A Proteomic Approach Reveals That miR-423-5p Modulates Glucidic and Amino Acid Metabolism in Prostate Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Proteomic Profiling of LNCaP Cells Overexpressing miR-423-5p
2.2. Prediction of Functional Targets by Integrative Analysis of miRNA Predicted Targets and Protein Expression Data
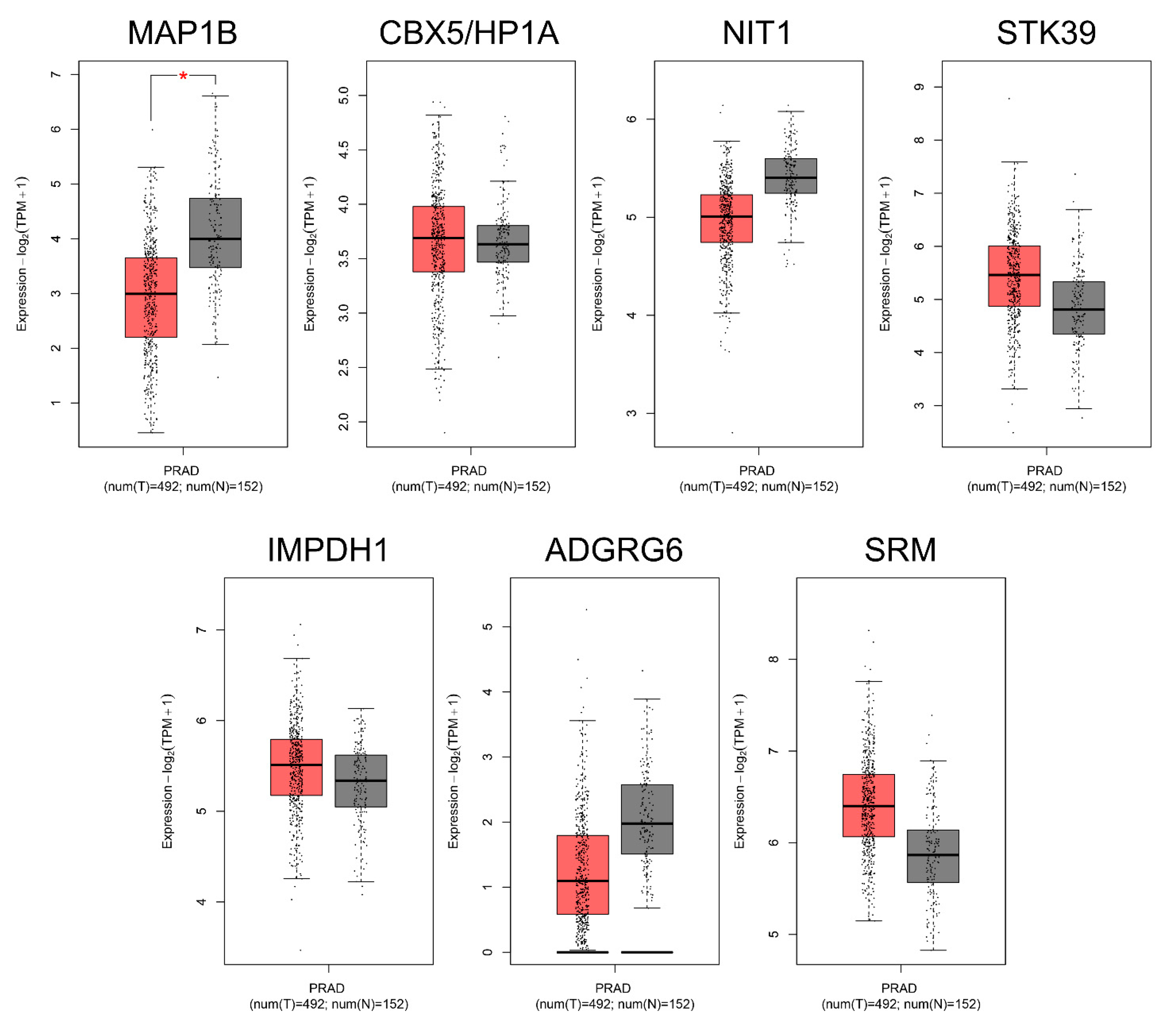
2.3. Protein Expression Analysis and Gene Expression Profiling of miR-423-5p Targets in Prostate Cancer
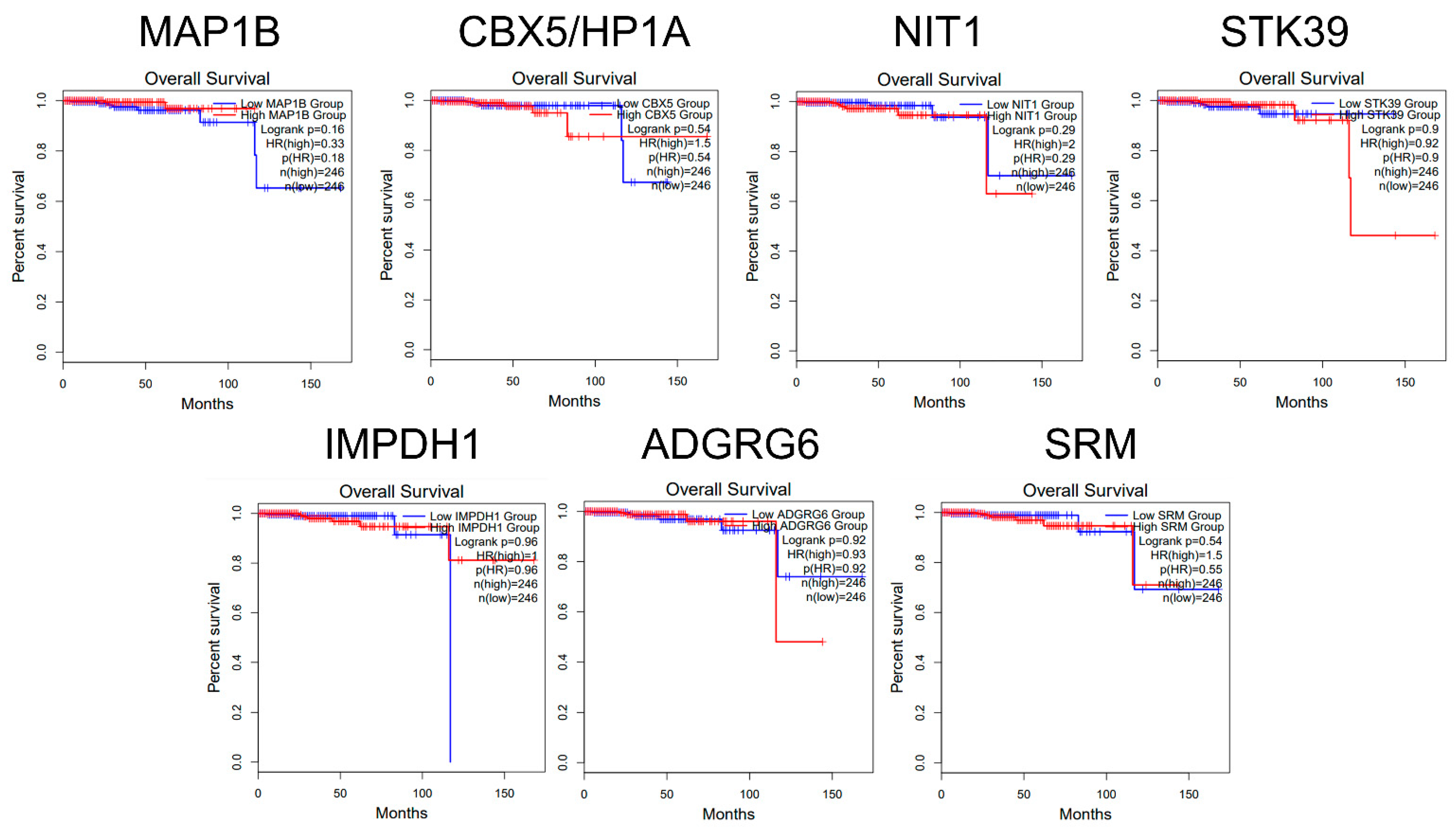
2.4. Overall Survival Analysis Based on the Expression of miR-423-5p Target Genes
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Growth Conditions
4.2. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
4.3. Retroviral Expression of Empty Vector and miR-423-5p-Mimic
4.4. Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) Analysis
4.5. Library Generation, Spectral Alignment, and Fold Change Analysis
4.6. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Dyba, T.; Randi, G.; Bray, F.; Martos, C.; Giusti, F.; Nicholson, N.; Gavin, A.; Flego, M.; Neamtiu, L.; Dimitrova, N.; et al. The European cancer burden in 2020: Incidence and mortality estimates for 40 countries and 25 major cancers. Eur. J. Cancer 2021, 157, 308–347. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Teply, B.A. Biomarkers for Treatment Response in Advanced Prostate Cancer. Cancers 2021, 13, 5723. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef] [Green Version]
- Luu, H.N.; Lin, H.-Y.; Sørensen, K.D.; Ogunwobi, O.O.; Kumar, N.; Chornokur, G.; Phelan, C.; Jones, D.; Kidd, L.; Batra, J.; et al. miRNAs associated with prostate cancer risk and progression. BMC Urol. 2017, 17, 18. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Feng, G. MiR-423-5p aggravates lung adenocarcinoma via targeting CADM1. Thorac. Cancer 2020, 12, 210–217. [Google Scholar] [CrossRef]
- Sun, G.; Ding, X.; Bi, N.; Wu, L.; Wang, J.; Zhang, W.; Dong, X.; Lv, N.; Song, Y.; Zhan, Q.; et al. MiR-423-5p in brain metastasis: Potential role in diagnostics and molecular biology. Cell Death Dis. 2018, 9, 936. [Google Scholar] [CrossRef]
- Li, S.; Zeng, A.; Hu, Q.; Yan, W.; Liu, Y.; You, Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro-Oncology 2016, 19, 55–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Wang, X.; Yang, X.; Liu, Y.; Shi, Y.; Ren, J.; Guleng, B. miRNA423-5p regulates cell proliferation and invasion by targeting trefoil factor 1 in gastric cancer cells. Cancer Lett. 2014, 347, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xue, S.; Zhang, X.; Li, F.; Bei, S.; Feng, L. CircRNA PVT1 modulated cell migration and invasion through Epithelial-Mesenchymal Transition (EMT) mediation in gastric cancer through miR-423-5p/Smad3 pathway. Regen. Ther. 2022, 21, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zeng, X.; Huang, Y.; Chen, S.; Lin, F.; Yang, G.; Yang, N. miR-423-5p serves as a diagnostic indicator and inhibits the proliferation and invasion of ovarian cancer. Exp. Ther. Med. 2018, 15, 4723–4730. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Yu, T.; An, Q.; Cao, X.; Pan, H. MicroRNA-423-5p inhibits colon cancer growth by promoting caspase-dependent apoptosis. Exp. Ther. Med. 2018, 16, 1225–1231. [Google Scholar] [CrossRef] [Green Version]
- Ferri, C.; Di Biase, A.; Bocchetti, M.; Zappavigna, S.; Wagner, S.; Le Vu, P.; Luce, A.; Cossu, A.M.; Vadakekolathu, J.; Miles, A.; et al. MiR-423-5p prevents MALAT1-mediated proliferation and metastasis in prostate cancer. J. Exp. Clin. Cancer Res. 2022, 41, 20. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [Green Version]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—Integrative database of human microRNA target predictions. Nucleic Acids Res. 2017, 46, D360–D370. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2017, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, I.; Yang, X.; Brown, J.; Ramakrishnan, A.; Torok-Storb, B.; Kabos, P.; Hesselberth, J.R.; Pillai, M.M. Genome-Wide Analysis of miRNA-mRNA Interactions in Marrow Stromal Cells. Stem Cells 2013, 32, 662–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karginov, F.V.; Hannon, G.J. Remodeling of Ago2–mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013, 27, 1624–1632. [Google Scholar] [CrossRef] [Green Version]
- Whisnant, A.; Bogerd, H.P.; Flores, O.; Ho, P.; Powers, J.G.; Sharova, N.; Stevenson, M.; Chen, C.-H.; Cullen, B.R. In-Depth Analysis of the Interaction of HIV-1 with Cellular microRNA Biogenesis and Effector Mechanisms. mBio 2013, 4, e00193-13. [Google Scholar] [CrossRef] [Green Version]
- Haecker, I.; Gay, L.A.; Yang, Y.; Hu, J.; Morse, A.M.; McIntyre, L.M.; Renne, R. Ago HITS-CLIP Expands Understanding of Kaposi’s Sarcoma-associated Herpesvirus miRNA Function in Primary Effusion Lymphomas. PLoS Pathog. 2012, 8, e1002884. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct Conversion of Fibroblasts to Neurons by Reprogramming PTB-Regulated MicroRNA Circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef] [Green Version]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, A.; Barrera, E.; Bazant, W.; Burke, M.; Füllgrabe, A.; Fuentes, A.M.-P.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2017, 46, D246–D251. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Yang, W.; Wang, J.; Chen, Z.; Chen, J.; Meng, Y.; Chen, L.; Chang, Y.; Geng, B.; Sun, L.; Dou, L.; et al. NFE2 Induces miR-423-5p to Promote Gluconeogenesis and Hyperglycemia by Repressing the Hepatic FAM3A-ATP-Akt Pathway. Diabetes 2017, 66, 1819–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Z.; Wang, S.; Huang, X.; Chen, P.; Deng, L.; Li, S.; Lin, S.; Wang, Z.; Liu, B. Plasma exosomal microRNAs Associated with Metabolism as Early Predictor of Gestational Diabetes Mellitus. Diabetes 2022, 71, 2272–2283. [Google Scholar] [CrossRef] [PubMed]
- Bin Yu, B.; Zhu, H.-D.; Shi, X.-L.; Chen, P.-P.; Sun, X.-M.; Xia, G.-Y.; Fang, M.; Zhong, Y.-X.; Tang, X.-L.; Zhang, T.; et al. iTRAQ-based quantitative proteomic analysis of thoracic aortas from adult rats born to preeclamptic dams. Clin. Proteom. 2021, 18, 22. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; Elf, S.; Shan, C.; Kang, H.-B.; Ji, Q.; Zhou, L.; Hitosugi, T.; Zhang, L.; Zhang, S.; Seo, J.H.; et al. 6-Phosphogluconate dehydrogenase links oxidative PPP, lipogenesis and tumour growth by inhibiting LKB1–AMPK signalling. Nature 2015, 17, 1484–1496. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Du, W.; Yang, X. A critical role of glucose-6-phosphate dehydrogenase in TAp73-mediated cell proliferation. Cell Cycle 2013, 12, 3720–3726. [Google Scholar] [CrossRef] [Green Version]
- Jones, N.P.; Schulze, A. Targeting cancer metabolism—Aiming at a tumour’s sweet-spot. Drug Discov. Today 2012, 17, 232–241. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Gregory, M.A.; D’Alessandro, A.; Alvarez-Calderon, F.; Kim, J.; Nemkov, T.; Adane, B.; Rozhok, A.I.; Kumar, A.; Kumar, V.; Pollyea, D.A.; et al. ATM/G6PD-driven redox metabolism promotes FLT3 inhibitor resistance in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2016, 113, E6669–E6678. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Ke, M.; Qi, M.; Han, Z.; Cao, Y.; Deng, Z.; Qian, J.; Yang, Y.; Gu, C. G6PD promotes cell proliferation and dexamethasone resistance in multiple myeloma via increasing anti-oxidant production and activating Wnt/β-catenin pathway. Exp. Hematol. Oncol. 2022, 11, 77. [Google Scholar] [CrossRef]
- Mele, L.; Paino, F.; Papaccio, F.; Regad, T.; Boocock, D.; Stiuso, P.; Lombardi, A.; Liccardo, D.; Aquino, G.; Barbieri, A.; et al. A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 2018, 9, 572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mele, L.; la Noce, M.; Paino, F.; Regad, T.; Wagner, S.; Liccardo, D.; Papaccio, G.; Lombardi, A.; Caraglia, M.; Tirino, V.; et al. Glucose-6-phosphate dehydrogenase blockade potentiates tyrosine kinase inhibitor effect on breast cancer cells through autophagy perturbation. J. Exp. Clin. Cancer Res. 2019, 38, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiuso, P.; Potenza, N.; Lombardi, A.; Ferrandino, I.; Monaco, A.; Zappavigna, S.; Vanacore, D.; Mosca, N.; Castiello, F.; Porto, S.; et al. MicroRNA-423-5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated with Sorafenib. Mol. Ther. Nucleic Acids 2015, 4, e233. [Google Scholar] [CrossRef]
- Zhu, X.; Lu, X. MiR-423-5p inhibition alleviates cardiomyocyte apoptosis and mitochondrial dysfunction caused by hypoxia/reoxygenation through activation of the wnt/β-catenin signaling pathway via targeting MYBL2. J. Cell. Physiol. 2019, 234, 22034–22043. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.-P.; Liu, L.-S.; Chen, C.-B.; Zhou, J.; Zheng, Y.-T.; Wang, X.-P.; Han, M.; Wang, C.-X. MicroRNA-423-5p facilitates hypoxia/reoxygenation-induced apoptosis in renal proximal tubular epithelial cells by targeting GSTM1 via endoplasmic reticulum stress. Oncotarget 2017, 8, 82064–82077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holbert, C.E.; Cullen, M.T.; Casero, R.A.; Stewart, T.M. Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 2022, 22, 467–480. [Google Scholar] [CrossRef]
- Halpain, S.; Dehmelt, L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006, 7, 224. [Google Scholar] [CrossRef]
- Dehmelt, L.; Halpain, S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2004, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Bhat, K.M.; Setaluri, V. Microtubule-Associated Proteins as Targets in Cancer Chemotherapy. Clin. Cancer Res. 2007, 13, 2849–2854. [Google Scholar] [CrossRef] [Green Version]
- Chien, T.-M.; Chan, T.-C.; Huang, S.K.-H.; Yeh, B.-W.; Li, W.-M.; Huang, C.-N.; Li, C.-C.; Wu, W.-J.; Li, C.-F. Role of Microtubule-Associated Protein 1b in Urothelial Carcinoma: Overexpression Predicts Poor Prognosis. Cancers 2020, 12, 630. [Google Scholar] [CrossRef]
- Balahmar, R.M.; Boocock, D.J.; Coveney, C.; Ray, S.; Vadakekolathu, J.; Regad, T.; Ali, S.; Sivasubramaniam, S. Identification and characterisation of NANOG+/ OCT-4high/SOX2+ doxorubicin-resistant stem-like cells from transformed trophoblastic cell lines. Oncotarget 2018, 9, 7054–7065. [Google Scholar] [CrossRef] [PubMed]
- Vyas, F.S.; Hargreaves, A.J.; Bonner, P.L.; Boocock, D.J.; Coveney, C.; Dickenson, J.M. A1 adenosine receptor-induced phosphorylation and modulation of transglutaminase 2 activity in H9c2 cells: A role in cell survival. Biochem. Pharmacol. 2016, 107, 41–58. [Google Scholar] [CrossRef] [PubMed]

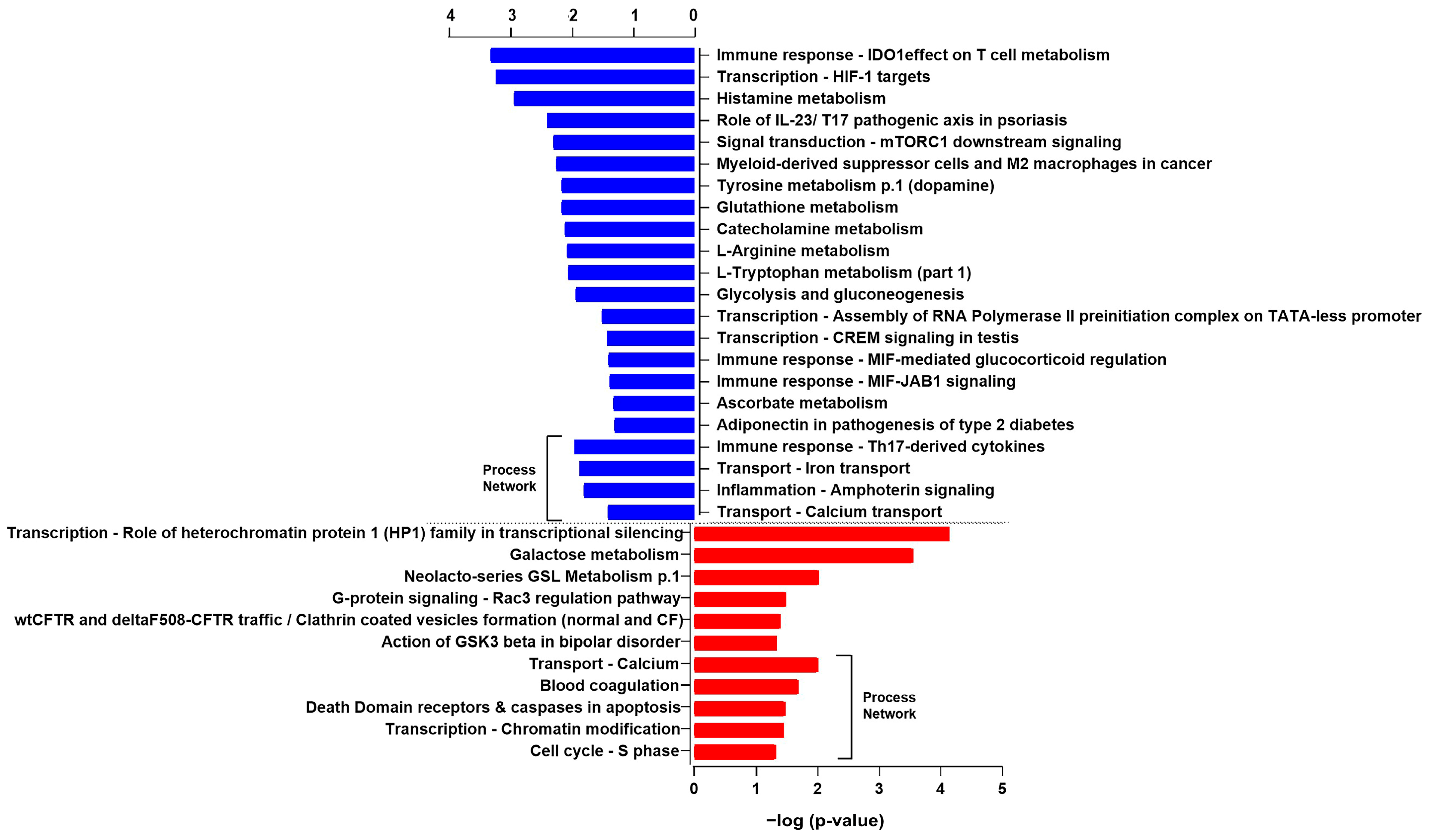


| N° | Pathways Modulated by Downregulated Proteins | Total | In Data | p-Value | Network Objects from Active Data |
|---|---|---|---|---|---|
| 1 | Immune response_The effect of IDO1 on T cell metabolism | 89 | 3 | 4.910 × 10−4 | LDHA, HX2, G6PD |
| 2 | Transcription_HIF-1 targets | 95 | 3 | 5.945 × 10−4 | LDHA, TfR1, HX2 |
| 3 | Histamine metabolism | 29 | 2 | 1.185 × 10−3 | ALDHA1, ALDH2 |
| 4 | Role of IL-23/T17 pathogenic axis in psoriasis | 54 | 2 | 4.067 × 10−3 | Calgranulin A, Calgranulin B |
| 5 | Signal transduction_mTORC1 downstream signaling | 61 | 2 | 5.163 × 10−3 | GRB10, G6PD |
| 6 | Myeloid-derived suppressor cells and M2 macrophages in cancer | 64 | 2 | 5.669 × 10−3 | Calgranulin A, Calgranulin B |
| 7 | Tyrosine metabolism p.1 (dopamine) | 71 | 2 | 6.937 × 10−3 | ALDHA1, ALDH2 |
| 8 | Glutathione metabolism | 71 | 2 | 6.937 × 10−3 | GSTT2, G6PD |
| 9 | Catecholamine metabolism | 76 | 2 | 7.914 × 10−3 | ALDHA1, ALDH2 |
| 10 | L-Arginine metabolism | 79 | 2 | 8.529 × 10−3 | ALDHA1, ALDH2 |
| 11 | L-Tryptophan metabolism (part 1) | 81 | 2 | 8.950 × 10−3 | ALDHA1, ALDH2 |
| 12 | Glycolysis and gluconeogenesis | 94 | 2 | 1.191 × 10−2 | LDHA, HX2 |
| 13 | Transcription_Assembly of RNA Polymerase II preinitiation complex on TATA-less promoters | 18 | 1 | 3.164 × 10−2 | AMH |
| 14 | Transcription_CREM signaling in testis | 22 | 1 | 3.854 × 10−2 | HX2 |
| 15 | Immune response_MIF-mediated glucocorticoid regulation | 23 | 1 | 4.026 × 10−2 | MIF |
| 16 | Immune response_MIF-JAB1 signaling | 24 | 1 | 4.197 × 10−2 | MIF |
| 17 | Ascorbate metabolism | 28 | 1 | 4.880 × 10−2 | ALDH2 |
| 18 | Adiponectin in pathogenesis of type 2 diabetes | 29 | 1 | 5.050 × 10−2 | ACADM |
| N° | Pathways Modulated by Upregulated Proteins | Total | In Data | p-Value | Network Objects from Active Data |
|---|---|---|---|---|---|
| 1 | Transcription_Role of heterochromatin protein 1 (HP1) family in transcriptional silencing | 40 | 3 | 7.294 × 10−5 | HP1 alpha, HP1 beta, HP1 |
| 2 | Galactose metabolism | 63 | 3 | 2.843 × 10−4 | GALK1, GALT |
| 3 | Neolacto-series GSL Metabolism p.1 | 72 | 2 | 9.649 × 10−3 | ASNA1 |
| 4 | G-protein signaling_Rac3 regulation pathway | 16 | 1 | 3.279 × 10−2 | Calmyrin |
| 5 | wtCFTR and deltaF508-CFTR traffic/Clathrin coated vesicles formation (normal and CF) | 20 | 1 | 4.083 × 10−2 | Myosin I |
| 6 | Action of GSK3 beta in bipolar disorder | 23 | 1 | 4.681 × 10−2 | MAP-1B |
| N° | Process Networks Modulated by Downregulated Proteins | Total | In Data | p-Value | Network Objects from Active Data |
|---|---|---|---|---|---|
| 1 | Immune response_Th17-derived cytokines | 98 | 2 | 1.128 × 10−2 | Calgranulin A, Calgranulin B |
| 2 | Transport_Iron transport | 108 | 2 | 1.359 × 10−2 | TfR1, Junctin |
| 3 | Inflammation_Amphoterin signaling | 118 | 2 | 1.608 × 10−2 | Calgranulin A, Calgranulin B |
| 4 | Transport_Calcium transport | 192 | 2 | 3.987 × 10−2 | Calgranulin A, Junctin |
| N° | Process Networks Modulated by Upregulated Proteins | Total | In Data | p-Value | Network Objects from Active Data |
|---|---|---|---|---|---|
| 1 | Transport_Calcium | 192 | 3 | 9.894 × 10−3 | Annexin V, Calmyrin |
| 2 | Blood coagulation | 94 | 2 | 2.056 × 10−2 | Annexin V, Annexin IV |
| 3 | Apoptosis Death Domain receptors & caspases in apoptosis | 123 | 2 | 3.390 × 10−2 | PEA15 |
| 4 | Transcription_Chromatin modification | 127 | 2 | 3.595 × 10−2 | HP1 alpha, HP1 beta |
| 5 | Cell cycle_S phase | 149 | 2 | 4.806 × 10−2 | HP1 alpha, HP1 |
| Prediction Tools | Predicted miTGs (miRNA Target Genes) |
|---|---|
| DIANA microT-CDS | 935 |
| TargetScan_Human_v8.0 | 232 |
| mirDIP | 6413 |
| mirDB | 698 |
| Targets predicted by all 4 tools | 6695 |
| Targets predicted by a minimum 2/4 prediction tools | 1100 |
| Gene Name | Protein Name | Protein Description | FC 1 miR-423-5p/ctr |
|---|---|---|---|
| MAP1B | MAP1B | Microtubule-associated protein 1B | 2.27 |
| CBX5/HP1A | CBX5 | Chromobox protein homolog 5 | 1.62 |
| NIT1 | NIT1 | Deaminated glutathione amidase | 1.62 |
| STK39 | STK39 | STE20/SPS1-related proline-alanine-rich protein kinase | 1.56 |
| IMPDH1 | IMDH1 | Inosine-5′-monophosphate dehydrogenase 1 | −2.49 |
| ADGRG6 | AGRG6 | Adhesion G-protein coupled receptor G6 | −2.75 |
| SRM | SPEE | Spermidine synthase | −2.75 |
| Protein Name | Protein Description | Protein Expression Status LNCaP Cell Line with miR-423-5p/ctr | Protein Expression Level LNCaP Cell Line 1 | miTGs 2 Name | Transcriptomic Expression Status PRAD 3 (T/N) |
|---|---|---|---|---|---|
| MAP1B | Microtubule-associated protein 1B | Upregulated | High | MAP1B | Downregulated |
| CBX5 | Chromobox protein homolog 5 | Upregulated | High | CBX5/HP1A | Upregulated |
| NIT1 | Deaminated glutathione amidase | Upregulated | High | NIT1 | Downregulated |
| STK39 | STE20/SPS1-related proline-alanine-rich protein kinase | Upregulated | High | STK39 | Upregulated |
| IMDH1 | Inosine-5′-monophosphate dehydrogenase 1 | Downregulated | High | IMPDH1 | Upregulated |
| AGRG6 | Adhesion G-protein coupled receptor G6 | Downregulated | High | ADGRG6 | Downregulated |
| SPEE | Spermidine Synthase | Downregulated | High | SRM | Upregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luce, A.; Lombardi, A.; Ferri, C.; Zappavigna, S.; Tathode, M.S.; Miles, A.K.; Boocock, D.J.; Vadakekolathu, J.; Bocchetti, M.; Alfano, R.; et al. A Proteomic Approach Reveals That miR-423-5p Modulates Glucidic and Amino Acid Metabolism in Prostate Cancer Cells. Int. J. Mol. Sci. 2023, 24, 617. https://doi.org/10.3390/ijms24010617
Luce A, Lombardi A, Ferri C, Zappavigna S, Tathode MS, Miles AK, Boocock DJ, Vadakekolathu J, Bocchetti M, Alfano R, et al. A Proteomic Approach Reveals That miR-423-5p Modulates Glucidic and Amino Acid Metabolism in Prostate Cancer Cells. International Journal of Molecular Sciences. 2023; 24(1):617. https://doi.org/10.3390/ijms24010617
Chicago/Turabian StyleLuce, Amalia, Angela Lombardi, Carmela Ferri, Silvia Zappavigna, Madhura S. Tathode, Amanda K. Miles, David J. Boocock, Jayakumar Vadakekolathu, Marco Bocchetti, Roberto Alfano, and et al. 2023. "A Proteomic Approach Reveals That miR-423-5p Modulates Glucidic and Amino Acid Metabolism in Prostate Cancer Cells" International Journal of Molecular Sciences 24, no. 1: 617. https://doi.org/10.3390/ijms24010617






