et al., 2016 [151] |
| miR-21, miR-30d 3, miR-34a 3, miR-148a | Up | Plasma | RT-qPCR | T2DM (31) HC (27) | American | 52.9 ± 2.0 25.3 ± 2.2 | 15/16 15/12 | 34.1 ± 1.3 24.1 ± 0.9 | Diagnosed | 6.56 ± 0.11 5.24 ± 0.06 | 0.928 3 90.32; 88.89 | Seyhan et al., 2016 [216] |
| miR-571, miR-661, miR-770-5p, miR-892b, miR-1303 4 | Up | Serum | TLDA, RT-qPCR | T2DM (92) HC (92) | Chinese | 47.7 ± 13.9 50.2 ± 14.2 | 58/34 56/36 | 25.6 ± 4.5 23.6 ± 2.0 | 2.1 ± 2.7 – | 9.8 ± 2.9 5.3 ± 0.4 | 0.71 (0.64–0.79) 3 | Wang et al., 2016 [166] |
| miR-125b, miR-126, miR-221 5 | N/A | |||||||||||
| miR-572 | Up | Plasma | Microarray, RT-qPCR | T2DM (50) HC (50) | Han Chinese | 46.22 ± 6.90 45.52 ± 6.22 | 27/23 22/28 | 25.41 ± 0.32 25.36 ± 0.38 | Newly diagnosed | 8.69 ± 0.36 5.41 ± 0.29 | 0.843 (0.766–0.920) 87.8; 71.4 | Yan et al., 2016 [179] |
| miR-320b | Down | 0.946 (0.906–0.985) 92.0; 85.7 | ||||||||||
| miR-1249 | 0.784 (0.685–0.883) 86.0; 77.55 | |||||||||||
| miR-15a | Down | Whole blood | RT-qPCR | T2DM (24) HC (24) | Bahrainis | 52 ± 6.0 49 ± 9.1 | 10/14 13/11 | 25.3 ± 1.8 24.2 ± 1.0 | Diagnosed | 7.5 ± 0.8 4.8 ± 0.6 | 0.864 (0.751–0.977) | Al Kafaji et al., 2015 [222] |
| miR-34c-5p, miR-576-3p | Up | PBMC | Microarray, RT-qPCR | T2DM (64) HC (44) | Ecuadorian | 61 (37–85) 53 (32–87) | 24/40 13/31 | 29.5 (22–49) 28.7 (23–42) | Diagnosed | 7.0 (3.2–12.5) 5.6 (3.9–6.9) | N/A | Baldeón et al., 2015 [181] |
| miR-185 | Down | Plasma | qPCR | T2DM (34) HC (30) | Mongolian (Chinese) | N/A | N/A | N/A | Diagnosed | N/A | N/A | Bao et al., 2015, [215] |
| miR-142, miR-143, miR-155, miR-223 | Down | Platelet | RT-qPCR | T2DM (22) HC (22) | German | 45.7 ± 3.1 41.6 ± 7.5 | 10/12 10/12 | N/A | Diagnosed | 9.01 ± 0.37 4.98 ± 0.58 | N/A | Elgheznawy et al., 2015 [148] |
| miR-101, miR-375, miR-802 | Up | Serum | sRNA-Seq (mice), RT-qPCR | T2DM (155) HC (49) | Japanese | 62.3 ± 13.2 46.0 ± 9.67 | 96/59 25/24 | 25.9 ± 4.97 23.6 ± 4.05 | Diagnosed | 7.31 ± 1.08 6.03 ± 0.39 | N/A | Higuchi et al., 2015 [167] |
| miR-10b, miR-130a, miR-143 | Down | Whole blood | Microarray, RT-qPCR | T2DM (12) HC (24) | Xinjiang Uygurian | 56 ± 10 49 ± 13 | N/A | 30.9 ± 5.8 26.3 ± 3.6 | Diagnosed | N/A | N/A | Jiao et al., 2015 [182] |
| miR-146a 6 | Down | PBMC | qPCR | T2DM (35) HC (35) | Indian | 47.3 ± 7 44.7 ± 6 | 19/16 17/18 | 24.6 ± 2 23.9 ± 2 | Diagnosed | 7.8 ± 1.5 5.5 ± 0.4 | N/A | Lenin et al., 2015 [204] |
| miR-103b | Down | Platelet | RT-qPCR | T2DM (43) HC (46) | Han Chinese | 59 ± 9.3 51.4 ± 9.4 | 19/24 17/29 | 23.3 ± 5.4 21.9 ± 2.9 | Newly diagnosed | 7.0 ± 1.3 5.1 ± 0.5 | N/A | Luo et al., 2015 [210] |
| miR-155 | Down | PBMC | RT-qPCR | T2DM (20) HC (20) | Iranian | 46.5 ± 5.8 47.5 ± 4.4 | 10/10 10/10 | 28.7 ± 4.9 26.2 ± 4.0 | Diagnosed | 7.02 ± 0.5 5.7 ± 0.7 | N/A | Mazloom et al., 2015 [209] |
| miR-21-5p, miR-126-3p | Down | Plasma | RT-qPCR | T2DM (76) HC (107) | Italian | 65.56 ± 6.96 64.25 ± 7.56 | 36/40 49/58 | 28.47 ± 4.34 26.67 ± 5.4 | Diagnosed | 7.34 ± 1.28 5.96 ± 0.41 | N/A | Olivieri et al., 2015 [203] |
| miR-130b-3p, miR-374a-5p | Up | Serum | miRNA PCR assay, RT-qPCR | T2DM (49) HC (49) | Asian Indian | 44.4 ± 8.1 44.3 ± 6.9 | 25/24 26/23 | 25.7 ± 3.5 24.5 ± 2.6 | Newly diagnosed | 7.8 ± 1.6 5.6 ± 0.4 | N/A | Prabu et al., 2015 [168] |
| miR-126 | Down | Plasma | qPCR | T2DM (20) HC (20) | Han Chinese | 61.20 ± 10.62 57.25 ± 9.64 | 13/7 9/11 | 24.53 ± 2.87 23.90 ± 2.34 | Newly diagnosed | N/A | 0.806 77.78; 66.67 | Zhang et al., 2015 [197] |
| miR-146a | Down | Serum | RT-qPCR | T2DM (56) HC (40) | Ecuadorian | 62 (38–85) 54 (32–87) | 22/34 12/28 | 29.2 (22–39) 29.3 (23–42) | Diagnosed | 7.1 (4.8–12.5) 5.7 (3.9–6.7) | N/A | Baldeón et al., 2014 [202] |
| miR-126 | Down | Serum | RT-qPCR | T2DM (160) HC (138) | Chinese | 50.2 ± 6.7 46.7 ± 7.2 | 78/82 67/71 | 23.32 ± 0.31 22.87 ± 0.32 | Newly diagnosed | 9.16 ± 1.64 4.69 ± 0.57 | 0.792 (0.707–0.877) | Liu et al., 2014 [196] |
| miR-140-5p, miR-142-3p 3, miR-222 | Up | Plasma | Microarray, RT-qPCR | T2DM (48) HC (45) | Spanish | 54 ± 10 7a 57.7 ± 8 7b 48.1 ± 10.1 8a 50.6 ± 14.4 8b | Only male | 26.4 ± 2.4 7a 33.4 ± 3.3 7b 25.2 ± 1.8 8a 32.2 ± 2.4 8b | Diagnosed | 7.67 ± 1.46 7a 7.06 ± 2.14 7b 4.73 ± 0.35 8a 4.81 ± 0.33 8b | 0.975 3 | Ortega et al., 2014 [169] |
| miR-125b, miR-126 3, miR-130b, miR-192, miR-195 3, miR-423-5p 3, miR-532-5p | Down | |||||||||||
| miR-326 | Up | Plasma (exosomes) | Microarray, RT-qPCR | T2DM (18) HC (12) | Italian | 57.2 ± 9.6 49.5 ± 12.4 | 12/6 6/6 | 31.6 ± 5.1 32.9 ± 5.4 | Newly diagnosed | 9.6 ± 1.5 5.7 ± 0.5 | 0.912 (0.799–1.000) 3 | Santovito et al., 2014 [170] |
| let-7a, let-7f | Down | |||||||||||
| miR-375 | Up | Plasma | RT-qPCR | T2DM (100) HC (100) | Chinese Kazak | 51.33 ± 11.75 48.55 ± 12.41 | 54/46 44/56 | 26.30 ± 4.08 24.44 ± 4.63 | Diagnosed | N/A | N/A | Sun et al., 2014 [184] |
| miR-199a | Up | Plasma | RT-qPCR | T2DM (64) HC (64) | Han Chinese | 46–62 | N/A | N/A | Newly diagnosed | N/A | N/A | Yan et al., 2014 [186] |
| miR-23a | Down | Serum | Solexa sequencing, RT-qPCR | T2DM (24) HC (20) | Han Chinese | 51.13 ± 9.21 46.65 ± 16.18 | 16/8 8/12 | 25.27 ± 2.90 25.55 ± 5.27 | Newly diagnosed | 9.49 ± 2.45 5.98 ± 0.80 | 0.835 (0.717–0.954) 79.2; 75.0 | Yang et al., 2014 [171] |
| miR-486 | 0.698 (0.540–0.856) 79.2; 60.0 | |||||||||||
| let-7i | 0.771 (0.629–0.913) 75.0; 70.0 | |||||||||||
| miR-96, miR-146a, miR-186, miR-191, miR-192 | N/A | |||||||||||
| miR-146a, miR-155 | Down | PBMC | RT-qPCR | T2DM (20) HC (20) | Méxican | 40–60 18–28 | 11/9 11/9 | 31.9 ± 7.4 23.1 ± 2.5 | 0–20 | 7.9 ± 1.7 4.8 ± 0.7 | N/A | Corral-Fernández et al., 2013 [213] |
| miR-146a | Up | Plasma | RT-qPCR | T2DM (90) HC (90) | Han Chinese | 48.5 (42.0–56.0) 48.00 (41.8–55.0) | 47/43 47/43 | 24.58 ± 3.66 23.38 ± 2.95 | Newly diagnosed | N/A | 0.725 (0.651–0.799) | Rong et al., 2013 [195] |
| miR-126 | Down | Plasma | qPCR | T2DM (30) HC (30) | Han Chinese | 63 ± 8.6 61 ± 9 | 16/14 16/14 | N/A | Newly diagnosed | N/A | N/A | Zhang et al., 2013 [201] |
| miR-27a, miR-150, miR-192, miR-320a | Up | Whole blood | Microarray, RT-qPCR | T2DM (29) HC (29) | Singaporean | 44.2 ± 8.4 45.7 ± 11.3 | N/A | 26.5 ± 5.9 23.7 ± 3.2 | Newly diagnosed | N/A | N/A | Karolina et al., 2012 [172] |
| miR-17, miR-92a, miR-130a, miR-195, miR-197, miR-509-5p, miR-652 | Down | |||||||||||
| miR-146a | Down | PBMC | qPCR | T2DM (20) HC (20) | Indian | 43.7 ± 5.1 42.0 ± 4.7 | N/A | 26.4 ± 3.7 25.8 ± 4.0 | Diagnosed | 7.9 ± 1.8 5.5 ± 0.2 | N/A | Balasubramanyam et al., 2011 [208] |
| miR-29a, miR-144, miR-150, miR-192, miR-320 | Up | Whole blood | Microarray, RT-qPCR | T2DM (8) HC (7) | Singaporean | 46.7 ± 3.4 46.3 ± 7.5 | Only male | 24.5 ± 1.1 22.4 ± 2.3 | Diagnosed | N/A | N/A | Karolina et al., 2011 [180] |
| miR-30d, miR-146a, miR-182 | Down | |||||||||||
| miR-29a, miR-144, miR-150, miR-192, miR-320 | Up | T2DM (13) HC (8) | 41.0 ± 12.1 43.3 ± 5.7 | 28.0 ± 4.9 24.4 ± 3.1 | Newly diagnosed | |||||||
| miR-30d, miR-146a, miR-182 | Down | |||||||||||
| miR-9, miR-29a, miR-30d, miR-34a, miR-124a, miR-146a, miR-375 | Up | Serum | RT-qPCR | T2DM (18) HC (19) | Han Chinese | 47.33 ± 2.62 41.00 ± 2.62 | 9/9 12/7 | 26.26 ± 0.79 26.63 ± 0.80 | Newly diagnosed | N/A | N/A | Kong et al., 2011 [183] |
| miR-28-3p | Up | Plasma | Microarray, RT-qPCR | T2DM (80) HC (80) | Italian (Bruneck cohort) | 66.3 ± 8.9 66.3 ± 8.9 | 30/50 30/50 | 28.0 ± 4.4 25.0 ± 4.0 | Diagnosed | 6.5 ± 1.4 5.4 ± 0.3 | N/A | Zampetaki et al., 2010 [174] |
| miR-15a, miR-20b, miR-21, miR-24, miR-29b, miR-126, miR-150, miR-191, miR-197, miR-223, miR-320, miR-486 | Down |
4.2. MicroRNAs as Potential Biomarkers for CAD
4.3. MicroRNAs as Potential Biomarkers for CAD Related to T2DM
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples | Ethnicity | Age [Years] | Gender (Male/Female, n) | BMI [kg/m2] | Duration of T2DM [Years] | HbA1c [%] | Value of Biomarker AUC (95% CI); SV [%]; SP [%] | Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-499 | Down | Serum | RT-qPCR | T2DM+CAD (60) T2DM (60) HC (60) | Egyptian | Age-matched | Sex-matched | BMI-matched | N/A | N/D | 0.720 73.0; 70.0 | Oraby et al., 2022 [226] |
| miR-19a-3p | Up | Plasma (EVs) | sRNA-Seq, RT-qPCR | T2DM+CAD (32) HC (20) | Chinese | Age-matched | Sex-matched | N/A | N/A | N/D | 0.698 (0.530–0.866) 2 53.9; 85.7 | Zhang et al., 2022 [303] |
| miR-15a-3p | Down | 0.874 (0.765–0.982) 2 88.5; 71.4 | ||||||||||
| miR-18a-5p | 0.871 (0.760–0.982) 2 80.8; 78.6 | |||||||||||
| miR-133a-3p | 0.745 (0.567–0.922) 2 88.5; 57.1 | |||||||||||
| miR-155-5p | 0.901 (0.800–1.000) 2 92.3; 78.6 | |||||||||||
| miR-210-3p | 0.786 (0.647–0.925) 2 61.5; 92.9 | |||||||||||
| miR-1, miR-133 | Up | Whole blood | RT-qPCR | T2DM+CAD (30) T2DM (30) HC (30) | Bahrainis | 60 ± 12 58 ± 11.5 56 ± 5.1 | 15/15 12/18 14/16 | 25.35 ± 4.4 25.7 ± 5.2 24.2 ± 4.6 | 15 ± 4.4 14 ± 9.3 – | 8.68 ± 2.6 7.09 ± 1.06 5.03 ± 0.7 | 0.752 (0.626–0.879) 1,3 | Al-Muhtaresh et al., 2019 [301] |
| miR-1 | 0.912 (0.828–0.995) 2 | |||||||||||
| miR-133 | 0.920 (0.842–0.998) 2 | |||||||||||
| miR-30c | Down | Plasma | qPCR | T2DM+CAD (27) T2DM (47) CAD (34) HC (32) | Han Chinese | 64.5 ± 6.5 60.5 ± 11.1 60.9 ± 5.3 58.6 ± 8.1 | 17/10 23/24 18/16 17/15 | 25.02 ± 3.12 24.76 ± 3.29 24.57 ± 3.01 24.49 ± 2.30 | Newly diagnosed | 9.13 ± 1.01 9.15 ± 1.02 6.28 ± 0.69 5.36 ± 0.35 | 0.474 (0.355–0.593) 1 70.2; 52.0 0.972 (0.940–1.000) 2 90.9; 85.2 | Luo et al., 2019 [224] |
| miR-342 | Up | Serum | RT-qPCR | T2DM+CAD (50) T2DM (50) CAD (50) HC (50) | Egyptian | 62.30 ± 0.61 62.06 ± 1.26 62.32 ± 0.56 62.22 ± 0.69 | Only female | 28.87 ± 0.33 27.58 ± 0.28 27.88 ± 0.23 23.82 ± 0.14 | 12.06 ± 0.30 12.06 ± 0.30 – – | 11.92 ± 0.18 10.75 ± 0.17 9.73 ± 0.17 4.10 ± 0.68 | 0.781 1 80.0; 72.0 | Seleem et al., 2019 [233] |
| miR-450 | Down | 0.824 1 72.0; 78.0 | ||||||||||
| miR-92a | Up | Serum | RT-qPCR | T2DM+CAD (117) T2DM (69) HC (68) | Chinese | 64.73 ± 8.22 64.29 ± 3.77 62.98 ± 7.42 | 79/38 48/21 45/23 | 26.44 ± 3.31 25.61 ± 5.76 24.08 ± 2.42 | N/A | 8.22 ± 2.64 6.80 ± 2.41 5.29 ± 0.33 | 0.866 1 76.9; 88.4 0.958 2 78.6; 98.5 | Wang et al., 2019 [300] |
| miR-210 | Up | Plasma | RT-qPCR | T2DM+CAD (46) T2DM (54) HC (20) | Egyptian | 57.0 ± 6.2 56.5 ± 7.7 58.1 ± 1.1 | 23/23 29/25 11/9 | 29.7 ± 3.5 30.7 ± 5.3 23.2 ± 0.2 | 11.2 ± 5.2 10.8 ± 7.8 – | 9.4 ± 1.0 8.3 ± 1.1 4.8 ± 0.4 | 0.980 1 93.5; 100.0 0.980 2 97.8; 100.0 | Amr et al., 2018 [223] |
| miR-126 | Down | 0.970 1 91.3; 100.0 0.980 2 97.8; 95.0 | ||||||||||
| miR-126 | Down | Whole blood | RT-qPCR | T2DM+CAD (45) T2DM (45) HC (45) | Bahrainis | 64 ± 11.7 61 ± 12 53 ± 8.6 | 24/21 23/22 21/24 | 26.1 ± 4.3 25.4 ± 4.8 24 ± 4.5 | 18 ± 9.3 16 ± 6 – | 9.6 ± 3.2 7.4 ± 8.3 3.64 ± 1.1 | 0.807 (0.714–0.900) 1 0.948 (0.894–1.000) 2 | Al-Kafaji et al., 2017 [198] |
| miR-9 4, miR-370 | Up | Serum | RT-qPCR | T2DM+CAD (50) T2DM (50) CAD (50) HC (50) | Egyptian | 62.30 ± 0.45 62.06 ± 1.26 62.32 ± 0.56 62.22 ± 0.69 | 35/15 32/18 38/12 36/14 | 28.87 ± 0.32 27.58 ± 0.27 27.88 ± 0.23 23.82 ± 0.13 | 12.06 ± 0.30 12.22 ± 0.30 – – | 11.0–12.5 5 | N/A 84.0; 84.0 3 | Motawae et al., 2015 [302] |
5. MicroRNAs as Potential Therapeutic Targets in T2DM and CAD
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABCA1 | ATP-binding cassette: sub-family A, member 1 |
| ACS | acute coronary syndrome |
| ADAM10 | a disintegrin and metalloproteinase domain-containing protein 10 |
| AGEs | advanced-glycation end-products |
| AGO | Argonaute |
| Agr-1 | argininase-1 |
| AMPK | adenosine monophosphate-activated protein kinase |
| ApoA-1 | apolipoprotein A-1 |
| AUC | area under the curve |
| BMI | body mass index |
| CAC | coronary artery calcification |
| CAD | coronary artery disease |
| CAV-1 | caveolin 1 |
| CCL-2 | chemokine CC-motif ligand 2 |
| CCS | chronic coronary syndrome |
| CI | confidence interval |
| circRNA | circular RNA |
| COX | cyclooxygenase |
| DAXX | death-domain associated protein |
| DGCR8 | DiGeorge syndrome Critical Region 8 |
| EC | endothelial cell |
| ECM | extracellular matrix |
| EDCF | endothelium-derived cyclooxygenase-dependent contracting factor |
| EMP2 | epithelial membrane protein 2 |
| EndMT | endothelial-to-mesenchymal transition |
| eNOS | endothelial nitric oxide synthase |
| EPC | endothelial progenitor cell |
| ERK | extracellular signal-regulated kinase |
| ET-1 | endothelin-1 |
| EVs | extracellular vesicles |
| EXP5 | Exportin-5 |
| FSP-1 | fibroblast specific protein-1 |
| GLUT-4 | glucose transporter-4 |
| GS | Gensini score |
| HbA1c | glycated hemoglobin A1c |
| HDL-C | high-density lipoprotein cholesterol |
| HMGB1 | high mobility group box-1 |
| HSC70 | heat shock cognate 70 |
| HSP90 | heat shock protein 90 |
| ICAM-1 | intercellular adhesion molecule-1 |
| IGF-1R | insulin-like growth factor-1 receptor |
| IFN-γ | interferon-γ |
| IL | interleukin |
| IRAK1 | interleukin 1 receptor associated kinase 1 |
| IRS-1 | insulin receptor substrate 1 |
| JNK | c-Jun N-terminal kinase |
| KLF | Krüppel-like factor |
| KRAS | Kirsten Rat Sarcoma Viral Oncogene Homolog |
| LDL-C | low-density lipoprotein cholesterol |
| LNA | locked nucleic acid |
| lncRNA | long non-coding RNA |
| α-SMA | alpha smooth muscle actin |
| MAPK | mitogen-activated protein kinase |
| MCP-1 | monocyte chemoattractant protein-1 |
| MEK | mitogen-activated protein kinase |
| MGO | methylglyoxal |
| MPO | myeloperoxidase |
| mRNA | messenger RNA |
| miRNA, miR | microRNA |
| mTOR | mammalian target of rapamycin |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| ncRNA | non-coding RNA |
| NF-κB | nuclear factor-kappa B |
| NGS | next-generation sequencing |
| NO | nitric oxide |
| NPM1 | nucleophosmin 1 |
| OR | odds ratio |
| ox-LDL | oxidized low-density lipoprotein |
| PAI-1 | plasminogen activator inhibitor-1 |
| PBMC | peripheral blood mononuclear cell |
| PDGF | platelet-derived growth factor |
| PDGFRβ | platelet-derived growth factor receptor beta |
| PECAM-1 | platelet endothelial cell adhesion molecule-1 |
| PHLPP2 | PH domain leucine-rich repeat protein phosphatase 2 |
| PI3K | phosphatidylinositol 3-kinase |
| piRNA | PIWI-interacting RNA |
| PKC | protein kinase C |
| pre-miRNA | precursor miRNA |
| pri-miRNA | primary miRNA |
| RAGE | receptor for advanced glycation end-products |
| RISC | RNA-induced silencing complex |
| RNA pol II | RNA polymerase II |
| ROC | receiver operating characteristic |
| ROCK1 | Rho-associated coiled-coil forming protein kinase 1 |
| ROS | reactive oxygen species |
| RT-qPCR | real-time polymerase chain reaction |
| siRNA | small interfering RNA |
| SM22α | smooth muscle protein 22 alpha |
| sncRNA | small non-coding RNA |
| Spred-1 | sprouty-related, EVH1 domain-containing protein 1 |
| sRAGE | soluble receptor for advanced glycation end-products |
| SREBP | sterol regulatory element-binding protein |
| SRF | serum-response factor |
| SYNTAX | Synergy between Percutaneous Coronary Intervention with Taxus and Cardiac Surgery |
| T2DM | type 2 diabetes mellitus |
| TC | total cholesterol |
| TG | triglycerides |
| TGF-β | transforming growth factor-β |
| TNF-α | tumor necrosis factor α |
| TPM1 | tropomyosin 1 |
| TRAF6 | tumor necrosis factor receptor associated factor 6 |
| TRBP | trans-activation response RNA binding protein |
| 3′ UTR | 3′ untranslated region |
| WHR | waist-to-hip ratio |
| VCAM-1 | vascular cell adhesion molecule-1 |
| VE-cadherin | vascular endothelial cadherin |
| VEGF | vascular endothelial growth factor |
| VSMC | vascular smooth muscle cell |
| vWF | von Willebrand Factor |
References
- Wild, S.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care 2004, 27, 1047–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Magliano, D.J.; Zimmet, P.Z. The worldwide epidemiology of type 2 diabetes mellitus—Present and future perspectives. Nat. Rev. Endocrinol. 2011, 8, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Lascar, N.; Brown, J.; Pattison, H.; Barnett, A.H.; Bailey, C.J.; Bellary, S. Type 2 diabetes in adolescents and young adults. Lancet Diabetes Endocrinol. 2018, 6, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2021, 183, 109119. [Google Scholar] [CrossRef]
- Henning, R.J. Type-2 diabetes mellitus and cardiovascular disease. Future Cardiol. 2018, 14, 491–509. [Google Scholar] [CrossRef] [PubMed]
- Peer, N.; Balakrishna, Y.; Durao, S. Screening for type 2 diabetes mellitus. Cochrane Database Syst. Rev. 2020, 5, CD005266. [Google Scholar] [CrossRef]
- Ogurtsova, K.; Guariguata, L.; Barengo, N.C.; Ruiz, P.L.-D.; Sacre, J.W.; Karuranga, S.; Sun, H.; Boyko, E.J.; Magliano, D.J. IDF diabetes Atlas: Global estimates of undiagnosed diabetes in adults for 2021. Diabetes Res. Clin. Pract. 2021, 183, 109118. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Chawla, R.; Jaggi, S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J. Endocrinol. Metab. 2016, 20, 546–553. [Google Scholar] [CrossRef]
- Gedebjerg, A.; Almdal, T.P.; Berencsi, K.; Rungby, J.; Nielsen, J.S.; Witte, D.R.; Friborg, S.; Brandslund, I.; Vaag, A.; Beck-Nielsen, H.; et al. Prevalence of micro- and macrovascular diabetes complications at time of type 2 diabetes diagnosis and associated clinical characteristics: A cross-sectional baseline study of 6958 patients in the Danish DD2 cohort. J. Diabetes Complicat. 2018, 32, 34–40. [Google Scholar] [CrossRef]
- Dal Canto, E.; Ceriello, A.; Rydén, L.; Ferrini, M.; Hansen, T.B.; Schnell, O.; Standl, E.; Beulens, J.W. Diabetes as a cardiovascular risk factor: An overview of global trends of macro and micro vascular complications. Eur. J. Prev. Cardiol. 2019, 26, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Goldfine, A.B.; Phua, E.-J.; Abrahamson, M.J. Glycemic Management in Patients With Coronary Artery Disease and Prediabetes or Type 2 Diabetes Mellitus. Circulation 2014, 129, 2567–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartnik, M.; Rydén, L.; Ferrari, R.; Malmberg, K.; Pyörälä, K.; Simoons, M.; Standl, E.; Soler-Soler, J.; Öhrvik, J.; The Euro Heart Survey Investigators. The prevalence of abnormal glucose regulation in patients with coronary artery disease across Europe. The Euro Heart Survey on diabetes and the heart. Eur. Heart J. 2004, 25, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- Khot, U.N.; Khot, M.B.; Bajzer, C.T.; Sapp, S.K.; Ohman, E.M.; Brener, S.J.; Ellis, S.G.; Lincoff, A.M.; Topol, E.J. Prevalence of Conventional Risk Factors in Patients With Coronary Heart Disease. JAMA 2003, 290, 898–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munkhaugen, J.; Hjelmesæth, J.; Otterstad, J.E.; Helseth, R.; Sollid, S.T.; Gjertsen, E.; Gullestad, L.; Perk, J.; Moum, T.; Husebye, E.; et al. Managing patients with prediabetes and type 2 diabetes after coronary events: Individual tailoring needed—A cross-sectional study. BMC Cardiovasc. Disord. 2018, 18, 160. [Google Scholar] [CrossRef]
- Laakso, M. Cardiovascular disease in type 2 diabetes from population to man to mechanisms: The Kelly West Award Lecture 2008. Diabetes Care 2010, 33, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Einarson, T.R.; Acs, A.; Ludwig, C.; Panton, U.H. Prevalence of cardiovascular disease in type 2 diabetes: A systematic literature review of scientific evidence from across the world in 2007–2017. Cardiovasc. Diabetol. 2018, 17, 83. [Google Scholar] [CrossRef] [Green Version]
- Duan, L.; Liu, C.; Hu, J.; Liu, Y.; Wang, J.; Chen, G.; Li, Z.; Chen, H. Epigenetic mechanisms in coronary artery disease: The current state and prospects. Trends Cardiovasc. Med. 2018, 28, 311–319. [Google Scholar] [CrossRef]
- Poznyak, A.; Grechko, A.V.; Poggio, P.; Myasoedova, V.A.; Alfieri, V.; Orekhov, A.N. The Diabetes Mellitus–Atherosclerosis Connection: The Role of Lipid and Glucose Metabolism and Chronic Inflammation. Int. J. Mol. Sci. 2020, 21, 1835. [Google Scholar] [CrossRef] [Green Version]
- Vazzana, N.; Ranalli, P.; Cuccurullo, C.; Davì, G. Diabetes mellitus and thrombosis. Thromb. Res. 2012, 129, 371–377. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- He, Y.; Ding, Y.; Liang, B.; Lin, J.; Kim, T.-K.; Yu, H.; Hang, H.; Wang, K. A Systematic Study of Dysregulated MicroRNA in Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2017, 18, 456. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Chen, Y.; Liao, Y.; Li, R.; Lin, C.; Xiu, L.; Yu, H.; Ding, Y. Research Status of Differentially Expressed Noncoding RNAs in Type 2 Diabetes Patients. Biomed Res. Int. 2020, 2020, 3816056. [Google Scholar] [CrossRef] [PubMed]
- González-Sánchez, L.E.; Ortega-Camarillo, C.; Contreras-Ramos, A.; Barajas-Nava, L.A. miRNAs as biomarkers for diagnosis of type 2 diabetes: A systematic review. J. Diabetes 2021, 13, 792–816. [Google Scholar] [CrossRef] [PubMed]
- Romaine, S.P.R.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Meola, N.; Gennarino, V.A.; Banfi, S. microRNAs and genetic diseases. Pathogenetics 2009, 2, 7. [Google Scholar] [CrossRef]
- Niaz, S. The AGO proteins: An overview. Biol. Chem. 2018, 399, 525–547. [Google Scholar] [CrossRef] [PubMed]
- Witkos, T.M.; Koscianska, E.; Krzyzosiak, W.J. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera-Carrillo, E.; Berkhout, B. Dicer-independent processing of small RNA duplexes: Mechanistic insights and applications. Nucleic Acids Res. 2017, 45, 10369–10379. [Google Scholar] [CrossRef] [Green Version]
- Gareev, I.; Beylerli, O.; Yang, G.; Sun, J.; Pavlov, V.; Izmailov, A.; Shi, H.; Zhao, S. The current state of MiRNAs as biomarkers and therapeutic tools. Clin. Exp. Med. 2020, 20, 349–359. [Google Scholar] [CrossRef]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Gennarino, V.A.; Sardiello, M.; Mutarelli, M.; Dharmalingam, G.; Maselli, V.; Lago, G.; Banfi, S. HOCTAR database: A unique resource for microRNA target prediction. Gene 2011, 480, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [Green Version]
- Yuan, T.; Yang, T.; Chen, H.; Fu, D.; Hu, Y.; Wang, J.; Yuan, Q.; Yu, H.; Xu, W.; Xie, X. New insights into oxidative stress and inflammation during diabetes mellitus-accelerated atherosclerosis. Redox Biol. 2019, 20, 247–260. [Google Scholar] [CrossRef]
- Hansson, G.K.; Libby, P.; Tabas, I. Inflammation and plaque vulnerability. J. Intern. Med. 2015, 278, 483–493. [Google Scholar] [CrossRef] [Green Version]
- Aronson, D.; Edelman, E.R. Revascularization for coronary artery disease in diabetes mellitus: Angioplasty, stents and coronary artery bypass grafting. Rev. Endocr. Metab. Disord. 2010, 11, 75–86. [Google Scholar] [CrossRef] [Green Version]
- La Sala, L.; Prattichizzo, F.; Ceriello, A. The link between diabetes and atherosclerosis. Eur. J. Prev. Cardiol. 2019, 26, 15–24. [Google Scholar] [CrossRef]
- Giordo, R.; Ahmed, Y.M.A.; Allam, H.; Abusnana, S.; Pappalardo, L.; Nasrallah, G.K.; Mangoni, A.A.; Pintus, G. EndMT Regulation by Small RNAs in Diabetes-Associated Fibrotic Conditions: Potential Link With Oxidative Stress. Front. Cell Dev. Biol. 2021, 9, 683594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, Q.; Thakur, A.; Alfred, M.O.; Chakraborty, M.; Ghosh, A.; Yu, X. Endothelial dysfunction in diabetes and hypertension: Role of microRNAs and long non-coding RNAs. Life Sci. 2018, 213, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Petrie, J.R.; Guzik, T.J.; Touyz, R.M. Diabetes, Hypertension, and Cardiovascular Disease: Clinical Insights and Vascular Mechanisms. Can. J. Cardiol. 2018, 34, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigro, C.; Mirra, P.; Prevenzano, I.; Leone, A.; Fiory, F.; Longo, M.; Cabaro, S.; Oriente, F.; Beguinot, F.; Miele, C. miR-214-Dependent Increase of PHLPP2 Levels Mediates the Impairment of Insulin-Stimulated Akt Activation in Mouse Aortic Endothelial Cells Exposed to Methylglyoxal. Int. J. Mol. Sci. 2018, 19, 522. [Google Scholar] [CrossRef] [Green Version]
- Mirra, P.; Nigro, C.; Prevenzano, I.; Procopio, T.; Leone, A.; Raciti, G.A.; Andreozzi, F.; Longo, M.; Fiory, F.; Beguinot, F.; et al. The role of miR-190a in methylglyoxal-induced insulin resistance in endothelial cells. Biochim. Biophys. Acta-Mol. Basis Dis. 2017, 1863, 440–449. [Google Scholar] [CrossRef]
- Piperi, C.; Goumenos, A.; Adamopoulos, C.; Papavassiliou, A.G. AGE/RAGE signalling regulation by miRNAs: Associations with diabetic complications and therapeutic potential. Int. J. Biochem. Cell Biol. 2015, 60, 197–201. [Google Scholar] [CrossRef]
- Geroldi, D.; Falcone, C.; Emanuele, E. Soluble Receptor for Advanced Glycation End Products: From Disease Marker to Potential Therapeutic Target. Curr. Med. Chem. 2006, 13, 1971–1978. [Google Scholar] [CrossRef]
- Shi, Y.; Vanhoutte, P.M. Macro- and microvascular endothelial dysfunction in diabetes. J. Diabetes 2017, 9, 434–449. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Wang, F.; Shao, M.; Wang, Y.; Zhu, H. MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1. Vascul. Pharmacol. 2017, 88, 48–55. [Google Scholar] [CrossRef]
- Meng, S.; Cao, J.-T.; Zhang, B.; Zhou, Q.; Shen, C.-X.; Wang, C.-Q. Downregulation of microRNA-126 in endothelial progenitor cells from diabetes patients, impairs their functional properties, via target gene Spred-1. J. Mol. Cell. Cardiol. 2012, 53, 64–72. [Google Scholar] [CrossRef]
- Jansen, F.; Yang, X.; Hoelscher, M.; Cattelan, A.; Schmitz, T.; Proebsting, S.; Wenzel, D.; Vosen, S.; Franklin, B.S.; Fleischmann, B.K.; et al. Endothelial Microparticle–Mediated Transfer of MicroRNA-126 Promotes Vascular Endothelial Cell Repair via SPRED1 and Is Abrogated in Glucose-Damaged Endothelial Microparticles. Circulation 2013, 128, 2026–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhou, Q.; Pei, C.; Liu, B.; Li, M.; Fang, L.; Sun, Y.; Li, Y.; Meng, S. Hyperglycemia and Advanced Glycation End Products Regulate miR-126 Expression in Endothelial Progenitor Cells. J. Vasc. Res. 2016, 53, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Cao, J.; Zhang, X.; Fan, Y.; Fang, L.; Wang, C.; Lv, Z.; Fu, D.; Li, Y. Downregulation of MicroRNA-130a Contributes to Endothelial Progenitor Cell Dysfunction in Diabetic Patients via Its Target Runx3. PLoS ONE 2013, 8, e68611. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Meng, S.; Liu, B.; Li, M.-Q.; Li, Y.; Fang, L.; Li, Y.-G. MicroRNA-130a regulates autophagy of endothelial progenitor cells through Runx3. Clin. Exp. Pharmacol. Physiol. 2014, 41, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Li, D.; Yang, J.; Xie, J.; Yu, F.; Ma, Y.; Zhu, X.; Zhao, J.; Lv, Z. MicroRNA-130a Targets MAP3K12 to Modulate Diabetic Endothelial Progenitor Cell Function. Cell. Physiol. Biochem. 2015, 36, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Xiong, Y.; Li, G.; Liu, M.; He, T.; Tang, Y.; Chen, Y.; Cai, L.; Jiang, R.; Tao, J. MiR-21 is Overexpressed in Response to High Glucose and Protects Endothelial Cells from Apoptosis. Exp. Clin. Endocrinol. Diabetes 2013, 121, 425–430. [Google Scholar] [CrossRef] [PubMed]
- La Sala, L.; Mrakic-Sposta, S.; Micheloni, S.; Prattichizzo, F.; Ceriello, A. Glucose-sensing microRNA-21 disrupts ROS homeostasis and impairs antioxidant responses in cellular glucose variability. Cardiovasc. Diabetol. 2018, 17, 105. [Google Scholar] [CrossRef] [Green Version]
- Shao, M.; Yu, M.; Zhao, J.; Mei, J.; Pan, Y.; Zhang, J.; Wu, H.; Yu, M.; Liu, F.; Chen, G. miR-21-3p regulates AGE/RAGE signalling and improves diabetic atherosclerosis. Cell Biochem. Funct. 2020, 38, 965–975. [Google Scholar] [CrossRef]
- Liu, X.; Wang, D.; Yang, X.; Lei, L. Huayu Tongmai Granules protects against vascular endothelial dysfunction via up-regulating miR-185 and down-regulating RAGE. Biosci. Rep. 2018, 38, BSR20180674. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Ailifeire, M.; Wang, C.; Luo, L.; Zhang, J.; Yuan, L.; Zhang, L.; Li, X.; Wang, M. miR-320/VEGFA axis affects high glucose-induced metabolic memory during human umbilical vein endothelial cell dysfunction in diabetes pathology. Microvasc. Res. 2020, 127, 103913. [Google Scholar] [CrossRef]
- Jiang, Z.; Wu, J.; Ma, F.; Jiang, J.; Xu, L.; Du, L.; Huang, W.; Wang, Z.; Jia, Y.; Lu, L.; et al. MicroRNA-200a improves diabetic endothelial dysfunction by targeting KEAP1/NRF2. J. Endocrinol. 2020, 245, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, J.; Qu, D.; Wang, L.; Luo, J.-Y.; Lau, C.W.; Liu, P.; Gao, Z.; Tipoe, G.L.; Lee, H.K.; et al. Inhibition of miR-200c Restores Endothelial Function in Diabetic Mice Through Suppression of COX-2. Diabetes 2016, 65, 1196–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Guan, Q.; Jin, X. miR-200c serves an important role in H5V endothelial cells in high glucose by targeting Notch1. Mol. Med. Rep. 2017, 16, 2149–2155. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Kim, Y.-R.; Vikram, A.; Kumar, S.; Kassan, M.; Gabani, M.; Lee, S.K.; Jacobs, J.S.; Irani, K. P66Shc-Induced MicroRNA-34a Causes Diabetic Endothelial Dysfunction by Downregulating Sirtuin1. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2394–2403. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liang, W.; Tian, Y.; Ma, F.; Huang, W.; Jia, Y.; Jiang, Z.; Wu, H. Inhibition of P53/miR-34a improves diabetic endothelial dysfunction via activation of SIRT1. J. Cell. Mol. Med. 2019, 23, 3538–3548. [Google Scholar] [CrossRef] [Green Version]
- Gou, L.; Zhao, L.; Song, W.; Wang, L.; Liu, J.; Zhang, H.; Huang, Y.; Lau, C.W.; Yao, X.; Tian, X.Y.; et al. Inhibition of miR-92a Suppresses Oxidative Stress and Improves Endothelial Function by Upregulating Heme Oxygenase-1 in db/db Mice. Antioxid. Redox Signal. 2018, 28, 358–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Li, Y.; Sun, G.; Guo, D.; Bai, X. miR-181c-3p and -5p promotes high-glucose-induced dysfunction in human umbilical vein endothelial cells by regulating leukemia inhibitory factor. Int. J. Biol. Macromol. 2018, 115, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Collado, A.; Sun, C.; Tratsiakovich, Y.; Mahdi, A.; Winter, H.; Chernogubova, E.; Seime, T.; Narayanan, S.; Jiao, T.; et al. Downregulation of Erythrocyte miR-210 Induces Endothelial Dysfunction in Type 2 Diabetes. Diabetes 2022, 71, 285–297. [Google Scholar] [CrossRef]
- Brennan, E.; Wang, B.; McClelland, A.; Mohan, M.; Marai, M.; Beuscart, O.; Derouiche, S.; Gray, S.; Pickering, R.; Tikellis, C.; et al. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes 2017, 66, 2266–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Chen, M.; Xu, Q.; Liang, J.; Chen, R.; Xiao, Y.; Fang, M.; Chen, L. Effect of the Diabetic Environment On the Expression of MiRNAs in Endothelial Cells: Mir-149-5p Restoration Ameliorates the High Glucose-Induced Expression of TNF-α and ER Stress Markers. Cell. Physiol. Biochem. 2017, 43, 120–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X.; Zheng, J.; Li, M.; Yu, M.; Ping, F.; Wang, T.; Wang, X. Vildagliptin, a dipeptidyl peptidase-4 inhibitor, attenuated endothelial dysfunction through miRNAs in diabetic rats. Arch. Med. Sci. 2019, 17, 1378–1387. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Liu, Q.; Hu, N.; Zheng, F.; Zhang, X.; Ni, Y.; Liu, J. Downregulation of hsa_circ_0068087 ameliorates TLR4/NF-κB/NLRP3 inflammasome-mediated inflammation and endothelial cell dysfunction in high glucose conditioned by sponging miR-197. Gene 2019, 709, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Cao, Y.; Chen, S.; Ruiz, M.; Chakrabarti, S. miRNA-1 regulates endothelin-1 in diabetes. Life Sci. 2014, 98, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wu, Y.; Ye, S. MiR-181c restrains nitration stress of endothelial cells in diabetic db/db mice through inhibiting the expression of FoxO1. Biochem. Biophys. Res. Commun. 2017, 486, 29–35. [Google Scholar] [CrossRef]
- Silambarasan, M.; Tan, J.R.; Karolina, D.S.; Armugam, A.; Kaur, C.; Jeyaseelan, K. MicroRNAs in Hyperglycemia Induced Endothelial Cell Dysfunction. Int. J. Mol. Sci. 2016, 17, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, W.; Hu, X.; Shi, R.; Han, S.; Cao, C.; Wang, K.; Li, M. MiR-31 regulates the function of diabetic endothelial progenitor cells by targeting Satb2. Acta Biochim. Biophys. Sin. 2018, 50, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Xiao, X.; Zheng, J.; Li, M. A glucagon-like peptide-1 analog, liraglutide, ameliorates endothelial dysfunction through miRNAs to inhibit apoptosis in rats. PeerJ 2019, 7, e6567. [Google Scholar] [CrossRef] [Green Version]
- Luo, E.; Wang, D.; Yan, G.; Qiao, Y.; Zhu, B.; Liu, B.; Hou, J.; Tang, C. The NF-κB/miR-425-5p/MCT4 axis: A novel insight into diabetes-induced endothelial dysfunction. Mol. Cell. Endocrinol. 2020, 500, 110641. [Google Scholar] [CrossRef]
- Huang, Z.; Li, N.; Shan, Y.; Liang, C. Hsa-miRNA-29a protects against high glucose-induced damage in human umbilical vein endothelial cells. J. Cell. Biochem. 2019, 120, 5860–5868. [Google Scholar] [CrossRef]
- Chen, H.; Feng, Z.; Li, L.; Fan, L. MicroRNA-9 rescues hyperglycemia-induced endothelial cell dysfunction and promotes arteriogenesis through downregulating Notch1 signaling. Mol. Cell. Biochem. 2021, 476, 2777–2789. [Google Scholar] [CrossRef]
- Rawal, S.; Munasinghe, P.E.; Shindikar, A.; Paulin, J.; Cameron, V.; Manning, P.; Williams, M.J.A.; Jones, G.T.; Bunton, R.; Galvin, I.; et al. Down-regulation of proangiogenic microRNA-126 and microRNA-132 are early modulators of diabetic cardiac microangiopathy. Cardiovasc. Res. 2017, 113, 90–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.-F.; Wan, X.-X.; Zhao, L.-L.; Guo, Z.; Shen, R.-T.; Zeng, P.-Y.; Wang, L.-H.; Yuan, J.-J.; Yang, W.-J.; Yue, C.; et al. MicroRNA-139-5p upregulation is associated with diabetic endothelial cell dysfunction by targeting c-jun. Aging 2020, 13, 1186–1211. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, H.; Liu, C.; Mu, X.; Cheng, S. Hyperglycemia inhibition of endothelial miR-140-3p mediates angiogenic dysfunction in diabetes mellitus. J. Diabetes Complicat. 2019, 33, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yin, L.; Kuang, H. miR-181a/b-5p regulates human umbilical vein endothelial cell angiogenesis by targeting PDGFRA. Cell Biochem. Funct. 2020, 38, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, Y.-H.; Li, F.; Yang, T.; Lu, Y.W.; Geng, Y.-J. microRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009, 381, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Cui, Y.; Fan, L.; Mu, X.; Hua, Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 71–78. [Google Scholar] [CrossRef]
- Kim, J. MicroRNAs as critical regulators of the endothelial to mesenchymal transition in vascular biology. BMB Rep. 2018, 51, 65–72. [Google Scholar] [CrossRef]
- Souilhol, C.; Harmsen, M.C.; Evans, P.C.; Krenning, G. Endothelial–mesenchymal transition in atherosclerosis. Cardiovasc. Res. 2018, 114, 565–577. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.-H.; Li, R.; Zeng, Y.; Zhou, T.; Xiong, F.; Zhu, M. MicroRNA-142-3p inhibits high-glucose-induced endothelial-to-mesenchymal transition through targeting TGF-β1/Smad pathway in primary human aortic endothelial cells. Int. J. Clin. Exp. Pathol. 2018, 11, 1208–1217. [Google Scholar]
- Guan, G.; Wei, N.; Song, T.; Zhao, C.; Sun, Y.; Pan, R.; Zhang, L.; Xu, Y.; Dai, Y.; Han, H. miR-448-3p alleviates diabetic vascular dysfunction by inhibiting endothelial–mesenchymal transition through DPP-4 dysregulation. J. Cell. Physiol. 2020, 235, 10024–10036. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Q.; Zhan, Y.; Ke, J.; Lv, P.; Huang, J. The role of miR-328 in high glucose-induced endothelial-to-mesenchymal transition in human umbilical vein endothelial cells. Life Sci. 2018, 207, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-C.; Sung, M.-L.; Kuo, H.-C.; Chien, S.-J.; Yen, C.-K.; Chen, C.-N. Differential Regulation of Human Aortic Smooth Muscle Cell Proliferation by Monocyte-Derived Macrophages from Diabetic Patients. PLoS ONE 2014, 9, e113752. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wu, N.; Leong, M.C.; Zhang, W.; Ye, Z.; Li, R.; Huang, J.; Zhang, Z.; Li, L.; Yao, X.; et al. miR-145 improves metabolic inflammatory disease through multiple pathways. J. Mol. Cell Biol. 2020, 12, 152–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Zhou, Z.; Wang, J.; Li, S. MiR-130b promotes obesity associated adipose tissue inflammation and insulin resistance in diabetes mice through alleviating M2 macrophage polarization via repression of PPAR-γ. Immunol. Lett. 2016, 180, 1–8. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, X.; Hu, Q.; Zhang, R.; Yin, Y. Suppression of TLR4 by miR-448 is involved in Diabetic development via regulating Macrophage polarization. J. Pharm. Pharmacol. 2019, 71, 806–815. [Google Scholar] [CrossRef]
- Sun, J.; Huang, Q.; Li, S.; Meng, F.; Li, X.; Gong, X. miR-330-5p/Tim-3 axis regulates macrophage M2 polarization and insulin resistance in diabetes mice. Mol. Immunol. 2018, 95, 107–113. [Google Scholar] [CrossRef]
- Hu, F.; Tong, J.; Deng, B.; Zheng, J.; Lu, C. MiR-495 regulates macrophage M1/M2 polarization and insulin resistance in high-fat diet-fed mice via targeting FTO. Pflügers Arch.-Eur. J. Physiol. 2019, 471, 1529–1537. [Google Scholar] [CrossRef]
- Jaiswal, A.; Reddy, S.S.; Maurya, M.; Maurya, P.; Barthwal, M.K. MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFα. Cell. Mol. Immunol. 2019, 16, 495–507. [Google Scholar] [CrossRef]
- Kuschnerus, K.; Straessler, E.T.; Müller, M.F.; Lüscher, T.F.; Landmesser, U.; Kränkel, N. Increased Expression of miR-483-3p Impairs the Vascular Response to Injury in Type 2 Diabetes. Diabetes 2019, 68, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Kimura, Y.; Tamasawa, N.; Matsumura, K.; Murakami, H.; Yamashita, M.; Matsuki, K.; Tanabe, J.; Murakami, H.; Matsui, J.; Daimon, M. Clinical Significance of Determining Plasma MicroRNA33b in Type 2 Diabetic Patients with Dyslipidemia. J. Atheroscler. Thromb. 2016, 23, 1276–1285. [Google Scholar] [CrossRef] [Green Version]
- Distel, E.; Barrett, T.J.; Chung, K.; Girgis, N.M.; Parathath, S.; Essau, C.C.; Murphy, A.J.; Moore, K.J.; Fisher, E.A. miR33 Inhibition Overcomes Deleterious Effects of Diabetes Mellitus on Atherosclerosis Plaque Regression in Mice. Circ. Res. 2014, 115, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Guan, P.; Li, D.; Hang, Y.; Ye, X.; Han, L.; Lu, Y.; Bai, X.; Zhang, P.; Hu, W. Intermedin attenuates macrophage phagocytosis via regulation of the long noncoding RNA Dnm3os/miR-27b-3p/SLAMF7 axis in a mouse model of atherosclerosis in diabetes. Biochem. Biophys. Res. Commun. 2021, 583, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.E.; Riches, K. The vascular smooth muscle cell: A therapeutic target in Type 2 diabetes? Clin. Sci. 2013, 125, 167–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shyu, K.-G.; Cheng, W.-P.; Wang, B.-W. Angiotensin II Downregulates MicroRNA-145 to Regulate Kruppel-like Factor 4 and Myocardin Expression in Human Coronary Arterial Smooth Muscle Cells under High Glucose Conditions. Mol. Med. 2015, 21, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, Y.; Li, W.; Yang, J. MicroRNA-145 alleviates high glucose-induced proliferation and migration of vascular smooth muscle cells through targeting ROCK1. Biomed. Pharmacother. 2018, 99, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Riches, K.; Alshanwani, A.R.; Warburton, P.; O’Regan, D.J.; Ball, S.G.; Wood, I.C.; Turner, N.A.; Porter, K.E. Elevated expression levels of miR-143/5 in saphenous vein smooth muscle cells from patients with Type 2 diabetes drive persistent changes in phenotype and function. J. Mol. Cell. Cardiol. 2014, 74, 240–250. [Google Scholar] [CrossRef] [Green Version]
- Chettimada, S.; Ata, H.; Rawat, D.K.; Gulati, S.; Kahn, A.G.; Edwards, J.G.; Gupte, S.A. Contractile protein expression is upregulated by reactive oxygen species in aorta of Goto-Kakizaki rat. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H214–H224. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Ye, S.; Wang, W. miR-217 alleviates high-glucose-induced vascular smooth muscle cell dysfunction via regulating ROCK1. J. Biochem. Mol. Toxicol. 2021, 35, e22668. [Google Scholar] [CrossRef]
- Xu, Q.; Liang, Y.; Liu, X.; Zhang, C.; Liu, X.; Li, H.; Liang, J.; Yang, G.; Ge, Z. miR-132 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting E2F5. Mol. Med. Rep. 2019, 20, 2012–2020. [Google Scholar] [CrossRef]
- Yang, J.; Chen, L.; Ding, J.; Fan, Z.; Li, S.; Wu, H.; Zhang, J.; Yang, C.; Wang, H.; Zeng, P.; et al. MicroRNA-24 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting HMGB1. Gene 2016, 586, 268–273. [Google Scholar] [CrossRef]
- Yang, J.; Zeng, P.; Yang, J.; Liu, X.; Ding, J.; Wang, H.; Chen, L. MicroRNA-24 regulates vascular remodeling via inhibiting PDGF-BB pathway in diabetic rat model. Gene 2018, 659, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Zhang, J.; Yang, J.; Fan, Z.; Liu, X.; Gao, W.; Zeng, P.; Xiong, M.; Ma, C.; Yang, J. MicroRNA-24 attenuates vascular remodeling in diabetic rats through PI3K/Akt signaling pathway. Nutr. Metab. Cardiovasc. Dis. 2019, 29, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Fan, Z.; Yang, J.; Ding, J.; Yang, C.; Chen, L. MicroRNA-24 Attenuates Neointimal Hyperplasia in the Diabetic Rat Carotid Artery Injury Model by Inhibiting Wnt4 Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cai, W.; Fan, Z.; Yang, C.; Wang, W.; Xiong, M.; Ma, C.; Yang, J. MicroRNA-24 inhibits the oxidative stress induced by vascular injury by activating the Nrf2/Ho-1 signaling pathway. Atherosclerosis 2019, 290, 9–18. [Google Scholar] [CrossRef]
- Fan, Z.; Yang, J.; Yang, C.; Zhang, J.; Cai, W.; Huang, C. MicroRNA-24 attenuates diabetic vascular remodeling by suppressing the NLRP3/caspase-1/IL-1β signaling pathway. Int. J. Mol. Med. 2020, 45, 1534–1542. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Xia, L.; Fan, X.; Ostriker, A.C.; Yarovinsky, T.; Su, M.; Zhang, Y.; Peng, X.; Xie, Y.; Pi, L.; et al. Platelet-derived miR-223 promotes a phenotypic switch in arterial injury repair. J. Clin. Investig. 2019, 129, 1372–1386. [Google Scholar] [CrossRef]
- Su, M.; Fan, S.; Ling, Z.; Fan, X.; Xia, L.; Liu, Y.; Li, S.; Zhang, Y.; Zeng, Z.; Tang, W.H. Restoring the Platelet miR-223 by Calpain Inhibition Alleviates the Neointimal Hyperplasia in Diabetes. Front. Physiol. 2020, 11, 742. [Google Scholar] [CrossRef]
- Lu, X.; Ma, S.-T.; Zhou, B.; Li, T. MiR-9 promotes the phenotypic switch of vascular smooth muscle cells by targeting KLF5. Turk. J. Med. Sci. 2019, 49, 928–938. [Google Scholar] [CrossRef]
- Ye, D.; Lou, G.; Li, A.; Dong, F.; Chen, G.; Xu, W.; Liu, Y.; Hu, S. MicroRNA-125a-mediated regulation of the mevalonate signaling pathway contributes to high glucose-induced proliferation and migration of vascular smooth muscle cells. Mol. Med. Rep. 2020, 21, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Chen, S.; Zhang, Z.; Wang, C.; Liu, C. FAM3B mediates high glucose-induced vascular smooth muscle cell proliferation and migration via inhibition of miR-322-5p. Sci. Rep. 2017, 7, 2298. [Google Scholar] [CrossRef] [Green Version]
- Jansen, F.; Zietzer, A.; Stumpf, T.; Flender, A.; Schmitz, T.; Nickenig, G.; Werner, N. Endothelial microparticle-promoted inhibition of vascular remodeling is abrogated under hyperglycaemic conditions. J. Mol. Cell. Cardiol. 2017, 112, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Martinez, M.; Cidad, P.; Moreno-Estar, S.; Fernández, M.; Albinsson, S.; Cózar-Castellano, I.; López-López, J.R.; Pérez-Garcia, M.T. miR-126 contributes to the epigenetic signature of diabetic vascular smooth muscle and enhances antirestenosis effects of Kv1.3 blockers. Mol. Metab. 2021, 53, 101306. [Google Scholar] [CrossRef] [PubMed]
- Torella, D.; Iaconetti, C.; Tarallo, R.; Marino, F.; Giurato, G.; Veneziano, C.; Aquila, I.; Scalise, M.; Mancuso, T.; Cianflone, E.; et al. miRNA Regulation of the Hyperproliferative Phenotype of Vascular Smooth Muscle Cells in Diabetes. Diabetes 2018, 67, 2554–2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hien, T.T.; Garcia-Vaz, E.; Stenkula, K.G.; Sjögren, J.; Nilsson, J.; Gomez, M.F.; Albinsson, S. MicroRNA-dependent regulation of KLF4 by glucose in vascular smooth muscle. J. Cell. Physiol. 2018, 233, 7195–7205. [Google Scholar] [CrossRef]
- Sun, G.; Song, H.; Wu, S. miR-19a promotes vascular smooth muscle cell proliferation, migration and invasion through regulation of Ras homolog family member B. Int. J. Mol. Med. 2019, 44, 1991–2002. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, L.; Yun, H.; Han, Y. MiR-138 promotes smooth muscle cells proliferation and migration in db/db mice through down-regulation of SIRT1. Biochem. Biophys. Res. Commun. 2015, 463, 1159–1164. [Google Scholar] [CrossRef]
- Reddy, M.A.; Das, S.; Zhuo, C.; Jin, W.; Wang, M.; Lanting, L.; Natarajan, R. Regulation of Vascular Smooth Muscle Cell Dysfunction Under Diabetic Conditions by miR-504. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 864–873. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Gao, X.; Sun, Y.; Zhao, J.; Chen, J.; Gao, L.; Zhao, L.; Li, Y. Dihydroartemisinin alleviates high glucose-induced vascular smooth muscle cells proliferation and inflammation by depressing the miR-376b-3p/KLF15 pathway. Biochem. Biophys. Res. Commun. 2020, 530, 574–580. [Google Scholar] [CrossRef]
- Coleman, C.B.; Lightell, D.J.; Moss, S.C.; Bates, M.; Parrino, P.E.; Woods, T.C. Elevation of miR-221 and -222 in the internal mammary arteries of diabetic subjects and normalization with metformin. Mol. Cell. Endocrinol. 2013, 374, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Lightell, D.J.; Moss, S.C.; Woods, T.C. Upregulation of miR-221 and -222 in response to increased extracellular signal-regulated kinases 1/2 activity exacerbates neointimal hyperplasia in diabetes mellitus. Atherosclerosis 2018, 269, 71–78. [Google Scholar] [CrossRef]
- Jia, S.; Ma, W.; Zhang, C.; Zhang, Y.; Yao, Z.; Quan, X.; Guo, X.; Wang, C. Tanshinone IIA attenuates high glucose induced human VSMC proliferation and migration through miR-21-5p-mediated tropomyosin 1 downregulation. Arch. Biochem. Biophys. 2019, 677, 108154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, R.; Lv, J.; Wang, X.; Ye, J.; Lu, N.; Liang, X.; Yang, L. MicroRNA-99a inhibits insulin-induced proliferation, migration, dedifferentiation, and rapamycin resistance of vascular smooth muscle cells by inhibiting insulin-like growth factor-1 receptor and mammalian target of rapamycin. Biochem. Biophys. Res. Commun. 2017, 486, 414–422. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, M.; Singh, J. Endothelial dysfunction and platelet hyperactivity in type 2 diabetes mellitus: Molecular insights and therapeutic strategies. Cardiovasc. Diabetol. 2018, 17, 121. [Google Scholar] [CrossRef]
- Randriamboavonjy, V.; Fleming, I. Platelet Function and Signaling in Diabetes Mellitus. Curr. Vasc. Pharmacol. 2012, 10, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Pordzik, J.; Pisarz, K.; De Rosa, S.; Jones, A.D.; Eyileten, C.; Indolfi, C.; Malek, L.; Postula, M. The Potential Role of Platelet-Related microRNAs in the Development of Cardiovascular Events in High-Risk Populations, Including Diabetic Patients: A Review. Front. Endocrinol. 2018, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Pordzik, J.; Jakubik, D.; Jarosz-Popek, J.; Wicik, Z.; Eyileten, C.; De Rosa, S.; Indolfi, C.; Siller-Matula, J.M.; Czajka, P.; Postula, M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: Bioinformatic analysis and review. Cardiovasc. Diabetol. 2019, 18, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fejes, Z.; Póliska, S.; Czimmerer, Z.; Káplár, M.; Penyige, A.; Gál Szabó, G.; Beke Debreceni, I.; Kunapuli, S.P.; Kappelmayer, J.; Nagy Jr, B. Hyperglycaemia suppresses microRNA expression in platelets to increase P2RY12 and SELP levels in type 2 diabetes mellitus. Thromb. Haemost. 2017, 117, 529–542. [Google Scholar] [CrossRef]
- Elgheznawy, A.; Shi, L.; Hu, J.; Wittig, I.; Laban, H.; Pircher, J.; Mann, A.; Provost, P.; Randriamboavonjy, V.; Fleming, I. Dicer Cleavage by Calpain Determines Platelet microRNA Levels and Function in Diabetes. Circ. Res. 2015, 117, 157–165. [Google Scholar] [CrossRef]
- Zhou, M.; Gao, M.; Luo, Y.; Gui, R.; Ji, H. Long non-coding RNA metallothionein 1 pseudogene 3 promotes p2y12 expression by sponging miR-126 to activate platelet in diabetic animal model. Platelets 2019, 30, 452–459. [Google Scholar] [CrossRef] [Green Version]
- De Boer, H.C.; van Solingen, C.; Prins, J.; Duijs, J.M.G.J.; Huisman, M.V.; Rabelink, T.J.; van Zonneveld, A.J. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur. Heart J. 2013, 34, 3451–3457. [Google Scholar] [CrossRef]
- Luo, M.; Li, R.; Ren, M.; Chen, N.; Deng, X.; Tan, X.; Li, Y.; Zeng, M.; Yang, Y.; Wan, Q.; et al. Hyperglycaemia-induced reciprocal changes in miR-30c and PAI-1 expression in platelets. Sci. Rep. 2016, 6, 36687. [Google Scholar] [CrossRef] [PubMed]
- Stratz, C.; Nührenberg, T.; Fiebich, B.L.; Amann, M.; Kumar, A.; Binder, H.; Hoffmann, I.; Valina, C.; Hochholzer, W.; Trenk, D.; et al. Controlled type II diabetes mellitus has no major influence on platelet micro-RNA expression. Results from micro-array profiling in a cohort of 60 patients. Thromb. Haemost. 2014, 111, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhan, J.-K.; Zhong, J.-Y.; Wang, Y.-J.; Wang, Y.; Li, S.; He, J.-Y.; Tan, P.; Chen, Y.-Y.; Liu, X.-B.; et al. lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating high-glucose-induced calcification/senescence of VSMCs. Aging 2019, 11, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Pei, Y.; Rui-Wang; Yang, J.; Zhao, Y.; Liu, X.; Shen, H.; Ma, Q.; Zhang, S.; Ge, H. Association of Plasma MiRNA-204 and the Presence and Severity of Coronary Artery Calcification in Patients With Type 2 Diabetes. Angiology 2021, 72, 451–458. [Google Scholar] [CrossRef]
- Zhong, J.-Y.; Cui, X.-J.; Zhan, J.-K.; Wang, Y.-J.; Li, S.; Lin, X.; Xiang, Q.-Y.; Ni, Y.-Q.; Liu, L.; Liu, Y.-S. LncRNA-ES3 inhibition by Bhlhe40 is involved in high glucose–induced calcification/senescence of vascular smooth muscle cells. Ann. N. Y. Acad. Sci. 2020, 1474, 61–72. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Zhang, X.-Z.; Li, F.; Ji, Q.-R. MiR-128-3p accelerates cardiovascular calcification and insulin resistance through ISL1-dependent Wnt pathway in type 2 diabetes mellitus rats. J. Cell. Physiol. 2019, 234, 4997–5010. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, P.; Raina, S.; Chugh, J.; Sharma, S. miRNAs: Early prognostic biomarkers for Type 2 diabetes mellitus? Biomark. Med. 2015, 9, 1025–1040. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Mayer, J.; Pospisilova, S. MicroRNA isolation and stability in stored RNA samples. Biochem. Biophys. Res. Commun. 2009, 390, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Tijsen, A.J.; Pinto, Y.M.; Creemers, E.E. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H1085–H1095. [Google Scholar] [CrossRef] [Green Version]
- Ali Sheikh, M.S.; Alduraywish, A.; Almaeen, A.; Alruwali, M.; Alruwaili, R.; Alomair, B.M.; Salma, U.; Hedeab, G.M.; Bugti, N.; Abdulhabeeb, I.A.M. Therapeutic Value of miRNAs in Coronary Artery Disease. Oxid. Med. Cell. Longev. 2021, 2021, 8853748. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wan, S.; Yang, T.; Niu, D.; Zhang, A.; Yang, C.; Cai, J.; Wu, J.; Song, J.; Zhang, C.-Y.; et al. Increased serum microRNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci. Rep. 2016, 6, 20032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef] [Green Version]
- Prabu, P.; Rome, S.; Sathishkumar, C.; Aravind, S.; Mahalingam, B.; Shanthirani, C.S.; Gastebois, C.; Villard, A.; Mohan, V.; Balasubramanyam, M. Circulating MiRNAs of ‘Asian Indian Phenotype’ Identified in Subjects with Impaired Glucose Tolerance and Patients with Type 2 Diabetes. PLoS ONE 2015, 10, e0128372. [Google Scholar] [CrossRef] [Green Version]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Rovira, O.; Guerra, E.; Esteve, E.; Xifra, G.; Martínez, C.; Ricart, W.; Rieusset, J.; et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care 2014, 37, 1375–1383. [Google Scholar] [CrossRef] [Green Version]
- Santovito, D.; De Nardis, V.; Marcantonio, P.; Mandolini, C.; Paganelli, C.; Vitale, E.; Buttitta, F.; Bucci, M.; Mezzetti, A.; Consoli, A.; et al. Plasma Exosome MicroRNA Profiling Unravels a New Potential Modulator of Adiponectin Pathway in Diabetes: Effect of Glycemic Control. J. Clin. Endocrinol. Metab. 2014, 99, E1681–E1685. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.T.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA Profiles in Patients with Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.-Z.; Dong, J.; Zhang, J.; Wang, S.; He, Y.; Yan, Y.-X. Identification of Neuroendocrine Stress Response-Related Circulating MicroRNAs as Biomarkers for Type 2 Diabetes Mellitus and Insulin Resistance. Front. Endocrinol. 2018, 9, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [Green Version]
- Avgeris, M.; Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Scorilas, A.; Fragoulis, E.G.; Christodoulou, M.-I. Blood-based analysis of 84 microRNAs identifies molecules deregulated in individuals with type-2 diabetes, risk factors for the disease or metabolic syndrome. Diabetes Res. Clin. Pract. 2020, 164, 108187. [Google Scholar] [CrossRef]
- Yang, Z.-M.; Chen, L.-H.; Hong, M.; Chen, Y.-Y.; Yang, X.-R.; Tang, S.-M.; Yuan, Q.-F.; Chen, W.-W. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol. Med. Rep. 2017, 15, 2143–2153. [Google Scholar] [CrossRef] [Green Version]
- Matsha, T.E.; Kengne, A.P.; Hector, S.; Mbu, D.L.; Yako, Y.Y.; Erasmus, R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget 2018, 9, 30485–30498. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Ai, D.; Wu, R.; Zhang, T.; Jing, L.; Lu, J.; Zhong, L. Identification of the differential expression of serum microRNA in type 2 diabetes. Biosci. Biotechnol. Biochem. 2016, 80, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Wang, T.; Huang, S.; Di, Y.; Huang, Y.; Liu, X.; Luo, Z.; Han, W.; An, B. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 2016, 53, 693–702. [Google Scholar] [CrossRef]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Baldeón, R.L.; Weigelt, K.; de Wit, H.; Ozcan, B.; van Oudenaren, A.; Sempértegui, F.; Sijbrands, E.; Grosse, L.; van Zonneveld, A.-J.; Drexhage, H.A.; et al. Type 2 Diabetes Monocyte MicroRNA and mRNA Expression: Dyslipidemia Associates with Increased Differentiation-Related Genes but Not Inflammatory Activation. PLoS ONE 2015, 10, e0129421. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zhu, M.; Mao, X.; Long, M.; Du, X.; Wu, Y.; Abudureyimu, K.; Zhang, C.; Wang, Y.; Tao, Y.; et al. MicroRNA-130a expression is decreased in Xinjiang Uygur patients with type 2 diabetes mellitus. Am. J. Transl. Res. 2015, 7, 1984–1991. [Google Scholar] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Chang, X.; Yin, L.; Li, J.; Zhou, T.; Zhang, C.; Chen, X. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol. Med. Rep. 2014, 9, 967–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aljaibeji, H.; Elemam, N.M.; Mohammed, A.K.; Hasswan, H.; Thahyabat, M.A.; Alkhayyal, N.; Sulaiman, N.; Taneera, J. Let7b-5p is Upregulated in the Serum of Emirati Patients with Type 2 Diabetes and Regulates Insulin Secretion in INS-1 Cells. Exp. Clin. Endocrinol. Diabetes 2022, 130, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.-T.; Li, C.-L.; Tian, H.; Li, J.; Pei, Y.; Liu, Y.; Gong, Y.-P.; Fang, F.-S.; Sun, B.-R. MiR-199a is overexpressed in plasma of type 2 diabetes patients which contributes to type 2 diabetes by targeting GLUT4. Mol. Cell. Biochem. 2014, 397, 45–51. [Google Scholar] [CrossRef]
- Sucharita, S.; Ashwini, V.; Prabhu, J.S.; Avadhany, S.T.; Ayyar, V.; Bantwal, G. The Role of Circulating MicroRNA in the Regulation of Beta Cell Function and Insulin Resistance among Indians with Type 2 Diabetes. Indian J. Endocrinol. Metab. 2018, 22, 770–773. [Google Scholar] [CrossRef]
- Ali Beg, M.M.; Verma, A.K.; Saleem, M.; Saud Alreshidi, F.; Alenazi, F.; Ahmad, H.; Joshi, P.C. Role and Significance of Circulating Biomarkers: miRNA and E2F1 mRNA Expression and Their Association with Type-2 Diabetic Complications. Int. J. Endocrinol. 2020, 2020, 6279168. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, C.; Zhang, X.; Qiu, T.; Ma, Y.; Li, X.; Pang, H.; Xiong, J.; Yang, X.; Pan, C.; et al. Correlation analysis of microribonucleic acid-155 and microribonucleic acid-29 with type 2 diabetes mellitus, and the prediction and verification of target genes. J. Diabetes Investig. 2021, 12, 165–175. [Google Scholar] [CrossRef]
- Wang, M.; Wei, J.; Ji, T.; Zang, K. miRNA-770-5p expression is upregulated in patients with type 2 diabetes and miRNA-770-5p knockdown protects pancreatic β-cell function via targeting BAG5 expression. Exp. Ther. Med. 2021, 22, 664. [Google Scholar] [CrossRef]
- Lu, C.; Wang, D.; Feng, Y.; Feng, L.; Li, Z. miR-720 Regulates Insulin Secretion by Targeting Rab35. Biomed Res. Int. 2021, 2021, 6662612. [Google Scholar] [CrossRef] [PubMed]
- Shahrokhi, S.Z.; Saeidi, L.; Sadatamini, M.; Jafarzadeh, M.; Rahimipour, A.; Kazerouni, F. Can miR-145-5p be used as a marker in diabetic patients? Arch. Physiol. Biochem. 2022, 128, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Masoudi, F.; Sharifi, M.R.; Pourfarzam, M. Investigation of the relationship between miR-33a, miR-122, erythrocyte membrane fatty acids profile, and serum lipids with components of metabolic syndrome in type 2 diabetic patients. Res. Pharm. Sci. 2022, 17, 242–251. [Google Scholar] [CrossRef]
- Zeinali, F.; Aghaei Zarch, S.M.; Jahan-Mihan, A.; Kalantar, S.M.; Vahidi Mehrjardi, M.Y.; Fallahzadeh, H.; Hosseinzadeh, M.; Rahmanian, M.; Mozaffari-Khosravi, H. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and type 2 diabetes mellitus: A case control study. PLoS ONE 2021, 16, e0251697. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Bao, W.; Shan, Z.; Liu, J.; Yu, X.; Xia, S.; Gao, H.; Wang, X.; Yao, P.; Hu, F.B.; et al. Increased MicroRNA-146a Levels in Plasma of Patients with Newly Diagnosed Type 2 Diabetes Mellitus. PLoS ONE 2013, 8, e73272. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Gao, G.; Yang, C.; Zhou, K.; Shen, B.; Liang, H.; Jiang, X. The Role of Circulating MicroRNA-126 (miR-126): A Novel Biomarker for Screening Prediabetes and Newly Diagnosed Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2014, 15, 10567–10577. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Li, L.; Shang, Q.; Lv, C.; Wang, C.; Su, B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015, 463, 60–63. [Google Scholar] [CrossRef]
- Al-Kafaji, G.; Al-Mahroos, G.; Abdulla Al-Muhtaresh, H.; Sabry, M.A.; Abdul Razzak, R.; Salem, A.H. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomarkers 2017, 22, 268–278. [Google Scholar] [CrossRef]
- Weale, C.J.; Matshazi, D.M.; Davids, S.F.G.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. MicroRNAs-1299, -126-3p and -30e-3p as Potential Diagnostic Biomarkers for Prediabetes. Diagnostics 2021, 11, 949. [Google Scholar] [CrossRef]
- Monfared, Y.K.; Mirzaii-Dizgah, M.-R.; Khodabandehloo, E.; Sarookhani, M.R.; Hashemipour, S.; Mirzaii-Dizgah, I. Salivary microRNA-126 and 135a: A potentially non-invasive diagnostic biomarkers of type- 2 diabetes. J. Diabetes Metab. Disord. 2021, 20, 1631–1638. [Google Scholar] [CrossRef]
- Zhang, T.; Lv, C.; Li, L.; Chen, S.; Liu, S.; Wang, C.; Su, B. Plasma miR-126 Is a Potential Biomarker for Early Prediction of Type 2 Diabetes Mellitus in Susceptible Individuals. Biomed Res. Int. 2013, 2013, 761617. [Google Scholar] [CrossRef] [PubMed]
- Baldeón, R.L.; Weigelt, K.; de Wit, H.; Ozcan, B.; van Oudenaren, A.; Sempértegui, F.; Sijbrands, E.; Grosse, L.; Freire, W.; Drexhage, H.A.; et al. Decreased Serum Level of miR-146a as Sign of Chronic Inflammation in Type 2 Diabetic Patients. PLoS ONE 2014, 9, e115209. [Google Scholar] [CrossRef] [Green Version]
- Olivieri, F.; Spazzafumo, L.; Bonafè, M.; Recchioni, R.; Prattichizzo, F.; Marcheselli, F.; Micolucci, L.; Mensà, E.; Giuliani, A.; Santini, G.; et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget 2015, 6, 35372–35382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenin, R.; Sankaramoorthy, A.; Mohan, V.; Balasubramanyam, M. Altered immunometabolism at the interface of increased endoplasmic reticulum (ER) stress in patients with type 2 diabetes. J. Leukoc. Biol. 2015, 98, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Giannella, A.; Radu, C.M.; Franco, L.; Campello, E.; Simioni, P.; Avogaro, A.; de Kreutzenberg, S.V.; Ceolotto, G. Circulating levels and characterization of microparticles in patients with different degrees of glucose tolerance. Cardiovasc. Diabetol. 2017, 16, 118. [Google Scholar] [CrossRef]
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Omrani, M.D.; Sharifi, Z.; Golmohammadi, T. The rs2910164 variant is associated with reduced miR-146a expression but not cytokine levels in patients with type 2 diabetes. J. Endocrinol. Investig. 2018, 41, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, M.; Aghaei Zarch, S.M.; Vahidi Mehrjardi, M.Y.; Nazari, M.; Babakhanzadeh, E.; Ghadimi, H.; Zeinali, F.; Talebi, M. Evaluation of miR-181b and miR-126-5p expression levels in T2DM patients compared to healthy individuals: Relationship with NF-κB gene expression. Endocrinol. Diabetes Nutr. 2020, 67, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanyam, M.; Aravind, S.; Gokulakrishnan, K.; Prabu, P.; Sathishkumar, C.; Ranjani, H.; Mohan, V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol. Cell. Biochem. 2011, 351, 197–205. [Google Scholar] [CrossRef]
- Mazloom, H.; Alizadeh, S.; Pasalar, P.; Esfahani, E.N.; Meshkani, R. Downregulated microRNA-155 expression in peripheral blood mononuclear cells of type 2 diabetic patients is not correlated with increased inflammatory cytokine production. Cytokine 2015, 76, 403–408. [Google Scholar] [CrossRef]
- Luo, M.; Li, R.; Deng, X.; Ren, M.; Chen, N.; Zeng, M.; Yan, K.; Xia, J.; Liu, F.; Ma, W.; et al. Platelet-derived miR-103b as a novel biomarker for the early diagnosis of type 2 diabetes. Acta Diabetol. 2015, 52, 943–949. [Google Scholar] [CrossRef]
- Baldeón Rojas, L.; Weigelt, K.; de Wit, H.; Ozcan, B.; van Oudenaren, A.; Sempértegui, F.; Sijbrands, E.; Grosse, L.; van Zonneveld, A.-J.; Drexhage, H.A.; et al. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol. Metab. Syndr. 2016, 8, 6. [Google Scholar] [CrossRef]
- Wang, S.-S.; Li, Y.-Q.; Liang, Y.-Z.; Dong, J.; He, Y.; Zhang, L.; Yan, Y.-X. Expression of miR-18a and miR-34c in circulating monocytes associated with vulnerability to type 2 diabetes mellitus and insulin resistance. J. Cell. Mol. Med. 2017, 21, 3372–3380. [Google Scholar] [CrossRef] [Green Version]
- Corral-Fernández, N.E.; Salgado-Bustamante, M.; Martínez-Leija, M.E.; Cortez-Espinosa, N.; García-Hernández, M.H.; Reynaga-Hernández, E.; Quezada-Calvillo, R.; Portales-Pérez, D.P. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Tong, A.; Xu, Y. Pancreatic β-cell function is inhibited by miR-3666 in type 2 diabetes mellitus by targeting adiponectin. Brazilian J. Med. Biol. Res. 2019, 52, e8344. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Fu, X.; Si, M.; Wang, Y.; Ma, R.; Ren, X.; Lv, H. MicroRNA-185 Targets SOCS3 to Inhibit Beta-Cell Dysfunction in Diabetes. PLoS ONE 2015, 10, e0116067. [Google Scholar] [CrossRef]
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef] [Green Version]
- Li, M.Y.; Pan, S.R.; Qiu, A.Y. Roles of microRNA-221/222 in type 2 diabetic patients with post-menopausal breast cancer. Genet. Mol. Res. 2016, 15, gmr.15027259. [Google Scholar] [CrossRef]
- Wan, S.; Wang, J.; Wang, J.; Wu, J.; Song, J.; Zhang, C.-Y.; Zhang, C.; Wang, C.; Wang, J.-J. Increased serum miR-7 is a promising biomarker for type 2 diabetes mellitus and its microvascular complications. Diabetes Res. Clin. Pract. 2017, 130, 171–179. [Google Scholar] [CrossRef]
- Shao, Y.; Ren, H.; Lv, C.; Ma, X.; Wu, C.; Wang, Q. Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine 2017, 55, 130–138. [Google Scholar] [CrossRef]
- De Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Yao, F.; Lang, Y.; Feng, X. Elevated Circulating LINC-P21 Serves as a Diagnostic Biomarker of Type 2 Diabetes Mellitus and Regulates Pancreatic β-cell Function by Sponging miR-766-3p to Upregulate NR3C2. Exp. Clin. Endocrinol. Diabetes 2022, 130, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Al-Kafaji, G.; Al-Mahroos, G.; Alsayed, N.A.; Hasan, Z.A.; Nawaz, S.; Bakhiet, M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol. Med. Rep. 2015, 12, 7485–7490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amr, K.S.; Abdelmawgoud, H.; Ali, Z.Y.; Shehata, S.; Raslan, H.M. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br. J. Biomed. Sci. 2018, 75, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, G.; Xu, C.; Zeng, M.; Lin, F.; Wu, J.; Wan, Q. Circulating miR-30c as a predictive biomarker of type 2 diabetes mellitus with coronary heart disease by regulating PAI-1/VN interactions. Life Sci. 2019, 239, 117092. [Google Scholar] [CrossRef]
- Luo, M.; Xu, C.; Luo, Y.; Wang, G.; Wu, J.; Wan, Q. Circulating miR-103 family as potential biomarkers for type 2 diabetes through targeting CAV-1 and SFRP4. Acta Diabetol. 2020, 57, 309–322. [Google Scholar] [CrossRef]
- Oraby, H.E.; Elshaer, S.S.; Rashed, L.A.; Eldesoky, N.A. MicroRNA-499 Gene Expression in Egyptian Type 2 Diabetes Mellitus Patients with and without Coronary Heart Disease. Azhar Int. J. Pharm. Med. Sci. 2022, 2, 73–81. [Google Scholar] [CrossRef]
- Yazdanpanah, Z.; Kazemipour, N.; Kalantar, S.M.; Vahidi Mehrjardi, M.Y. Plasma miR-21 as a potential predictor in prediabetic individuals with a positive family history of type 2 diabetes mellitus. Physiol. Rep. 2022, 10, e15163. [Google Scholar] [CrossRef]
- La Sala, L.; Mrakic-Sposta, S.; Tagliabue, E.; Prattichizzo, F.; Micheloni, S.; Sangalli, E.; Specchia, C.; Uccellatore, A.C.; Lupini, S.; Spinetti, G.; et al. Circulating microRNA-21 is an early predictor of ROS-mediated damage in subjects with high risk of developing diabetes and in drug-naïve T2D. Cardiovasc. Diabetol. 2019, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Monfared, Y.K.; Honardoost, M.; Sarookhani, M.R.; Farzam, S.A. Circulating miR-135 May Serve as a Novel Co-biomarker of HbA1c in Type 2 Diabetes. Appl. Biochem. Biotechnol. 2020, 191, 623–630. [Google Scholar] [CrossRef]
- Saeidi, L.; Shahrokhi, S.Z.; Sadatamini, M.; Jafarzadeh, M.; Kazerouni, F. Can circulating miR-7-1-5p, and miR-33a-5p be used as markers of T2D patients? Arch. Physiol. Biochem. 2021, 1–7. [Google Scholar] [CrossRef]
- Al-Muhtaresh, H.A.; Al-Kafaji, G. Evaluation of Two-Diabetes Related microRNAs Suitability as Earlier Blood Biomarkers for Detecting Prediabetes and type 2 Diabetes Mellitus. J. Clin. Med. 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Weale, C.J.; Matshazi, D.M.; Davids, S.F.G.; Raghubeer, S.; Erasmus, R.T.; Kengne, A.P.; Davison, G.M.; Matsha, T.E. Expression Profiles of Circulating microRNAs in South African Type 2 Diabetic Individuals on Treatment. Front. Genet. 2021, 12, 702410. [Google Scholar] [CrossRef] [PubMed]
- Seleem, M.; Shabayek, M.; Ewida, H.A. MicroRNAs 342 and 450 together with NOX-4 activity and their association with coronary artery disease in diabetes. Diabetes. Metab. Res. Rev. 2019, 35, e3130. [Google Scholar] [CrossRef] [PubMed]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.-I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Roy, S.; Dhas, Y.; Mishra, N. Senescence-associated miR-34a and miR-126 in middle-aged Indians with type 2 diabetes. Clin. Exp. Med. 2020, 20, 149–158. [Google Scholar] [CrossRef]
- Shen, Y.; Xu, H.; Pan, X.; Wu, W.; Wang, H.; Yan, L.; Zhang, M.; Liu, X.; Xia, S.; Shao, Q. miR-34a and miR-125b are upregulated in peripheral blood mononuclear cells from patients with type 2 diabetes mellitus. Exp. Ther. Med. 2017, 14, 5589–5596. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Zhan, Q.; Yuan, M.; Duan, X.; Zhou, J.; Lu, J.; Li, Z.; Yu, F.; Zhou, X.; Yang, Q.; et al. The Expression of microRNA-223 and FAM5C in Cerebral Infarction Patients with Diabetes Mellitus. Cardiovasc. Toxicol. 2017, 17, 42–48. [Google Scholar] [CrossRef]
- Yan, L.-N.; Zhang, X.; Xu, F.; Fan, Y.-Y.; Ge, B.; Guo, H.; Li, Z.-L. Four-microRNA signature for detection of type 2 diabetes. World J. Clin. Cases 2020, 8, 1923–1931. [Google Scholar] [CrossRef]
- Sadeghzadeh, S.; Dehghani Ashkezari, M.; Seifati, S.M.; Vahidi Mehrjardi, M.Y.; Dehghan Tezerjani, M.; Sadeghzadeh, S.; Ladan, S.A.B. Circulating miR-15a and miR-222 as Potential Biomarkers of Type 2 Diabetes. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 3461–3469. [Google Scholar] [CrossRef]
- Meerson, A.; Najjar, A.; Saad, E.; Sbeit, W.; Barhoum, M.; Assy, N. Sex Differences in Plasma MicroRNA Biomarkers of Early and Complicated Diabetes Mellitus in Israeli Arab and Jewish Patients. Non-Coding RNA 2019, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Sundquist, J.; Zöller, B.; Memon, A.A.; Palmér, K.; Sundquist, K.; Bennet, L. Determination of 14 Circulating microRNAs in Swedes and Iraqis with and without Diabetes Mellitus Type 2. PLoS ONE 2014, 9, e86792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šimonienė, D.; Stukas, D.; Daukša, A.; Veličkienė, D. Clinical Role of Serum miR107 in Type 2 Diabetes and Related Risk Factors. Biomolecules 2022, 12, 558. [Google Scholar] [CrossRef] [PubMed]
- Montalescot, G.; Sechtem, U.; Achenbach, S.; Andreotti, F.; Arden, C.; Budaj, A.; Bugiardini, R.; Crea, F.; Cuisset, T.; Di Mario, C.; et al. 2013 ESC guidelines on the management of stable coronary artery disease: The Task Force on the management of stable coronary artery disease of the European Society of Cardiology. Eur. Heart J. 2013, 34, 2949–3003. [Google Scholar] [CrossRef] [PubMed]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, R.; Pavasini, R.; Censi, S.; Squeri, A.; Rosano, G. The New ESC Guidelines for the Diagnosis and Management of Chronic Coronary Syndromes: The Good and the Not So Good. Curr. Probl. Cardiol. 2021, 46, 100554. [Google Scholar] [CrossRef]
- Bertolone, D.T.; Gallinoro, E.; Esposito, G.; Paolisso, P.; Bermpeis, K.; De Colle, C.; Fabbricatore, D.; Mileva, N.; Valeriano, C.; Munhoz, D.; et al. Contemporary Management of Stable Coronary Artery Disease. High Blood Press. Cardiovasc. Prev. 2022, 29, 207–219. [Google Scholar] [CrossRef]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Röxe, T.; Müller-Ardogan, M.; et al. Circulating MicroRNAs in Patients With Coronary Artery Disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [Green Version]
- Sanlialp, M.; Dodurga, Y.; Uludag, B.; Alihanoglu, Y.I.; Enli, Y.; Secme, M.; Bostanci, H.E.; Cetin Sanlialp, S.; Tok, O.O.; Kaftan, A.; et al. Peripheral blood mononuclear cell microRNAs in coronary artery disease. J. Cell. Biochem. 2020, 121, 3005–3009. [Google Scholar] [CrossRef]
- Qiu, X.-K.; Ma, J. Alteration in microRNA-155 level correspond to severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2018, 78, 219–223. [Google Scholar] [CrossRef]
- Chang, S.-N.; Chen, J.-J.; Wu, J.-H.; Chung, Y.-T.; Chen, J.-W.; Chiu, C.-H.; Liu, C.-J.; Liu, M.-T.; Chang, Y.-C.; Li, C.; et al. Association between Exosomal miRNAs and Coronary Artery Disease by Next-Generation Sequencing. Cells 2021, 11, 98. [Google Scholar] [CrossRef]
- Wang, X.; Dong, Y.; Fang, T.; Wang, X.; Chen, L.; Zheng, C.; Kang, Y.; Jiang, L.; You, X.; Gai, S.; et al. Circulating MicroRNA-423-3p Improves the Prediction of Coronary Artery Disease in a General Population―Six-Year Follow-up Results from the China-Cardiovascular Disease Study. Circ. J. 2020, 84, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cai, H.; Zhu, M.; Qian, Y.; Lin, S.; Li, X. Circulating microRNAs as biomarkers for severe coronary artery disease. Medicine 2020, 99, e19971. [Google Scholar] [CrossRef] [PubMed]
- Faccini, J.; Ruidavets, J.-B.; Cordelier, P.; Martins, F.; Maoret, J.-J.; Bongard, V.; Ferrières, J.; Roncalli, J.; Elbaz, M.; Vindis, C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017, 7, 42916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Yang, S.H.; Li, S.; Cui, C.J.; Zhang, Y.; Zhu, C.G.; Guo, Y.L.; Wu, N.Q.; Gao, Y.; Sun, J.; et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (≤40 years) Coronary Artery Disease. Biomed. Environ. Sci. 2016, 29, 545–554. [Google Scholar] [CrossRef]
- Zhou, J.; Shao, G.; Chen, X.; Yang, X.; Huang, X.; Peng, P.; Ba, Y.; Zhang, L.; Jehangir, T.; Bu, S.; et al. miRNA 206 and miRNA 574-5p are highly expression in coronary artery disease. Biosci. Rep. 2016, 36, e00295. [Google Scholar] [CrossRef]
- Han, H.; Qu, G.; Han, C.; Wang, Y.; Sun, T.; Li, F.; Wang, J.; Luo, S. MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: A pilot microarray study and confirmation in a 32 patient cohort. Exp. Mol. Med. 2015, 47, e138. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Pei, Y.; Zhong, Y.; Jiang, S.; Shao, J.; Gong, J. Altered Serum MicroRNAs as Novel Diagnostic Biomarkers for Atypical Coronary Artery Disease. PLoS ONE 2014, 9, e107012. [Google Scholar] [CrossRef]
- D’Alessandra, Y.; Carena, M.C.; Spazzafumo, L.; Martinelli, F.; Bassetti, B.; Devanna, P.; Rubino, M.; Marenzi, G.; Colombo, G.I.; Achilli, F.; et al. Diagnostic Potential of Plasmatic MicroRNA Signatures in Stable and Unstable Angina. PLoS ONE 2013, 8, e80345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sondermeijer, B.M.; Bakker, A.; Halliani, A.; de Ronde, M.W.J.; Marquart, A.A.; Tijsen, A.J.; Mulders, T.A.; Kok, M.G.M.; Battjes, S.; Maiwald, S.; et al. Platelets in Patients with Premature Coronary Artery Disease Exhibit Upregulation of miRNA340* and miRNA624*. PLoS ONE 2011, 6, e25946. [Google Scholar] [CrossRef]
- Gholipour, A.; Shakerian, F.; Zahedmehr, A.; Irani, S.; Mowla, S.J.; Malakootian, M. Downregulation of Talin-1 is associated with the increased expression of miR-182-5p and miR-9-5p in coronary artery disease. J. Clin. Lab. Anal. 2022, 36, e24252. [Google Scholar] [CrossRef]
- Ali, W.; Mishra, S.; Rizvi, A.; Pradhan, A.; Perrone, M.A. Circulating microRNA-126 as an Independent Risk Predictor of Coronary Artery Disease: A Case-Control Study. EJIFCC 2021, 32, 347–362. [Google Scholar]
- Kumar, D.; Narang, R.; Sreenivas, V.; Rastogi, V.; Bhatia, J.; Saluja, D.; Srivastava, K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes 2020, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liang, T.; Chen, Y.; Wang, X.; Wu, T.; Xie, Z.; Luo, J.; Yu, Y.; Yu, H. Circulating Exosomal miRNAs as Novel Biomarkers for Stable Coronary Artery Disease. Biomed Res. Int. 2020, 2020, 3593962. [Google Scholar] [CrossRef]
- Guo, J.-F.; Zhang, Y.; Zheng, Q.-X.; Zhang, Y.; Zhou, H.-H.; Cui, L.-M. Association between elevated plasma microRNA-223 content and severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2018, 78, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.G.; Isbir, S.; Kunt, A.T.; Isbir, T. Circulating microRNAs as Novel Biomarkers for Atherosclerosis. In Vivo 2018, 32, 561–565. [Google Scholar] [CrossRef]
- O′Sullivan, J.F.; Neylon, A.; McGorrian, C.; Blake, G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016, 224, 310–316. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Xue, S.; Ding, H.; Wang, Y.; Qi, H.; Wang, Y.; Zhu, W.; Li, P. Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J. Cell. Mol. Med. 2020, 24, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Hao, J.; Sun, X.; Zhang, Y.; Wei, Q. Circulating pro-angiogenic micro-ribonucleic acid in patients with coronary heart disease. Interact. Cardiovasc. Thorac. Surg. 2018, 27, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Liang, Y.-Z.; Zhang, J.; Wu, L.-J.; Wang, S.; Hua, Q.; Yan, Y.-X. Potential Role of Lipometabolism-Related MicroRNAs in Peripheral Blood Mononuclear Cells as Biomarkers for Coronary Artery Disease. J. Atheroscler. Thromb. 2017, 24, 430–441. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yang, N.; Fei, Z.; Qiu, J.; Ma, D.; Liu, X.; Cai, G.; Li, S. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. Biomed. Rep. 2016, 5, 332–336. [Google Scholar] [CrossRef] [Green Version]
- Madhavan, M.V.; Gersh, B.J.; Alexander, K.P.; Granger, C.B.; Stone, G.W. Coronary Artery Disease in Patients ≥80 Years of Age. J. Am. Coll. Cardiol. 2018, 71, 2015–2040. [Google Scholar] [CrossRef] [PubMed]
- Ali Sheikh, M.S.; Xia, K.; Li, F.; Deng, X.; Salma, U.; Deng, H.; Wei, L.; Yang, T.-L.; Peng, J. Circulating miR-765 and miR-149: Potential Noninvasive Diagnostic Biomarkers for Geriatric Coronary Artery Disease Patients. Biomed Res. Int. 2015, 2015, 740301. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Jiang, J.; Zhou, R.; Li, X.; Ye, J. MicroRNA-451b participates in coronary heart disease by targeting VEGFA. Open Med. 2019, 15, 1105–1111. [Google Scholar] [CrossRef]
- Ali Sheikh, M.S.; Xia, K.; Li, F.; Deng, X.; Salma, U.; Li, T.; Deng, H.; Yang, D.; Haoyang, Z.; Yang, T.-L.; et al. The diagnostic value of circulating microRNAs for middle-aged (40–60-year-old) coronary artery disease patients. Clinics 2015, 70, 257–263. [Google Scholar] [CrossRef]
- Lin, D.-C.; Lin, J.-B.; Chen, Z.; Chen, R.; Wan, C.-Y.; Lin, S.-W.; Ruan, Q.-S.; Li, H.-Y.; Wu, S.-Y. Independent and combined effects of environmental factors and miR-126, miR-143, and miR-145 on the risk of coronary heart disease. J. Geriatr. Cardiol. 2017, 14, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Tu, Y.-F.; Liu, H.-M.; Xu, M.-E. Diagnostic utility of circulating plasma microRNA-101a in severity of coronary heart disease. Ir. J. Med. Sci. 2021, 190, 1391–1396. [Google Scholar] [CrossRef]
- Arora, M.; Kaul, D.; Sharma, Y.P. Human coronary heart disease: Importance of blood cellular miR-2909 RNomics. Mol. Cell. Biochem. 2014, 392, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.-H.; Yang, R.; Yang, B.-J.; Gao, Z.-Y. Association between circulating microRNA-208a and severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2017, 77, 379–384. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, J. miR-126 in Peripheral Blood Mononuclear Cells Negatively Correlates with Risk and Severity and is Associated with Inflammatory Cytokines as well as Intercellular Adhesion Molecule-1 in Patients with Coronary Artery Disease. Cardiology 2018, 139, 110–118. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Hu, Q.; Yang, S.; Zhang, B.; Jiang, H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015, 197, 123–124. [Google Scholar] [CrossRef]
- Wang, M.; Li, J.; Cai, J.; Cheng, L.; Wang, X.; Xu, P.; Li, G.; Liang, X. Overexpression of MicroRNA-16 Alleviates Atherosclerosis by Inhibition of Inflammatory Pathways. Biomed Res. Int. 2020, 2020, 8504238. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, M.; Feng, Q.; Zhu, L.; Bai, Z.; Wang, B.; Guo, Z.; Hou, A.; Li, H. MicroRNA-34a in coronary heart disease: Correlation with disease risk, blood lipid, stenosis degree, inflammatory cytokines, and cell adhesion molecules. J. Clin. Lab. Anal. 2022, 36, e24138. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gao, F.; Wang, X.; Wu, J.; Lu, K.; Liu, M.; Li, R.; Ding, L.; Wang, R. Circulating microRNA-378 levels serve as a novel biomarker for assessing the severity of coronary stenosis in patients with coronary artery disease. Biosci. Rep. 2019, 39, BSR20182016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L. The correlations of circulating microRNA-133a with the risk and severity of coronary heart disease. Int. J. Clin. Exp. Med. 2017, 10, 972–978. [Google Scholar]
- Li, H.-Y.; Zhao, X.; Liu, Y.-Z.; Meng, Z.; Wang, D.; Yang, F.; Shi, Q.-W. Plasma MicroRNA-126-5p is Associated with the Complexity and Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Cell. Physiol. Biochem. 2016, 39, 837–846. [Google Scholar] [CrossRef]
- Gao, H.; Guddeti, R.R.; Matsuzawa, Y.; Liu, L.-P.; Su, L.-X.; Guo, D.; Nie, S.-P.; Du, J.; Zhang, M. Plasma Levels of microRNA-145 Are Associated with Severity of Coronary Artery Disease. PLoS ONE 2015, 10, e0123477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, L.L.; Shah, S.A.V.; Ponde, C.K.; Rajani, R.M.; Ashavaid, T.F. Circulating miRNA-33: A potential biomarker in patients with coronary artery disease. Biomarkers 2019, 24, 36–42. [Google Scholar] [CrossRef]
- Hosseinpor, S.; Khalvati, B.; Safari, F.; Mirzaei, A.; Hosseini, E. The association of plasma levels of miR-146a, miR-27a, miR-34a, and miR-149 with coronary artery disease. Mol. Biol. Rep. 2022, 49, 3559–3567. [Google Scholar] [CrossRef]
- Babaee, M.; Chamani, E.; Ahmadi, R.; Bahreini, E.; Balouchnejadmojarad, T.; Nahrkhalaji, A.S.; Fallah, S. The expression levels of miRNAs- 27a and 23a in the peripheral blood mononuclear cells (PBMCs) and their correlation with FOXO1 and some inflammatory and anti-inflammatory cytokines in the patients with coronary artery disease (CAD). Life Sci. 2020, 256, 117898. [Google Scholar] [CrossRef]
- Zehtabian, S.H.; Alibakhshi, R.; Seyedena, S.Y.; Rai, A.R. Relationship between microRNA-206 plasma levels with the severity of coronary artery conflicts in patients with coronary artery disease. Bratisl. Med. J. 2019, 120, 581–585. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Rizvi, A.; Pradhan, A.; Perrone, M.A.; Ali, W. Circulating microRNA-126 &122 in patients with coronary artery disease: Correlation with small dense LDL. Prostaglandins Other Lipid Mediat. 2021, 153, 106536. [Google Scholar] [CrossRef] [PubMed]
- Gorur, A.; Celik, A.; Yildirim, D.D.; Gundes, A.; Tamer, L. Investigation of possible effects of microRNAs involved in regulation of lipid metabolism in the pathogenesis of atherosclerosis. Mol. Biol. Rep. 2019, 46, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Zhang, S.; Yan, S.; Wang, Z.; Wang, C.; Zhang, X. MiR-30e and miR-92a are related to atherosclerosis by targeting ABCA1. Mol. Med. Rep. 2019, 19, 3298–3304. [Google Scholar] [CrossRef] [PubMed]
- Choteau, S.A.; Cuesta Torres, L.F.; Barraclough, J.Y.; Elder, A.M.M.; Martínez, G.J.; Chen Fan, W.Y.; Shrestha, S.; Ong, K.L.; Barter, P.J.; Celermajer, D.S.; et al. Transcoronary gradients of HDL-associated MicroRNAs in unstable coronary artery disease. Int. J. Cardiol. 2018, 253, 138–144. [Google Scholar] [CrossRef]
- Ahmadi, R.; Heidarian, E.; Fadaei, R.; Moradi, N.; Malek, M.; Fallah, S. miR-342-5p Expression Levels in Coronary Artery Disease Patients and its Association with Inflammatory Cytokines. Clin. Lab. 2018, 64, 603–609. [Google Scholar] [CrossRef]
- De Rosa, S.; Arcidiacono, B.; Chiefari, E.; Brunetti, A.; Indolfi, C.; Foti, D.P. Type 2 Diabetes Mellitus and Cardiovascular Disease: Genetic and Epigenetic Links. Front. Endocrinol. 2018, 9, 2. [Google Scholar] [CrossRef]
- Vesa, C.M.; Popa, L.; Popa, A.R.; Rus, M.; Zaha, A.A.; Bungau, S.; Tit, D.M.; Corb Aron, R.A.; Zaha, D.C. Current Data Regarding the Relationship between Type 2 Diabetes Mellitus and Cardiovascular Risk Factors. Diagnostics 2020, 10, 314. [Google Scholar] [CrossRef]
- Merino, J.; Jablonski, K.A.; Mercader, J.M.; Kahn, S.E.; Chen, L.; Harden, M.; Delahanty, L.M.; Araneta, M.R.G.; Walford, G.A.; Jacobs, S.B.R.; et al. Interaction Between Type 2 Diabetes Prevention Strategies and Genetic Determinants of Coronary Artery Disease on Cardiometabolic Risk Factors. Diabetes 2020, 69, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Jansen, F.; Wang, H.; Przybilla, D.; Franklin, B.S.; Dolf, A.; Pfeifer, P.; Schmitz, T.; Flender, A.; Endl, E.; Nickenig, G.; et al. Vascular endothelial microparticles-incorporated microRNAs are altered in patients with diabetes mellitus. Cardiovasc. Diabetol. 2016, 15, 49. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-Y.; Zheng, Y.-S.; Li, Z.-G.; Cui, Y.-M.; Jiang, J.-C. MiR-92a contributes to the cardiovascular disease development in diabetes mellitus through NF-κB and downstream inflammatory pathways. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3070–3079. [Google Scholar] [CrossRef]
- Al-Muhtaresh, H.A.; Salem, A.H.; Al-Kafaji, G. Upregulation of Circulating Cardiomyocyte-Enriched miR-1 and miR-133 Associate with the Risk of Coronary Artery Disease in Type 2 Diabetes Patients and Serve as Potential Biomarkers. J. Cardiovasc. Transl. Res. 2019, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Motawae, T.M.; Ismail, M.F.; Shabayek, M.I.; Seleem, M.M. MicroRNAs 9 and 370 Association with Biochemical Markers in T2D and CAD Complication of T2D. PLoS ONE 2015, 10, e0126957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Qin, Z.; Liu, N.; Zhang, Z.; Lu, Y.; Xu, Y.; Zhang, J.; Tang, J. Diagnostic and Predictive Values of Circulating Extracellular Vesicle-Carried microRNAs in Ischemic Heart Disease Patients With Type 2 Diabetes Mellitus. Front. Cardiovasc. Med. 2022, 9, 813310. [Google Scholar] [CrossRef] [PubMed]
- Zogg, H.; Singh, R.; Ro, S. Current Advances in RNA Therapeutics for Human Diseases. Int. J. Mol. Sci. 2022, 23, 2736. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-E.; Fu, T.; Seok, S.; Kim, D.-H.; Yu, E.; Lee, K.-W.; Kang, Y.; Li, X.; Kemper, B.; Kemper, J.K. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 2013, 12, 1062–1072. [Google Scholar] [CrossRef] [Green Version]
- Bijkerk, R.; Esguerra, J.L.S.; Ellenbroek, J.H.; Au, Y.W.; Hanegraaf, M.A.J.; de Koning, E.J.; Eliasson, L.; van Zonneveld, A.J. In Vivo Silencing of MicroRNA-132 Reduces Blood Glucose and Improves Insulin Secretion. Nucleic Acid Ther. 2019, 29, 67–72. [Google Scholar] [CrossRef]
- Hung, Y.-H.; Kanke, M.; Kurtz, C.L.; Cubitt, R.; Bunaciu, R.P.; Miao, J.; Zhou, L.; Graham, J.L.; Hussain, M.M.; Havel, P.; et al. Acute suppression of insulin resistance-associated hepatic miR-29 in vivo improves glycemic control in adult mice. Physiol. Genomics 2019, 51, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Li, C.; Qi, W.; Zhang, Y.; Zhang, F.; Wu, J.X.; Hu, Y.N.; Wu, D.M.; Liu, Y.; Yan, T.T.; et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012, 55, 2032–2043. [Google Scholar] [CrossRef] [Green Version]
- Thibonnier, M.; Esau, C.; Ghosh, S.; Wargent, E.; Stocker, C. Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care 2020, 8, e001478. [Google Scholar] [CrossRef]
- Thibonnier, M.; Esau, C. Metabolic Benefits of MicroRNA-22 Inhibition. Nucleic Acid Ther. 2020, 30, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Fischer, A.; Muhly-Reinholz, M.; Zeiher, A.M.; Dimmeler, S. Long-term Inhibition of miR-21 Leads to Reduction of Obesity in db/db Mice. Obesity 2014, 22, 2352–2360. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.-Z.; Yang, D.-G.; Zhang, Q.-Y.; Deng, Z.-N.; Wang, K.; Mao, X.-J. MiR-21 antagomir improves insulin resistance and lipid metabolism disorder in streptozotocin-induced type 2 diabetes mellitus rats. Ann. Palliat. Med. 2020, 9, 394–404. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Yin, L.; Qi, Y.; Sun, H.; Sun, P.; Xu, M.; Tang, Z.; Peng, J. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics 2018, 8, 5593–5609. [Google Scholar] [CrossRef]
- Liu, R.; Wang, M.; Li, E.; Yang, Y.; Li, J.; Chen, S.; Shen, W.-J.; Azhar, S.; Guo, Z.; Hu, Z. Dysregulation of microRNA-125a contributes to obesity-associated insulin resistance and dysregulates lipid metabolism in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158640. [Google Scholar] [CrossRef]
- Cheng, X.; Huang, Y.; Yang, P.; Bu, L. miR-383 ameliorates high glucose-induced β-cells apoptosis and hyperglycemia in high-fat induced diabetic mice. Life Sci. 2020, 263, 118571. [Google Scholar] [CrossRef]
- Tian, F.-J.; An, L.-N.; Wang, G.-K.; Zhu, J.-Q.; Li, Q.; Zhang, Y.-Y.; Zeng, A.; Zou, J.; Zhu, R.-F.; Han, X.-S.; et al. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc. Res. 2014, 103, 100–110. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, L.; Liang, X.; Zhu, G. MicroRNA-155 Promotes Atherosclerosis Inflammation via Targeting SOCS1. Cell. Physiol. Biochem. 2015, 36, 1371–1381. [Google Scholar] [CrossRef]
- Wei, Y.; Nazari-Jahantigh, M.; Chan, L.; Zhu, M.; Heyll, K.; Corbalán-Campos, J.; Hartmann, P.; Thiemann, A.; Weber, C.; Schober, A. The microRNA-342-5p Fosters Inflammatory Macrophage Activation Through an Akt1- and microRNA-155–Dependent Pathway During Atherosclerosis. Circulation 2013, 127, 1609–1619. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.-G.; Chen, B.-Y.; Sun, R.-H.; Mou, X.-Z.; Han, F.; Li, Q.; Huang, H.-J.; Liu, J.-Q.; Tu, Y.-X. miR-133b Downregulation Reduces Vulnerable Plaque Formation in Mice with AS through Inhibiting Macrophage Immune Responses. Mol. Ther. Nucleic Acids 2019, 16, 745–757. [Google Scholar] [CrossRef] [Green Version]
- Loyer, X.; Potteaux, S.; Vion, A.-C.; Guérin, C.L.; Boulkroun, S.; Rautou, P.-E.; Ramkhelawon, B.; Esposito, B.; Dalloz, M.; Paul, J.-L.; et al. Inhibition of MicroRNA-92a Prevents Endothelial Dysfunction and Atherosclerosis in Mice. Circ. Res. 2014, 114, 434–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Chen, X.; Xu, B.; Luo, Z.; Liang, Y.; Fang, S.; Li, M.; Wang, X.; Lin, X. Inhibition of MicroRNA-92 alleviates atherogenesis by regulation of macrophage polarization through targeting KLF4. J. Cardiol. 2022, 79, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science. 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and reduces VLDL triglycerides. Nature 2011, 478, 404–407. [Google Scholar] [CrossRef] [Green Version]
- Rayner, K.J.; Sheedy, F.J.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van Gils, J.M.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef] [Green Version]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33–dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015, 125, 4334–4348. [Google Scholar] [CrossRef] [Green Version]
- Goedeke, L.; Salerno, A.; Ramírez, C.M.; Guo, L.; Allen, R.M.; Yin, X.; Langley, S.R.; Esau, C.; Wanschel, A.; Fisher, E.A.; et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol. Med. 2014, 6, 1133–1141. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Goedeke, L.; Rotllan, N.; Canfrán-Duque, A.; Aranda, J.F.; Ramírez, C.M.; Araldi, E.; Lin, C.-S.; Anderson, N.N.; Wagschal, A.; de Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef] [Green Version]
- Soh, J.; Iqbal, J.; Queiroz, J.; Fernandez-Hernando, C.; Hussain, M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013, 19, 892–900. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.; Liu, L.; Jiang, Z.; Hao, X. Inhibition of microRNA-17-5p reduces the inflammation and lipid accumulation, and up-regulates ATP-binding cassette transporterA1 in atherosclerosis. J. Pharmacol. Sci. 2019, 139, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Yang, Y.; Yang, X.-Y.; Tong, Q. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb. Res. 2018, 171, 55–61. [Google Scholar] [CrossRef]
- Wang, J.; Li, P.; Xu, X.; Zhang, B.; Zhang, J. MicroRNA-200a Inhibits Inflammation and Atherosclerotic Lesion Formation by Disrupting EZH2-Mediated Methylation of STAT3. Front. Immunol. 2020, 11, 907. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Cao, Y.; Li, H.; Hu, Z.; Zhang, H.; Zhang, L.; Su, W.; Xu, Y.; Liang, L.; Melgiri, N.D.; et al. miR-532-3p-CSF2RA Axis as a Key Regulator of Vulnerable Atherosclerotic Plaque Formation. Can. J. Cardiol. 2020, 36, 1782–1794. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Hao, C.; Li, Z.; Zhang, P.; Wang, S.; Yang, S.; Wei, F.; Zhang, J. miR-449a induces EndMT, promotes the development of atherosclerosis by targeting the interaction between AdipoR2 and E-cadherin in Lipid Rafts. Biomed. Pharmacother. 2019, 109, 2293–2304. [Google Scholar] [CrossRef]
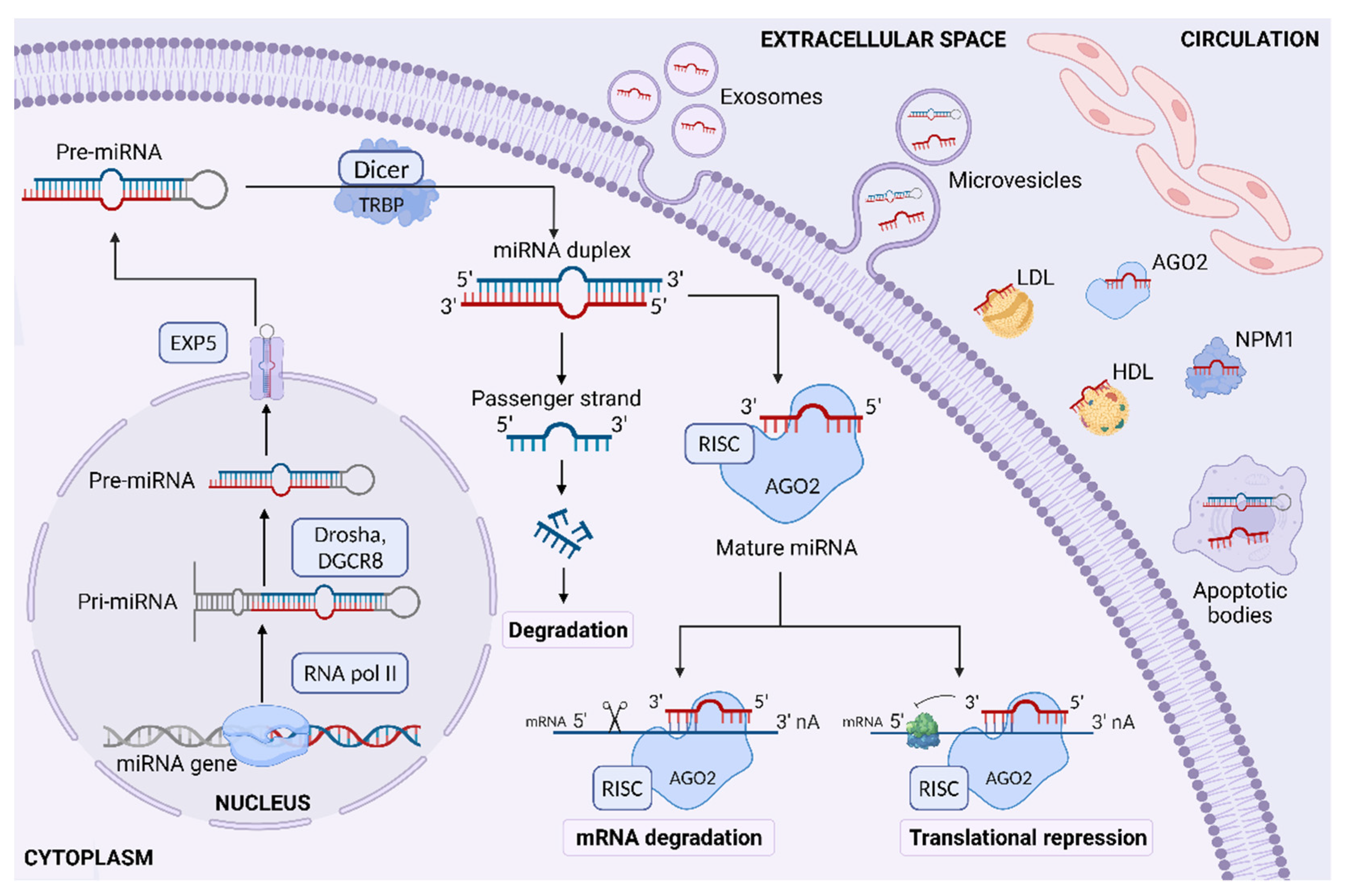

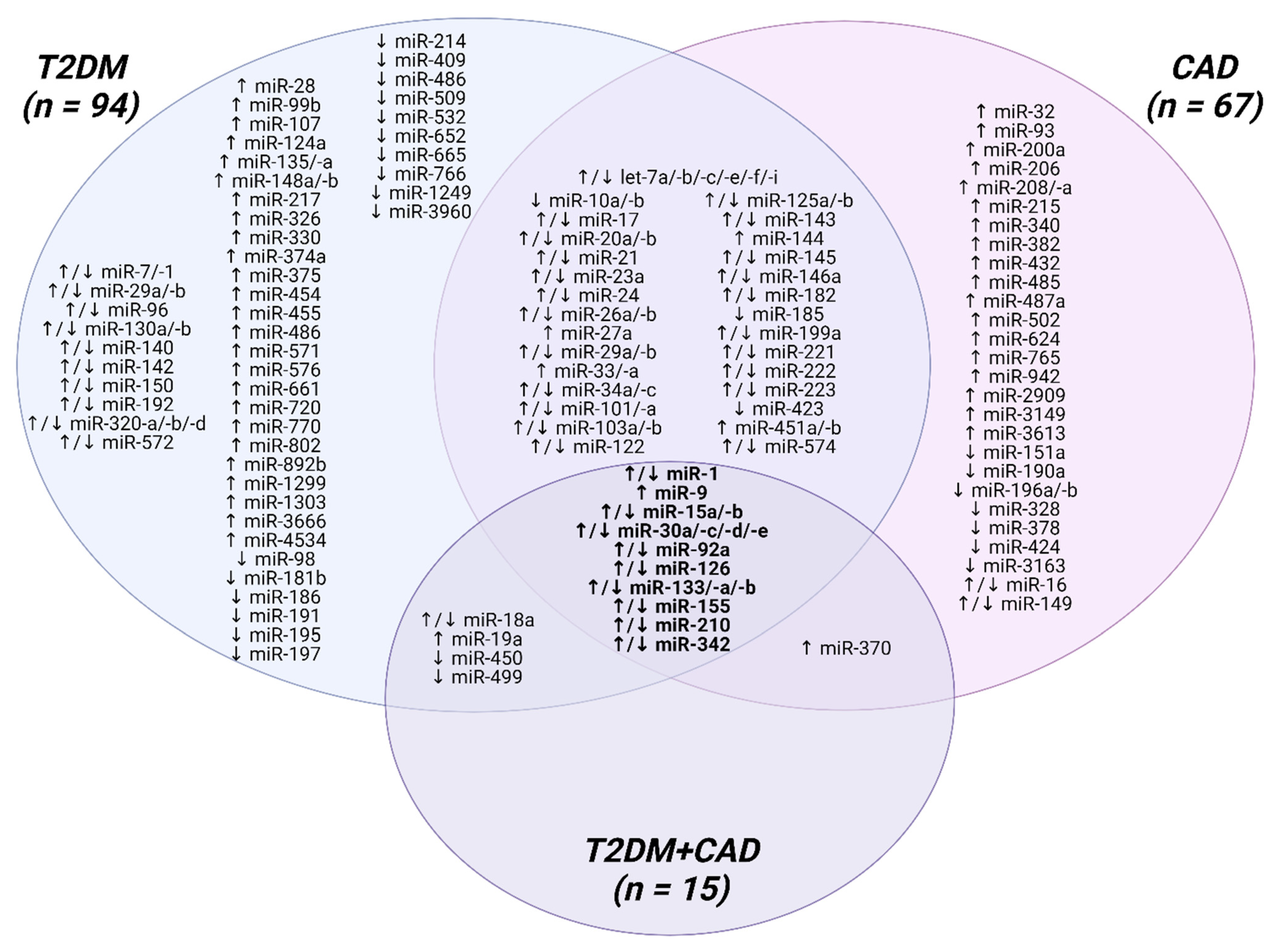
| miRNA | Expression Change | Sample Type | Assay Method | Number of Samples | Ethnicity | Age [Years] | Gender (Male/Female, n) | BMI [kg/m2] | Value of Biomarker AUC (95% CI); SV [%]; SP [%] | Author, Year (Reference) |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-9-5p | Up | Serum | RT-qPCR | CAD (40) HC (20) | Iranian | 42.36 ± 5.2 42.9 ± 4.6 | 19/21 4/16 | N/A | 0.693 (0.530–0.857) 74.07; 53.33 | Gholipour et al., 2022 [260] |
| miR-182-5p | 0.752 (0.608–0.897) 74.19; 66.67 | |||||||||
| miR-27a | Up | Plasma | qPCR | CAD (30) HC (30) | Iranian | 57.6 ± 20.32 55.30 ± 8.40 | Only male | 24.8–30.0 24.66–27.0 | 0.67 (0.54–0.81) 86.7; 46.7 | Hosseinpor et al., 2022 [288] |
| miR-146a | Down | N/A | ||||||||
| miR-34a | Up | Plasma | RT-qPCR | CAD (203) HC (100) | Chinese | 61.5 ± 9.4 62.0 ± 6.7 | 158/45 75/25 | 24.2 ± 3.0 23.8 ± 3.1 | 0.899 (0.865–0.934) 76.4; 90.0 | Li et al., 2022 [282] |
| miR-122 | Up | Serum | RT-qPCR | CAD (100) HC (100) | Indian | 52.15 ± 1.13 50.90 ± 2.08 | 75/25 74/26 | >26 (44.0%) >26 (33.0%) | 0.806 64.0; 84.0 | Ali et al., 2021 [261] |
| miR-126 | Down | 0.806 61.54; 80.0 | ||||||||
| miR-200a-3p, miR-382-3p, miR-432-5p, miR-3613-3p | Up | Plasma (exosomes) | NGS | CAD (52) HC (52) | Han Chinese | 65.04 ± 10.68 60.65 ± 11.26 | 43/9 34/18 | 26.87 ± 3.80 26.42 ± 3.57 | N/A | Chang et al., 2021 [250] |
| miR-125a-5p, miR-151a-3p, miR-185-5p, miR-328-3p | Down | |||||||||
| miR-122 | Down | Serum | RT-qPCR | CAD (78) HC (60) | Indian | 52.07 ± 9.94 50.13 ± 8.12 | N/A | 25.9 ± 4.8 25.2 ± 4.7 | N/A | Mishra et al., 2021 [291] |
| miR-101a | Down | Serum | RT-qPCR | CAD (200) HC (100) | Chinese | 62 (31–87) 1a 59 (37–80) 1c 60 (40–86) | 74/26 1a 78/22 1c 73/27 | N/A | N/A | Yu et al., 2021 [276] |
| miR-23a, miR-27a | Up | PBMC | qPCR | CAD (82) HC (80) | Iranian | 60.24 ± 0.91 57.40 ± 0.94 | 35/47 41/39 | 27.60 ± 0.48 26.23 ± 0.50 | N/A | Babaee et al., 2020 [289] |
| miR-21 | Up | Plasma | RT-qPCR | CAD (24) HC (54) | Indian | N/A | N/A | N/A | 0.780 (0.670–0.890) | Kumar et al., 2020 [262] |
| miR-133b | Down | CAD (28) HC (54) | 0.746 (0.620–0.870) | |||||||
| miR-21 | Up | PBMC | RT-qPCR | CAD (56) HC (29) | Turkish | 58.96 ± 8.95 56.93 ± 6.35 | 41/15 9/20 | N/A | N/A | Sanlialp et al., 2020 [248] |
| miR-155, miR-221 | Down | |||||||||
| miR-10a-5p | Down | Serum | sRNA-Seq, RT-qPCR | CAD (39) HC (39) | Chinese | 63.1 ± 7.4 59.3 ± 6.8 | 21/18 21/18 | 24.2 ± 2.3 23.2 ± 2.2 | 0.817 (0.715–0.918) | Wang et al., 2020 [251] |
| miR-423-3p | 0.656 (0.532–0.779) | |||||||||
| miR-423-3p | CAD (30) HC (21) | 63.9 ± 7.95 57.24 ± 5.35 | 15/15 9/12 | 24.57 ± 2.37 25.01 ± 2.02 | 0.808 (0.684–0.932) | |||||
| miR-16 | Down | Plasma, PBMC | RT-qPCR | CAD (40) HC (40) | Chinese | 63.33 ± 5.63 61.20 ± 5.82 | Only male | 26.69 ± 2.85 25.85 ± 3.12 | N/A | Wang et al., 2020 [281] |
| miR-29a-3p, miR-574-3p, miR-574-5p | Up | Plasma | qPCR | CAD (88) HC (67) | Chinese | 61.66 ± 1.32 63.72 ± 0.99 | 55/33 40/27 | 26.18 ± 0.50 24.89 ± 0.68 | 0.916 (0.856–0.957) 2 | Zhang et al., 2020 [267] |
| let-7i-5p, miR-26a-5p, miR-32-3p, miR-3149 | Up | Plasma | Microarray, RT-qPCR | CAD (40) HC (69) | Chinese | 56.2 ± 7.6 55.0 ± 6.5 | 30/10 43/26 | N/A | 0.837 (0.763–0.911) 2 | Zhang et al., 2020 [252] |
| miR-32-5p | Up | Serum (exosomes) | qPCR | CAD (20) HC (20) | Chinese | 64 (52–68) 57 (52–62) | 14/6 12/8 | 24.7 ± 2.9 24.0 ± 3.0 | 0.691 (0.525–0.858) 85.0; 55.0 | Zhang et al., 2020 [263] |
| miR-149-5p | 0.702 (0.536–0.869) 70.0; 75.0 | |||||||||
| miR-942-5p | 0.693 (0.527–0.858) 80.0; 60.0 | |||||||||
| miR-133a-5p, miR-144-3p, miR-222-5p | Up | Plasma | RT-qPCR | CAD (46) HC (43) | Turkish | 60.02 ± 10.01 55.26 ± 13.85 | 34/12 28/15 | 27.87 (25.04–30.43) 27.10 (24.38–29.42) | N/A | Gorur et al., 2019 [292] |
| miR-378 | Down | Plasma | RT-qPCR | CAD (215) HC (52) | Chinese | 61 ± 10 61 ± 12 | 153/62 30/22 | 23 ± 10 22 ± 10 | 0.789 (0.728–0.851) | Li et al., 2019 [283] |
| miR-451b | Up | Serum | RT-qPCR | CAD (30) HC (30) | Chinese | 46–59 45–58 | 15/15 15/15 | N/A | N/A | Lin et al., 2019 [273] |
| miR-30c | Down | Plasma | qPCR | CAD (34) HC (32) | Han Chinese | 60.9 ± 5.3 58.6 ± 8.1 | 18/16 17/15 | 24.57 ± 3.01 24.49 ± 2.30 | 0.895 (0.811–0.978) | Luo et al., 2019 [224] |
| miR-33 | Up | Plasma | RT-qPCR | CAD (30) HC (30) | Indian | 54.7 ± 8.7 56.17 ± 9.18 | 15/15 15/15 | 26.67 ± 3.7 27.47 ± 4.35 | N/A | Reddy et al., 2019 [287] |
| miR-30e, miR-92a | Up | Plasma (exosomes) | RT-qPCR | CAD (42) HC (42) | Chinese | 63 63 | 22/20 22/20 | N/A | N/A | Wang et al., 2019 [293] |
| miR-206 | Up | Plasma | qPCR | CAD (100) HC (30) | Iranian | 57 ± 9 55 ± 8 | 87/13 16/14 | 27.78 ± 3.45 27.45 ± 2.09 | N/A | Zehtabian et al., 2019 [290] |
| miR-342-5p | Up | PBMC | qPCR | CAD (82) HC (80) | Iranian | 60.10 ± 0.89 57.86 ± 0.97 | 35/47 41/39 | 27.56 ± 0.47 26.14 ± 0.49 | 0.702 (0.620–0.783) | Ahmadi et al., 2018 [295] |
| miR-20a, miR-92a, miR-223 | Up | Plasma | RT-qPCR | CAD (19) HC (6) | Australian | 65.2 ± 10.7 59.0 ± 5.1 | 19/0 5/1 | N/A | N/A | Choteau et al., 2018 [294] |
| miR-223 | Up | Plasma | RT-qPCR | CAD (300) HC (100) | Chinese | 56.2 | N/A | N/A | 0.933 (0.905–0.961)86.0; 91.3 | Guo et al., 2018 [264] |
| miR-155 | Up | Serum | RT-qPCR | CAD (300) HC (100) | Chinese | N/A | N/A | N/A | N/A | Qiu et al., 2018 [249] |
| miR-126 | Down | PBMC | qPCR | CAD (119) HC (96) | Chinese | 59 ± 11 57 ± 10 | 36/83 27/69 | 24.6 ± 3.9 23.8 ± 3.4 | 0.801 (0.740–0.861) 70.6; 85.4 | Wu at al., 2018 [279] |
| miR-221-3p | Down | Serum | qPCR | CAD (89) HC (93) | Turkish | 58.97 ± 13.79 57.07 ± 9.80 | N/A | NS | 0.623 (0.539–0.702) 76.27; 49.43 | Yilmaz et al., 2018 [265] |
| miR-222-3p | 0.654 (0.571–0.731) 69.49; 54.02 | |||||||||
| miR-17-5p, miR-92a, miR-126, miR-210, miR-378 | Down | Plasma | RT-qPCR | CAD (102) HC (92) | Chinese | 60.2 ± 11.4 57.9 ± 14.8 | 21/81 26/66 | 24.2 ± 3.7 23.6 ± 3.5 | 0.756 (0.687–0.725) 2 84.3; 60.9 | Zhang et al., 2018 [268] |
| miR-24, miR-33a, miR-103a, miR-122 | Up | PBMC | RT-qPCR | CAD (161) HC (149) | Chinese | 61.35 ± 7.10 61.08 ± 7.51 | 86/75 72/77 | 25.77 ± 3.06 24.81 ± 3.29 | 0.911 (0.880–0.942) 2 84.5; 81.9 | Dong et al., 2017 [269] |
| let-7c, miR-145, miR-155 | Down | Plasma | OpenArray RT-qPCR, RT-qPCR | CAD (69) HC (32) | French (South-western) | 58.4 ± 9.0 57.3 ± 11.6 | Only male | 27.3 ± 4.4 26.6 ± 3.1 | 0.708 (0.600–0.811) 2 75.76; 63.33 | Faccini et al., 2017 [253] |
| miR-126, miR-143, miR-145 3 | Up | PBMC | RT-qPCR | CAD (450) HC (450) | Chinese | < 60 (26.8%/25.6%) 60–80 (68.0%/67.8%) ≥ 80 (5.2%/6.7%) | 234/216 215/235 | 18.5–24.9 (89.9%/94.3%) 25.0–29.9 (10.1%/5.7%) | N/A | Lin et al., 2017 [275] |
| miR-208a | Up | Plasma | RT-qPCR | CAD (290) HC (110) | Chinese | N/A | N/A | N/A | 0.919 (0.893–0.945) 75.5; 93.6 | Zhang et al., 2017 [278] |
| miR-133a | Up | Plasma | RT-qPCR | CAD (79) HC (63) | Chinese | 58 ± 12 55 ± 11 | 57/22 39/24 | 24.8 ± 4.12 24.0 ± 3.7 | 0.597 (0.504–0.691) 29.1; 92.5 | Zhu, 2017 [284] |
| miR-145-3p | Down | Serum | miSript miRNA PCR Array, RT-qPCR | CAD (40) HC (40) | Han Chinese | 34.20 ± 5.93 36.58 ± 3.96 | 37/3 36/4 | 28.01 ± 4.90 27.33 ± 2.75 | 0.753 (0.643–0.863) 67.50; 82.10 | Du et al., 2016 [254] |
| miR-190a-5p | 0.782 (0.680–0.884) 70.00; 75.00 | |||||||||
| miR-196b-5p | 0.824 (0.731–0.917) 85.00; 72.50 | |||||||||
| miR-3163-3p | 0.758 (0.651–0.864) 57.50; 84.60 | |||||||||
| miR-126-5p | Down | Plasma | qPCR | CAD (110) HC (40) | Chinese | 66.5 ± 11.7 1a 67.4 ± 9.7 1b 68.9 ± 11.3 1c 64.0 ± 10.4 | 67/43 28/12 | 24.9 ± 2.7 1a 24.4 ± 3.0 1b 25.2 ± 3.2 1c 23.9 ± 3.5 | N/A | Li et al., 2016 [285] |
| miR-208a, miR-370 | Up | Plasma | RT-qPCR | CAD (95) HC (50) | Chinese | 65 (44–78) 65 (46–75) | 65/30 34/16 | 23 (20–26) 22 (20–24) | 0.856 (0.796–0.917) 2 73.7; 86.0 | Liu et al., 2016 [270] |
| miR-15a-5p | Up | Plasma | RT-qPCR | CAD (50) HC (50) | Irish | 65 ± 9 60 ± 13 | 43/7 42/8 | 27.69 ± 3.31 26.96 ± 2.96 | 0.67 | O’Sullivan et al., 2016 [266] |
| miR-16-5p | 0.68 | |||||||||
| miR-93-5p | 0.75 | |||||||||
| miR-146a-5p | Down | 0.65 | ||||||||
| miR-206 | Up | Plasma | Microarray, RT-qPCR | CAD (67) HC (67) | Chinese | 64.70 ± 6.79 63.69 ± 5.96 | 43/24 32/35 | N/A | 0.607 (0.508–0.706) | Zhou et al., 2016 [255] |
| miR-574-5p | 0.696 (0.609–0.787) | |||||||||
| miR-765 | Up | Plasma | RT-qPCR | CAD (37) HC (20) | Chinese | 72.97 ± 4.28 71.7 ± 5.2 | 25/12 10/10 | 23.08 ± 3.03 22.29 ± 1.49 | 0.959 | Ali Sheikh et al., 2015 [272] |
| miR-149 | Down | 0.938 | ||||||||
| miR-765 | Up | Plasma | RT-qPCR | CAD (65) HC (32) | Chinese | 53 (49–57) 53 (49–57) | 38/27 16/16 | 22 (19–25) 22 (20–23) | 0.968 (0.939–0.996) 81.5; 93.7 | Ali Sheikh et al., 2015 [274] |
| miR-149 | Down | 0.938 (0.894–0.983) 71.8; 95.3 | ||||||||
| miR-424 | 0.919 (0.863–0.975) 68.7; 92.3 | |||||||||
| miR-17-5p | Up | Plasma | qPCR | CAD (59) NS-CAD (33) HC (20) | Chinese | 65.07 ± 10.55 65.23 ± 7.46 55.90 ± 4.72 | 40/19 18/15 7/13 | N/A | 0.894 (0.780–0.968) | Chen et al., 2015 [280] |
| miR-145 | Down | Plasma | RT-qPCR | CAD (26) HC (28) | Chinese | N/A | N/A | N/A | N/A | Gao et al., 2015 [286] |
| miR-21, miR-34a | Up | Plasma | Microarray (mice), RT-qPCR | CAD (32) HC (20) | Chinese | 67 ± 11 62 ± 8 | Only male | 24.1 ± 3.7 23.6 ± 4.0 | N/A | Han et al., 2015 [256] |
| miR-23a | Down | |||||||||
| miR-2909 | Up | PBMC | RT-qPCR | CAD (80) HC (20) | Iranian | 50 ± 4 49 ± 8 | Only male | N/A | N/A | Arora et al., 2014 [277] |
| miR-208, miR-215, miR-487a, miR-502 | Up | Serum | TLDA, RT-qPCR | CAD (92) HC (34) | Chinese | 65.2 ± 10.5 59.4 ± 13.1 | 53/39 15/19 | 25.83 ± 1.48 24.82 ± 2.72 | 0.909 (0.858–0.960) 2 83.7; 82.4 | Wang et al., 2014 [257] |
| miR-29b | Down | |||||||||
| miR-1 | Up | Plasma | Microarray, RT-qPCR | CAD (34) HC (20) | Italian | 60.0 ± 10.6 62.5 ± 2.1 | 30/4 19/1 | N/A | 0.918 | D’Alessandra et al., 2013 [258] |
| miR-126 | 0.929 | |||||||||
| miR-485-3p | 0.851 | |||||||||
| miR-340, miR-624 | Up | Platelet | Microarray, RT-qPCR | CAD (40) HC (40) | Dutch | 51.4 ± 4.7 51.0 ± 4.6 | Only male | N/A | 0.71 (0.59–0.83) 2 (combined with miR-451, miR-454) | Sondermeijer et al., 2011 [259] |
| miR-133 | Up | Plasma | RT-qPCR | CAD (36) HC (17) | German | 67.69 ± 11.07 32.18 ± 8.78 | 25/11 6/11 | > 25 (38.7%) > 25 (23.5%) | N/A | Fichtlscherer et al., 2010 [247] |
| miR-17, miR-92a, miR-126, miR-145, miR-155, miR-199a | Down | |||||||||
| miR-17, miR-92a, miR-126, miR-145, miR-155 | Down | Serum | CAD (31) HC (14) | 68.06 ± 9.66 39.28 ± 17.52 | 21/10 5/9 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szydełko, J.; Matyjaszek-Matuszek, B. MicroRNAs as Biomarkers for Coronary Artery Disease Related to Type 2 Diabetes Mellitus—From Pathogenesis to Potential Clinical Application. Int. J. Mol. Sci. 2023, 24, 616. https://doi.org/10.3390/ijms24010616
Szydełko J, Matyjaszek-Matuszek B. MicroRNAs as Biomarkers for Coronary Artery Disease Related to Type 2 Diabetes Mellitus—From Pathogenesis to Potential Clinical Application. International Journal of Molecular Sciences. 2023; 24(1):616. https://doi.org/10.3390/ijms24010616
Chicago/Turabian StyleSzydełko, Joanna, and Beata Matyjaszek-Matuszek. 2023. "MicroRNAs as Biomarkers for Coronary Artery Disease Related to Type 2 Diabetes Mellitus—From Pathogenesis to Potential Clinical Application" International Journal of Molecular Sciences 24, no. 1: 616. https://doi.org/10.3390/ijms24010616
APA StyleSzydełko, J., & Matyjaszek-Matuszek, B. (2023). MicroRNAs as Biomarkers for Coronary Artery Disease Related to Type 2 Diabetes Mellitus—From Pathogenesis to Potential Clinical Application. International Journal of Molecular Sciences, 24(1), 616. https://doi.org/10.3390/ijms24010616






