The Role of Long Noncoding RNA (lncRNAs) Biomarkers in Renal Cell Carcinoma
Abstract
1. Introduction
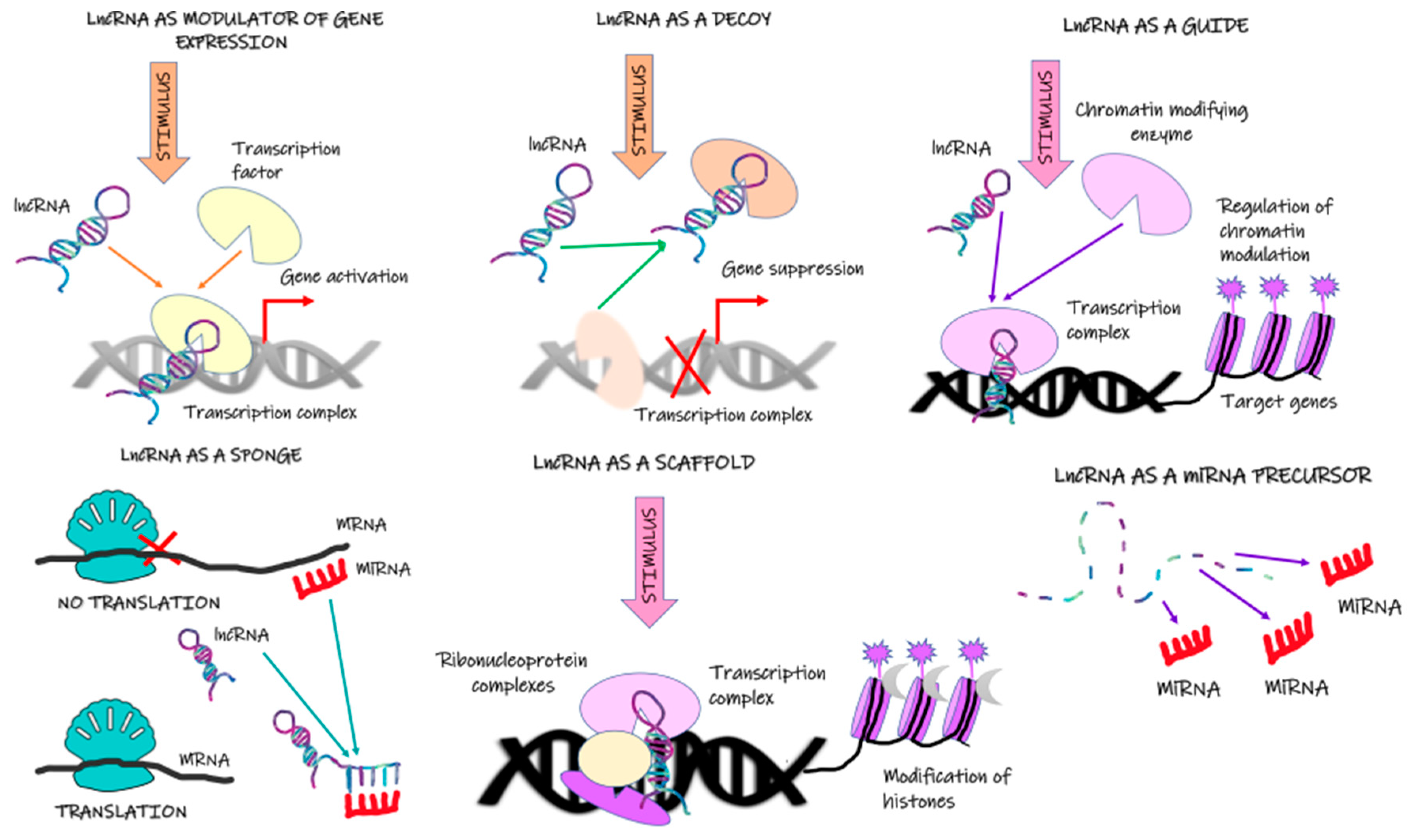
2. Long Noncoding RNA
2.1. MALAT1
2.2. RCAT1
2.3. DUXAP9
2.4. LncRNA TCL6 (lncTCL6)
2.5. LINC00342
2.6. AGAP2-AS1 (AGAP2 Antisense 1)
2.7. DLEU2
2.8. NNT-AS1
2.9. LINC00460
2.10. Lnc-LSG1
3. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Harada, K.i.; Miyake, H.; Kusuda, Y.; Fujisawa, M. Expression of epithelial–mesenchymal transition markers in renal cell carcinoma: Impact on prognostic outcomes in patients undergoing radical nephrectomy. BJU Int. 2012, 110, E1131–E1137. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of renal cell carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Brannon, A.R.; Reddy, A.; Seiler, M.; Arreola, A.; Moore, D.T.; Pruthi, R.S.; Wallen, E.M.; Nielsen, M.E.; Liu, H.; Nathanson, K.L.; et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer 2010, 1, 152–163. [Google Scholar] [CrossRef]
- Haake, S.M.; Brooks, S.A.; Welsh, E.; Fulp, W.J.; Chen, D.T.; Dhillon, J.; Haura, E.; Sexton, W.; Spiess, P.E.; Pow-Sang, J.; et al. Patients with ClearCode34-identified molecular subtypes of clear cell renal cell carcinoma represent unique populations with distinct comorbidities. Urol. Oncol. 2016, 34, 122.e1-7. [Google Scholar] [CrossRef]
- Ghatalia, P.; Rathmell, W.K. Systematic Review: ClearCode 34—A Validated Prognostic Signature in Clear Cell Renal Cell Carcinoma (ccRCC). Kidney Cancer 2018, 2, 23–29. [Google Scholar] [CrossRef]
- Wang, B.; Chen, D.; Hua, H. TBC1D3 family is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma. Mol. Oncolytics 2021, 22, 528–538. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2019; American Cancer Society: Atlanta, GA, USA, 2019; Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2019/cancer-facts-and-figures-2019.pdf (accessed on 22 December 2022).
- Pulikkottil, A.J.; Bamezai, S.; Ammer, T.; Mohr, F.; Feder, K.; Vegi, N.M.; Mandal, T.; Kohlhofer, U.; Quintanilla-Martinez, L.; Sinha, A. TET3 promotes AML growth and epigenetically regulates glucose metabolism and leukemic stem cell associated pathways. Leukemia 2021, 36, 416–425. [Google Scholar] [CrossRef]
- Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Lohse, C.M.; Frank, I.; Kwon, E.D.; Weaver, A.L.; Parker, A.S.; Zincke, H. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials. Cancer 2003, 97, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Ding, L.; Lu, Z.; Wang, R.; Yu, C.; Wang, H.; Zheng, Q.; Wang, X.; Xu, W.; Yu, H.; et al. METTL14-mediated Lnc-LSG1 m6A modification inhibits clear cell renal cell carcinoma metastasis via regulating ESRP2 ubiquitination. Mol. Nucleic Acids 2022, 27, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Wang, H.; Wu, Y.; Wang, K.; Li, G. Hub Long Noncoding RNAs with m6A Modification for Signatures and Prognostic Values in Kidney Renal Clear Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 682471. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.S.; Kabbinavar, F. Metastatic clear cell renal cell carcinoma: A review of current therapies and novel immunotherapies. Crit. Rev. Oncol. Hematol. 2015, 96, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed]
- Nakken, S.; Eikrem, Ø.; Marti, H.P.; Beisland, C.; Bostad, L.; Scherer, A.; Flatberg, A.; Beisvag, V.; Skandalou, E.; Furriol, J.; et al. AGAP2-AS1 as a prognostic biomarker in low-risk clear cell renal cell carcinoma patients with progressing disease. Cancer Cell Int. 2021, 21, 690. [Google Scholar] [CrossRef]
- Gallardo, E.; Méndez-Vidal, M.J.; Pérez-Gracia, J.L.; Sepúlveda-Sánchez, J.M.; Campayo, M.; Chirivella-González, I.; García-Del-Muro, X.; González-Del-Alba, A.; Grande, E.; Suárez, C. SEOM clinical guideline for treatment of kidney cancer (2017). Clin. Transl. Oncol. 2018, 20, 47–56. [Google Scholar] [CrossRef]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Z.; Wo, M.; Xu, W. The long non-coding RNA NNT-AS1 promotes clear cell renal cell carcinoma progression via regulation of the miR-137/Y-box binding protein 1 axis. Bioengineered 2021, 12, 8994–9005. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, F.; Niu, Y.; Yu, S. Aberration of lncRNA LINC00460 is a Promising Prognosis Factor and Associated with Progression of Clear Cell Renal Cell Carcinoma. Cancer Manag. Res. 2021, 13, 6489–6497. [Google Scholar] [CrossRef]
- Shima, H.; Kida, K.; Adachi, S.; Yamada, A.; Sugae, S.; Narui, K.; Miyagi, Y.; Nishi, M.; Ryo, A.; Murata, S. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res. Treat. 2018, 170, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.-X.; Koirala, P.; Mo, Y.-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Sun, W.; Guo, Z.; Zhang, J.; Yu, H.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900. [Google Scholar] [CrossRef]
- Li, T.; Tong, H.; Zhu, J.; Qin, Z.; Yin, S.; Sun, Y.; Liu, X.; He, W. Identification of a Three-Glycolysis-Related lncRNA Signature Correlated With Prognosis and Metastasis in Clear Cell Renal Cell Carcinoma. Front. Med. (Lausanne) 2021, 8, 777507. [Google Scholar] [CrossRef]
- Xia, R.; Geng, G.; Yu, X.; Xu, Z.; Guo, J.; Liu, H.; Li, N.; Li, Z.; Li, Y.; Dai, X. LINC01140 promotes the progression and tumor immune escape in lung cancer by sponging multiple microRNAs. J. Immunother. Cancer 2021, 9, e002746. [Google Scholar] [CrossRef]
- Barth, D.A.; Slaby, O.; Klec, C.; Juracek, J.; Drula, R.; Calin, G.A.; Pichler, M. Current concepts of non-coding RNAs in the pathogenesis of non-clear cell renal cell carcinoma. Cancers 2019, 11, 1580. [Google Scholar] [CrossRef]
- Bach, D.-H.; Lee, S.K. Long noncoding RNAs in cancer cells. Cancer Lett. 2018, 419, 152–166. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Chen, H.; Pan, Y.; Jin, X.; Chen, G. Identification of a four hypoxia-associated long non-coding RNA signature and establishment of a nomogram predicting prognosis of clear cell renal cell carcinoma. Front. Oncol. 2021, 11, 713346. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Doi, H.; Ishikawa, K.; Inada, M.; Tatsuno, S.; Wada, Y.; Oguma, Y.; Kawakami, H.; Nakamatsu, K.; Hosono, M. Serum lactate dehydrogenase is a predictive biomarker in patients with oropharyngeal cancer undergoing radiotherapy: Retrospective study on predictive factors. Head Neck 2021, 43, 3132–3141. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, Z.; Niu, Y.; Yang, Y.; Li, M.; Pang, Y.; Liu, M.; Cheng, X.; Xu, M.; Wang, Z. FBP1 regulates proliferation, metastasis, and chemoresistance by participating in C-MYC/STAT3 signaling axis in ovarian cancer. Oncogene 2021, 40, 5938–5949. [Google Scholar] [CrossRef]
- Zhu, S.; Guo, Y.; Zhang, X.; Liu, H.; Yin, M.; Chen, X.; Peng, C. Pyruvate kinase M2 (PKM2) in cancer and cancer therapeutics. Cancer Lett. 2021, 503, 240–248. [Google Scholar] [CrossRef]
- Yan, T.; Shen, C.; Jiang, P.; Yu, C.; Guo, F.; Tian, X.; Zhu, X.; Lu, S.; Han, B.; Zhong, M. Risk SNP-induced lncRNA-SLCC1 drives colorectal cancer through activating glycolysis signaling. Signal Transduct. Target. Ther. 2021, 6, 70. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; She, X.; Fan, L.; Li, P.; Feng, J.; Fu, H.; Liu, Q.; Liu, Q.; Zhao, C.; et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J. Hematol. Oncol. 2018, 11, 77. [Google Scholar] [CrossRef]
- Zhao, S.; Guan, B.; Mi, Y.; Shi, D.; Wei, P.; Gu, Y.; Cai, S.; Xu, Y.; Li, X.; Yan, D. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit. Oncogene 2021, 40, 4709–4724. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Malihi, P.D.; Graf, R.P.; Rodriguez, A.; Ramesh, N.; Lee, J.; Sutton, R.; Jiles, R.; Ruiz Velasco, C.; Sei, E.; Kolatkar, A.; et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin. Cancer Res. 2020, 26, 4143–4153. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Ohnami, S.; Tanabe, C.; Sasaki, H.; Yasuda, J.; Katai, H.; Yoshimura, K.; Terada, M.; Perucho, M.; Yoshida, T. The genomic damage estimated by arbitrarily primed PCR DNA fingerprinting is useful for the prognosis of gastric cancer. Gastroenterology 2003, 125, 1330–1340. [Google Scholar] [CrossRef]
- Yang, H.; Xiong, X.; Li, H. Development and Interpretation of a Genomic Instability Derived lncRNAs Based Risk Signature as a Predictor of Prognosis for Clear Cell Renal Cell Carcinoma Patients. Front. Oncol. 2021, 11, 678253. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 2013, 108, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018, 28, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Li, L.; Yang, W.; Hu, J.F.; Cui, J. Long noncoding RNA: A resident staff of genomic instability regulation in tumorigenesis. Cancer Lett. 2021, 503, 103–109. [Google Scholar] [CrossRef]
- Panda, S.; Setia, M.; Kaur, N.; Shepal, V.; Arora, V.; Singh, D.K.; Mondal, A.; Teli, A.; Tathode, M.; Gajula, R.; et al. Noncoding RNA Ginir functions as an oncogene by associating with centrosomal proteins. PLoS Biol. 2018, 16, e2004204. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Li, S.; Kong, F.; Ruan, H.; Xu, X.; Zhang, X.; Wu, Z.; Zhang, L.; Xu, Y.; Yuan, H.; et al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics 2020, 10, 265–280. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Xiang, C.; Molczan, A.; Baubet, V.; Conejo-Garcia, J.; Xu, X.; Lieberman, P.M.; Dahmane, N. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J. Cell Sci. 2012, 125, 4383–4394. [Google Scholar] [CrossRef]
- Lupiáñez, D.G.; Spielmann, M.; Mundlos, S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016, 32, 225–237. [Google Scholar] [CrossRef]
- Yamamoto, T.; Saitoh, N. Non-coding RNAs and chromatin domains. Curr. Opin. Cell Biol. 2019, 58, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, Q.; Liu, R.; Chen, Z.; Zhang, X.; Zhou, P.; Wang, Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018, 46, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, T.; Tang, J.; Zhang, L.; Fan, S.; Zhou, J.; Liang, C. Construction of Competitive Endogenous RNA Network and Verification of 3-Key LncRNA Signature Associated With Distant Metastasis and Poor Prognosis in Patients With Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 640150. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Han, L.; Yin, D.; He, X.; Hong, L.; Si, X.; Qiu, M.; Xu, T.; De, W.; Xu, L.; et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017, 45, 3086–3101. [Google Scholar] [CrossRef]
- Hadji, F.; Boulanger, M.C.; Guay, S.P.; Gaudreault, N.; Amellah, S.; Mkannez, G.; Bouchareb, R.; Marchand, J.T.; Nsaibia, M.J.; Guauque-Olarte, S.; et al. Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation 2016, 134, 1848–1862. [Google Scholar] [CrossRef]
- Xie, J.J.; Jiang, Y.Y.; Jiang, Y.; Li, C.Q.; Lim, M.C.; An, O.; Mayakonda, A.; Ding, L.W.; Long, L.; Sun, C.; et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology 2018, 154, 2137–2151.e2131. [Google Scholar] [CrossRef]
- Hämmerle, M.; Gutschner, T.; Uckelmann, H.; Ozgur, S.; Fiskin, E.; Gross, M.; Skawran, B.; Geffers, R.; Longerich, T.; Breuhahn, K.; et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 2013, 58, 1703–1712. [Google Scholar] [CrossRef]
- Jin, C.; Shi, L.; Li, K.; Liu, W.; Qiu, Y.; Zhao, Y.; Zhao, B.; Li, Z.; Li, Y.; Zhu, Q. Mechanism of tumor-derived extracellular vesicles in regulating renal cell carcinoma progression by the delivery of MALAT1. Oncol. Rep. 2021, 46, 187. [Google Scholar] [CrossRef]
- Watson, D.C.; Bayik, D.; Srivatsan, A.; Bergamaschi, C.; Valentin, A.; Niu, G.; Bear, J.; Monninger, M.; Sun, M.; Morales-Kastresana, A.; et al. Efficient production and enhanced tumor delivery of engineered extracellular vesicles. Biomaterials 2016, 105, 195–205. [Google Scholar] [CrossRef]
- Ghoroghi, S.; Mary, B.; Asokan, N.; Goetz, J.G.; Hyenne, V. Tumor extracellular vesicles drive metastasis (it’s a long way from home). FASEB Bioadv. 2021, 3, 930–943. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, Q.; Andre-Gregoire, G.; Guevel, L.; Gavard, J. Vesiclemia: Counting on extracellular vesicles for glioblastoma patients. Oncogene 2020, 39, 6043–6052. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, C.; D’Souza-Schorey, C. Tumor-derived extracellular vesicles: Molecular parcels that enable regulation of the immune response in cancer. J. Cell Sci. 2019, 132, jcs235085. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [PubMed]
- Ghoroghi, S.; Mary, B.; Larnicol, A.; Asokan, N.; Klein, A.; Osmani, N.; Busnelli, I.; Delalande, F.; Paul, N.; Halary, S.; et al. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife 2021, 10, e61539. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Duan, Z.; Zhang, C.; Wang, W.; He, H.; Liu, Y.; Wu, P.; Wang, S.; Song, M.; Chen, H.; et al. Mouse 4T1 Breast Cancer Cell-Derived Exosomes Induce Proinflammatory Cytokine Production in Macrophages via miR-183. J. Immunol. 2020, 205, 2916–2925. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Pan, Y.; Li, W.; Sun, C.; Liu, J.; Xu, T.; Shu, Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer-implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef]
- Wagener, N.; Holland, D.; Bulkescher, J.; Crnković-Mertens, I.; Hoppe-Seyler, K.; Zentgraf, H.; Pritsch, M.; Buse, S.; Pfitzenmaier, J.; Haferkamp, A.; et al. The enhancer of zeste homolog 2 gene contributes to cell proliferation and apoptosis resistance in renal cell carcinoma cells. Int. J. Cancer 2008, 123, 1545–1550. [Google Scholar] [CrossRef]
- Chen, S.; Ma, P.; Zhao, Y.; Li, B.; Jiang, S.; Xiong, H.; Wang, Z.; Wang, H.; Jin, X.; Liu, C. Biological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: Role of the MALAT-1-Livin protein interaction. J. Physiol. Sci. 2017, 67, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Ma, J.; Zhu, R.; Xu, C.; Zhang, J.; Chen, Y.; Chen, Z.; Gong, D.; Zheng, J.; Chen, C.; et al. MiR-532-5p suppresses renal cancer cell proliferation by disrupting the ETS1-mediated positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br. J. Cancer 2018, 119, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, F.; Yan, C.; Qu, W.; Zhu, L.; Guo, Z.; Zhou, F.; Zhang, W. Long Non-Coding RNA CASC19 Sponges microRNA-532 and Promotes Oncogenicity of Clear Cell Renal Cell Carcinoma by Increasing ETS1 Expression. Cancer Manag. Res. 2020, 12, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.J.; Lin, X.J.; Tang, X.Y.; Zheng, T.T.; Lin, Y.Y.; Hua, K.Q. Exosomal Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Angiogenesis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Int. J. Biol. Sci. 2018, 14, 1960–1973. [Google Scholar] [CrossRef]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R.V. Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Lab. Investig. 2018, 98, 1133–1142. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Z.; Xu, X. Long non-coding RNA MALAT1 correlates with cell viability and mobility by targeting miR-22-3p in renal cell carcinoma via the PI3K/Akt pathway. Oncol. Rep. 2019, 41, 1113–1121. [Google Scholar] [CrossRef]
- Syn, N.; Wang, L.; Sethi, G.; Thiery, J.P.; Goh, B.C. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharm. Sci. 2016, 37, 606–617. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, X.; Wang, H.; Wang, Y.; Yue, D.; Chen, R. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates renal cancer cell migration via cofilin-1. Oncol. Lett. 2020, 20, 53. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Chen, R.; Liu, Y.; Zhuang, H.; Yang, B.; Hei, K.; Xiao, M.; Hou, C.; Gao, H.; Zhang, X.; Jia, C.; et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017, 45, 9947–9959. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 2014, 159, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Magalhaes, M.A.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Eddy, R.; Condeelis, J. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 2007, 7, 429–440. [Google Scholar] [CrossRef]
- Guo, R.; Zou, B.; Liang, Y.; Bian, J.; Xu, J.; Zhou, Q.; Zhang, C.; Chen, T.; Yang, M.; Wang, H.; et al. LncRNA RCAT1 promotes tumor progression and metastasis via miR-214-5p/E2F2 axis in renal cell carcinoma. Cell Death Dis. 2021, 12, 689. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.R.; Wu, Y.S.; Wang, W.S.; Zhang, J.S.; Wu, Q.G. Upregulation of lncRNA DANCR functions as an oncogenic role in non-small lung cancer by regulating miR-214-5p/CIZ1 axis. Eur. Rev. Med. Pharm. Sci. 2020, 24, 2539–2547. [Google Scholar] [CrossRef]
- Guo, M.; Lin, B.; Li, G.; Lin, J.; Jiang, X. LncRNA TDRG1 promotes the proliferation, migration, and invasion of cervical cancer cells by sponging miR-214-5p to target SOX4. J. Recept. Signal Transduct. Res. 2020, 40, 281–293. [Google Scholar] [CrossRef]
- Apostolou, A.; Poreau, B.; Delrieu, L.; Thévenon, J.; Jouk, P.S.; Lallemand, G.; Emadali, A.; Sartelet, H. High Activation of the AKT Pathway in Human Multicystic Renal Dysplasia. Pathobiology 2020, 87, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Xu, W.-H.; Ren, F.; Wang, J.; Wang, H.-K.; Cao, D.-L.; Shi, G.-H.; Qu, Y.-Y.; Zhang, H.-L.; Ye, D.-W. Prognostic value of epithelial-mesenchymal transition markers in clear cell renal cell carcinoma. Aging 2020, 12, 866–883. [Google Scholar] [CrossRef]
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal cell carcinoma. BMJ 2014, 349, g4797. [Google Scholar] [CrossRef]
- Vaidya, A.M.; Sun, Z.; Ayat, N.; Schilb, A.; Liu, X.; Jiang, H.; Sun, D.; Scheidt, J.; Qian, V.; He, S.; et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjug. Chem. 2019, 30, 907–919. [Google Scholar] [CrossRef]
- Tan, L.; Tang, Y.; Li, H.; Li, P.; Ye, Y.; Cen, J.; Gui, C.; Luo, J.; Cao, J.; Wei, J. N6-Methyladenosine Modification of LncRNA DUXAP9 Promotes Renal Cancer Cells Proliferation and Motility by Activating the PI3K/AKT Signaling Pathway. Front. Oncol. 2021, 11, 641833. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, L.; Wang, Y.; Dai, G.; Xue, B.; Liu, Y.Y.; Zhu, J.; Zhu, J. BRD4 inhibition sensitizes renal cell carcinoma cells to the PI3K/mTOR dual inhibitor VS-5584. Aging 2020, 12, 19147–19158. [Google Scholar] [CrossRef] [PubMed]
- Merseburger, A.S.; Hennenlotter, J.; Kuehs, U.; Simon, P.; Kruck, S.; Koch, E.; Stenzl, A.; Kuczyk, M.A. Activation of PI3K is associated with reduced survival in renal cell carcinoma. Urol. Int. 2008, 80, 372–377. [Google Scholar] [CrossRef]
- Creighton, C.J.; Morgan, M.; Gunaratne, P.H.; Wheeler, D.A.; Gibbs, R.A.; Gordon Robertson, A.; Chu, A.; Beroukhim, R.; Cibulskis, K.; Signoretti, S.; et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Erin, N.; Grahovac, J.; Brozovic, A.; Efferth, T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist. Updates 2020, 53, 100715. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; An, S.; Choy, M.T.; Zhou, J.; Wu, S.; Liu, S.; Liu, B.; Yao, Z.; Zhu, X.; Wu, J.; et al. LncRNA DUXAP9-206 directly binds with Cbl-b to augment EGFR signaling and promotes non-small cell lung cancer progression. J. Cell Mol. Med. 2019, 23, 1852–1864. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Wang, C.; Liu, W.; Ai, Z.L. Emerging roles of the long non-coding RNA 01296/microRNA-143-3p/MSI2 axis in development of thyroid cancer. Biosci. Rep. 2019, 39, BSR20182376. [Google Scholar] [CrossRef]
- Chen, J.; Lou, W.; Ding, B.; Wang, X. Overexpressed pseudogenes, DUXAP8 and DUXAP9, promote growth of renal cell carcinoma and serve as unfavorable prognostic biomarkers. Aging 2019, 11, 5666–5688. [Google Scholar] [CrossRef]
- Kulkarni, P.; Dasgupta, P.; Hashimoto, Y.; Shiina, M.; Shahryari, V.; Tabatabai, Z.L.; Yamamura, S.; Tanaka, Y.; Saini, S.; Dahiya, R.; et al. A lncRNA TCL6-miR-155 Interaction Regulates the Src-Akt-EMT Network to Mediate Kidney Cancer Progression and Metastasis. Cancer Res. 2021, 81, 1500–1512. [Google Scholar] [CrossRef]
- Yang, K.; Lu, X.F.; Luo, P.C.; Zhang, J. Identification of Six Potentially Long Noncoding RNAs as Biomarkers Involved Competitive Endogenous RNA in Clear Cell Renal Cell Carcinoma. Biomed. Res. Int. 2018, 2018, 9303486. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; He, W.; Gou, X. Construction and comprehensive analysis of dysregulated long non-coding RNA-associated competing endogenous RNA network in clear cell renal cell carcinoma. J. Cell Biochem. 2018. [Google Scholar] [CrossRef]
- Liu, L.P.; Gong, Y.B. LncRNA-TCL6 promotes early abortion and inhibits placenta implantation via the EGFR pathway. Eur. Rev. Med. Pharm. Sci. 2018, 22, 7105–7112. [Google Scholar] [CrossRef]
- Roelants, C.; Giacosa, S.; Pillet, C.; Bussat, R.; Champelovier, P.; Bastien, O.; Guyon, L.; Arnoux, V.; Cochet, C.; Filhol, O. Combined inhibition of PI3K and Src kinases demonstrates synergistic therapeutic efficacy in clear-cell renal carcinoma. Oncotarget 2018, 9, 30066–30078. [Google Scholar] [CrossRef]
- Yonezawa, Y.; Nagashima, Y.; Sato, H.; Virgona, N.; Fukumoto, K.; Shirai, S.; Hagiwara, H.; Seki, T.; Ariga, T.; Senba, H.; et al. Contribution of the Src family of kinases to the appearance of malignant phenotypes in renal cancer cells. Mol. Carcinog. 2005, 43, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Sun, T.; Wang, H.; Shi, G.; Zhang, H.; Sun, F.; Ye, D. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget 2017, 8, 5789–5799. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.Y.; Wang, Y.; Wu, J.G.; Song, S.L.; Huang, G.; Xi, W.M.; Tan, L.L.; Wang, J.; Cao, Q. Analysis of long non-coding RNA expression profiles in clear cell renal cell carcinoma. Oncol. Lett. 2017, 14, 2757–2764. [Google Scholar] [CrossRef]
- Shen, P.; Qu, L.; Wang, J.; Ding, Q.; Zhou, C.; Xie, R.; Wang, H.; Ji, G. LncRNA LINC00342 contributes to the growth and metastasis of colorectal cancer via targeting miR-19a-3p/NPEPL1 axis. Cancer Cell Int. 2021, 21, 105. [Google Scholar] [CrossRef]
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic crosstalk in the breast cancer microenvironment. Eur. J. Cancer 2019, 121, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Nenkov, M.; Ma, Y.; Gaßler, N.; Chen, Y. Metabolic Reprogramming of Colorectal Cancer Cells and the Microenvironment: Implication for Therapy. Int. J. Mol. Sci. 2021, 22, 6262. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Fu, P.; Qu, L.; Liu, J.; Lin, A. Long Noncoding RNAs, New Critical Regulators in Cancer Immunity. Front. Oncol. 2020, 10, 550987. [Google Scholar] [CrossRef]
- Cai, C.F.; Ye, G.D.; Shen, D.Y.; Zhang, W.; Chen, M.L.; Chen, X.X.; Han, D.X.; Mi, Y.J.; Luo, Q.C.; Cai, W.Y.; et al. Chibby suppresses aerobic glycolysis and proliferation of nasopharyngeal carcinoma via the Wnt/β-catenin-Lin28/let7-PDK1 cascade. J. Exp. Clin. Cancer Res. 2018, 37, 104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Han, Q.; Zhao, H.; Zhang, J. Promotion of epithelial-mesenchymal transformation by hepatocellular carcinoma-educated macrophages through Wnt2b/β-catenin/c-Myc signaling and reprogramming glycolysis. J. Exp. Clin. Cancer Res. 2021, 40, 13. [Google Scholar] [CrossRef]
- Lee, S.Y.; Jeon, H.M.; Ju, M.K.; Kim, C.H.; Yoon, G.; Han, S.I.; Park, H.G.; Kang, H.S. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012, 72, 3607–3617. [Google Scholar] [CrossRef]
- Gao, L.; Zhao, A.; Wang, X. Upregulation of lncRNA AGAP2-AS1 is an independent predictor of poor survival in patients with clear cell renal carcinoma. Oncol. Lett. 2020, 19, 3993–4001. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Kristiansen, G.; Jung, K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat. Rev. Urol. 2016, 13, 734–752. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zheng, Y.; Dong, X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J. Cell Biochem. 2019, 120, 9056–9062. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.J.; Liu, Y.; Yang, B.; Tian, X.D.; Li, C.R.; Wang, B. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur. Rev. Med. Pharm. Sci. 2017, 21, 2392–2396. [Google Scholar]
- Luo, W.; Li, X.; Song, Z.; Zhu, X.; Zhao, S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging 2019, 11, 3811–3823. [Google Scholar] [CrossRef]
- Hui, B.; Ji, H.; Xu, Y.; Wang, J.; Ma, Z.; Zhang, C.; Wang, K.; Zhou, Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019, 10, 207. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Wang, L.; Yao, B.; Sun, L.; Liu, R.; Chen, T.; Niu, Y.; Tu, K.; Liu, Q. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16-5p and promotes proliferation and metastasis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 194. [Google Scholar] [CrossRef]
- Wang, W.; Yang, F.; Zhang, L.; Chen, J.; Zhao, Z.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget 2016, 7, 77225–77236. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Chen, M.; Xing, P.; Yan, X.; Xie, B. Increased Expression of Exosomal AGAP2-AS1 (AGAP2 Antisense RNA 1) In Breast Cancer Cells Inhibits Trastuzumab-Induced Cell Cytotoxicity. Med. Sci. Monit. 2019, 25, 2211–2220. [Google Scholar] [CrossRef]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lu, S.; Xu, Y.; Zheng, J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int. J. Biol. Macromol. 2019, 128, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Ramjiawan, R.R.; Griffioen, A.W.; Duda, D.G. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis 2017, 20, 185–204. [Google Scholar] [CrossRef]
- Chen, T.; You, Y.; Jiang, H.; Wang, Z.Z. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J. Cell Physiol. 2017, 232, 3261–3272. [Google Scholar] [CrossRef]
- Braune, E.B.; Lendahl, U. Notch -- a goldilocks signaling pathway in disease and cancer therapy. Discov. Med. 2016, 21, 189–196. [Google Scholar]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef]
- Chappell, J.C.; Payne, L.B.; Rathmell, W.K. Hypoxia, angiogenesis, and metabolism in the hereditary kidney cancers. J. Clin. Investig. 2019, 129, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Gong, B.; Wang, S.; Chen, Q.; Liu, Y.; Zhuang, C.; Li, Z.; Zhang, Z.; Ma, M.; Sun, T. Prognostic Value of Long Noncoding RNA DLEU2 and Its Relationship with Immune Infiltration in Kidney Renal Clear Cell Carcinoma and Liver Hepatocellular Carcinoma. Int. J. Gen. Med. 2021, 14, 8047–8064. [Google Scholar] [CrossRef]
- Xu, W.; Wang, B.; Cai, Y.; Guo, C.; Liu, K.; Yuan, C. DLEU2: A Meaningful Long Noncoding RNA in Oncogenesis. Curr. Pharm. Des. 2021, 27, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Mian, M.; Scandurra, M.; Chigrinova, E.; Shen, Y.; Inghirami, G.; Greiner, T.C.; Chan, W.C.; Vose, J.M.; Testoni, M.; Chiappella, A.; et al. Clinical and molecular characterization of diffuse large B-cell lymphomas with 13q14.3 deletion. Ann. Oncol. 2012, 23, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Qi, Y.; Xu, Y.; Wang, M.; Wang, J.; Wang, J.; Yuan, M.; Jia, Y.; Ma, X.; Wang, Y.; et al. lncRNA DLEU2 promotes gastric cancer progression through ETS2 via targeting miR-30a-5p. Cancer Cell Int. 2021, 21, 376. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wang, Y.; Cai, J.; Xie, Y.; Tao, C.; Jiang, Y.; Li, H.; Song, F. LncRNA DLEU2 promotes cervical cancer cell proliferation by regulating cell cycle and NOTCH pathway. Exp. Cell Res. 2021, 402, 112551. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Z.; Chen, Z.; Liu, B.; Wu, H. LncRNA DLEU2 is activated by STAT1 and induces gastric cancer development via targeting miR-23b-3p/NOTCH2 axis and Notch signaling pathway. Life Sci. 2021, 277, 119419. [Google Scholar] [CrossRef]
- Bao, S.; Jiang, X.; Jin, S.; Tu, P.; Lu, J. TGF-β1 induces immune escape by enhancing PD-1 and CTLA-4 expression on T lymphocytes in hepatocellular carcinoma. Front. Oncol. 2021, 11, 2516. [Google Scholar] [CrossRef]
- Poli, A.; Abdul-Hamid, S.; Zaurito, A.E.; Campagnoli, F.; Bevilacqua, V.; Sheth, B.; Fiume, R.; Pagani, M.; Abrignani, S.; Divecha, N. PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2010053118. [Google Scholar] [CrossRef]
- Sawant, A.; Hensel, J.A.; Chanda, D.; Harris, B.A.; Siegal, G.P.; Maheshwari, A.; Ponnazhagan, S. Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J. Immunol. 2012, 189, 4258–4265. [Google Scholar] [CrossRef]
- Xing, Z.; Zhang, M.; Liu, J.; Liu, G.; Feng, K.; Wang, X. LINC00337 induces tumor development and chemoresistance to paclitaxel of breast cancer by recruiting M2 tumor-associated macrophages. Mol. Immunol. 2021, 138, 1–9. [Google Scholar] [CrossRef]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Konno, Y.; Ihira, K.; Kobayashi, N.; Yue, J.; Watari, H. Long non-coding RNA DLEU2 drives EMT and glycolysis in endometrial cancer through HK2 by competitively binding with miR-455 and by modulating the EZH2/miR-181a pathway. J. Exp. Clin. Cancer Res. 2021, 40, 216. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qi, G.; Li, L. LncRNA NNT-AS1 promotes lung squamous cell carcinoma progression by regulating the miR-22/FOXM1 axis. Cell. Mol. Biol. Lett. 2020, 25, 34. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, R.; Qiao, Y.; Han, L.; Liu, M. LncRNA NNT-AS1 contributes to the cisplatin resistance of cervical cancer through NNT-AS1/miR-186/HMGB1 axis. Cancer Cell Int. 2020, 20, 190. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y. Circular RNAs in Hepatocellular Carcinoma: Emerging Functions to Clinical Significances. Front. Oncol. 2021, 11, 667428. [Google Scholar] [CrossRef]
- Li, K.; Sun, D.; Gou, Q.; Ke, X.; Gong, Y.; Zuo, Y.; Zhou, J.K.; Guo, C.; Xia, Z.; Liu, L.; et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018, 420, 80–90. [Google Scholar] [CrossRef]
- Yue, Q.Y.; Zhang, Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur. Rev. Med. Pharm. Sci. 2018, 22, 1003–1010. [Google Scholar] [CrossRef]
- Meng, X.; Sun, W.; Yu, J.; Zhou, Y.; Gu, Y.; Han, J.; Zhou, L.; Jiang, X.; Wang, C. LINC00460-miR-149-5p/miR-150-5p-Mutant p53 Feedback Loop Promotes Oxaliplatin Resistance in Colorectal Cancer. Mol. Nucleic Acids 2020, 22, 1004–1015. [Google Scholar] [CrossRef]
- Zhang, D.; Zeng, S.; Hu, X. Identification of a three-long noncoding RNA prognostic model involved competitive endogenous RNA in kidney renal clear cell carcinoma. Cancer Cell Int. 2020, 20, 319. [Google Scholar] [CrossRef]
- Yuan, B.; Yang, J.; Gu, H.; Ma, C. Down-regulation of LINC00460 represses metastasis of colorectal cancer via WWC2. Dig. Dis. Sci. 2020, 65, 442–456. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, J.; Wang, H.; Guo, H. Downregulation of long noncoding RNA LINC00460 expression suppresses tumor growth in vitro and in vivo in gastric cancer. Cancer Biomark. 2019, 24, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I. FOXM1 (Forkhead box M1) in tumorigenesis: Overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv. Cancer Res. 2013, 119, 191–419. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Li, Y.; Liu, J.; Yang, S.; Gui, Y.; Mao, X.; Nie, G.; Lai, Y. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol. Med. Rep. 2016, 13, 5386–5392. [Google Scholar] [CrossRef]
- Xie, M.; Lv, Y.; Liu, Z.; Zhang, J.; Liang, C.; Liao, X.; Liang, R.; Lin, Y.; Li, Y. Identification and validation of a four-miRNA (miRNA-21-5p, miRNA-9-5p, miR-149-5p, and miRNA-30b-5p) prognosis signature in clear cell renal cell carcinoma. Cancer Manag. Res. 2018, 10, 5759. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, M.; Agostini, F.; Masin, M.; Tartaglia, G.G. Predicting protein associations with long noncoding RNAs. Nat. Methods 2011, 8, 444–445. [Google Scholar] [CrossRef]
- Mizutani, A.; Koinuma, D.; Seimiya, H.; Miyazono, K. The Arkadia-ESRP2 axis suppresses tumor progression: Analyses in clear-cell renal cell carcinoma. Oncogene 2016, 35, 3514–3523. [Google Scholar] [CrossRef] [PubMed]
| Name | Plausible Mechanisms Involved | Effects |
|---|---|---|
| LINC00342 | Sponges miR-19a-3p [118] | |
| Lnc-LSG1 | Reduction of ESRP2 protein level [13] |
|
| AGAP2-AS1 | VEGF and Akt pathway Involvement of AGAP2-AS1 in angiogenesis, hypoxia, epithelial-mesenchymal transition, the notch signalling pathway, or stromal simulation [125,136,137,138] | |
| DLEU2 | Regulation of EMT, impact on Akt signalling pathway, stimulation of tumour cell proliferation via modulating Notch signalling pathway [144,145,146] |
|
| NNT-AS1 | Sponge of miR-22, miR-137 possible involvement in the regulation of gene expression at the post-transcriptional level [20] modulation of miR-137/YBX-1 axis | |
| LINC00460 | MiR-149-5p—the downstream target for LINC00460 modulation of FOXM1 |
|
| MALAT1 | Transcription factor (ETS1) Involvement in the regulation of RNA processing [90] Interaction with precofilin-1 (pre-CFL1) [92]. | |
| RCAT1 | Sponge of miR-214-5p | |
| DUXAP9 | m6A modifications, binding to IGF2BPS, the activation of the Akt-GSK3β-Snail signalling pathway, the interaction with E3 ubiquitin ligase Cbl-b [102] Induction of Akt/mTOR signalling via the activation of PI3K [102] |
|
| TCL6 | Regulation of EGFR/AKT pathway [113] lncTCL6-miR-155-Src/Akt/EMT network [110] |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rysz, J.; Konecki, T.; Franczyk, B.; Ławiński, J.; Gluba-Brzózka, A. The Role of Long Noncoding RNA (lncRNAs) Biomarkers in Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 643. https://doi.org/10.3390/ijms24010643
Rysz J, Konecki T, Franczyk B, Ławiński J, Gluba-Brzózka A. The Role of Long Noncoding RNA (lncRNAs) Biomarkers in Renal Cell Carcinoma. International Journal of Molecular Sciences. 2023; 24(1):643. https://doi.org/10.3390/ijms24010643
Chicago/Turabian StyleRysz, Jacek, Tomasz Konecki, Beata Franczyk, Janusz Ławiński, and Anna Gluba-Brzózka. 2023. "The Role of Long Noncoding RNA (lncRNAs) Biomarkers in Renal Cell Carcinoma" International Journal of Molecular Sciences 24, no. 1: 643. https://doi.org/10.3390/ijms24010643
APA StyleRysz, J., Konecki, T., Franczyk, B., Ławiński, J., & Gluba-Brzózka, A. (2023). The Role of Long Noncoding RNA (lncRNAs) Biomarkers in Renal Cell Carcinoma. International Journal of Molecular Sciences, 24(1), 643. https://doi.org/10.3390/ijms24010643








