Identification and Functional Validation of Auxin-Responsive Tabzip Genes from Wheat Leaves in Arabidopsis
Abstract
1. Introduction
2. Results
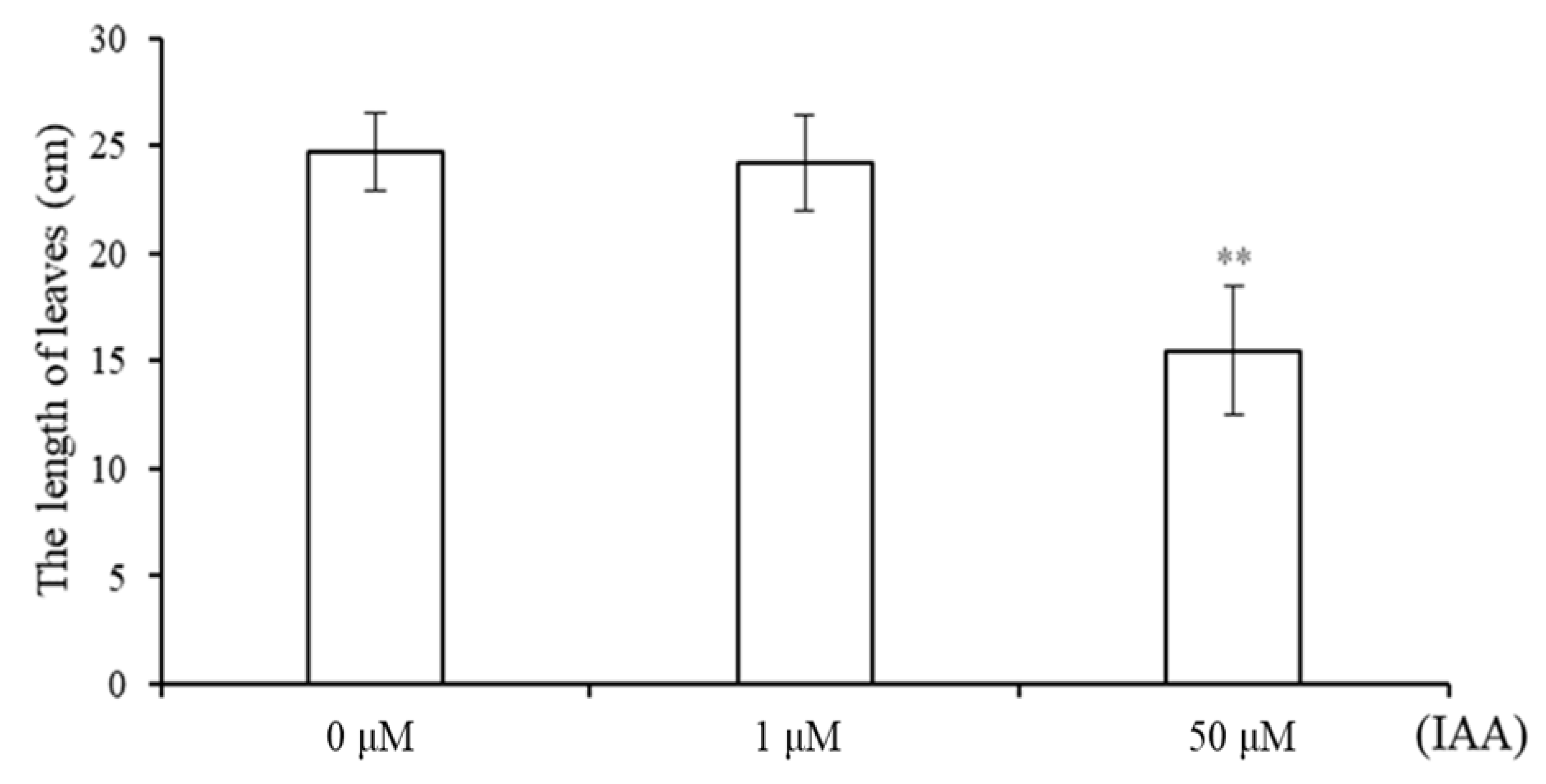
2.1. Phenotype of Wheat Seedlings under Different Concentrations of IAA
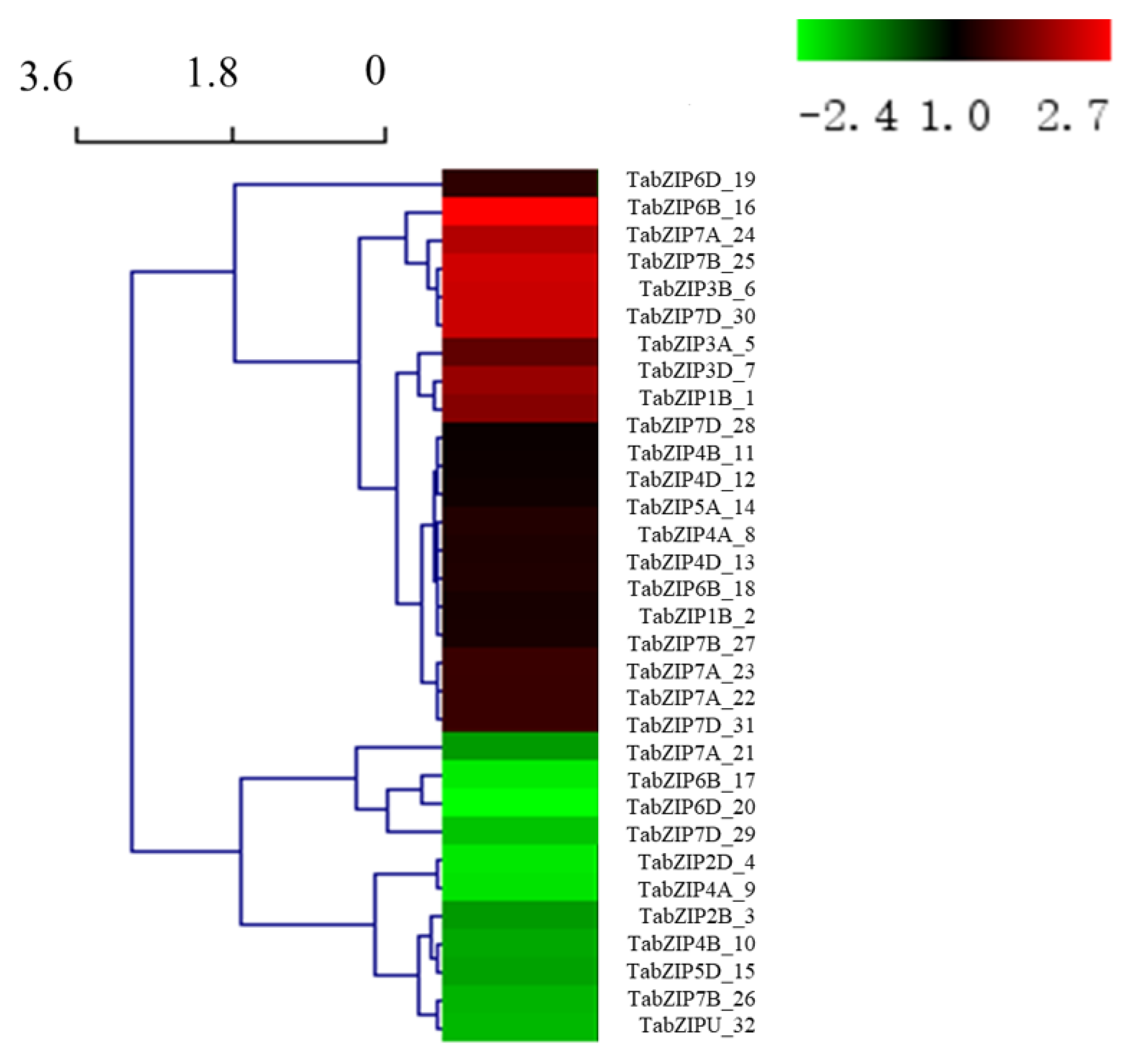
2.2. Identification of Auxin-Responsive TabZIP Genes in Wheat Leaves
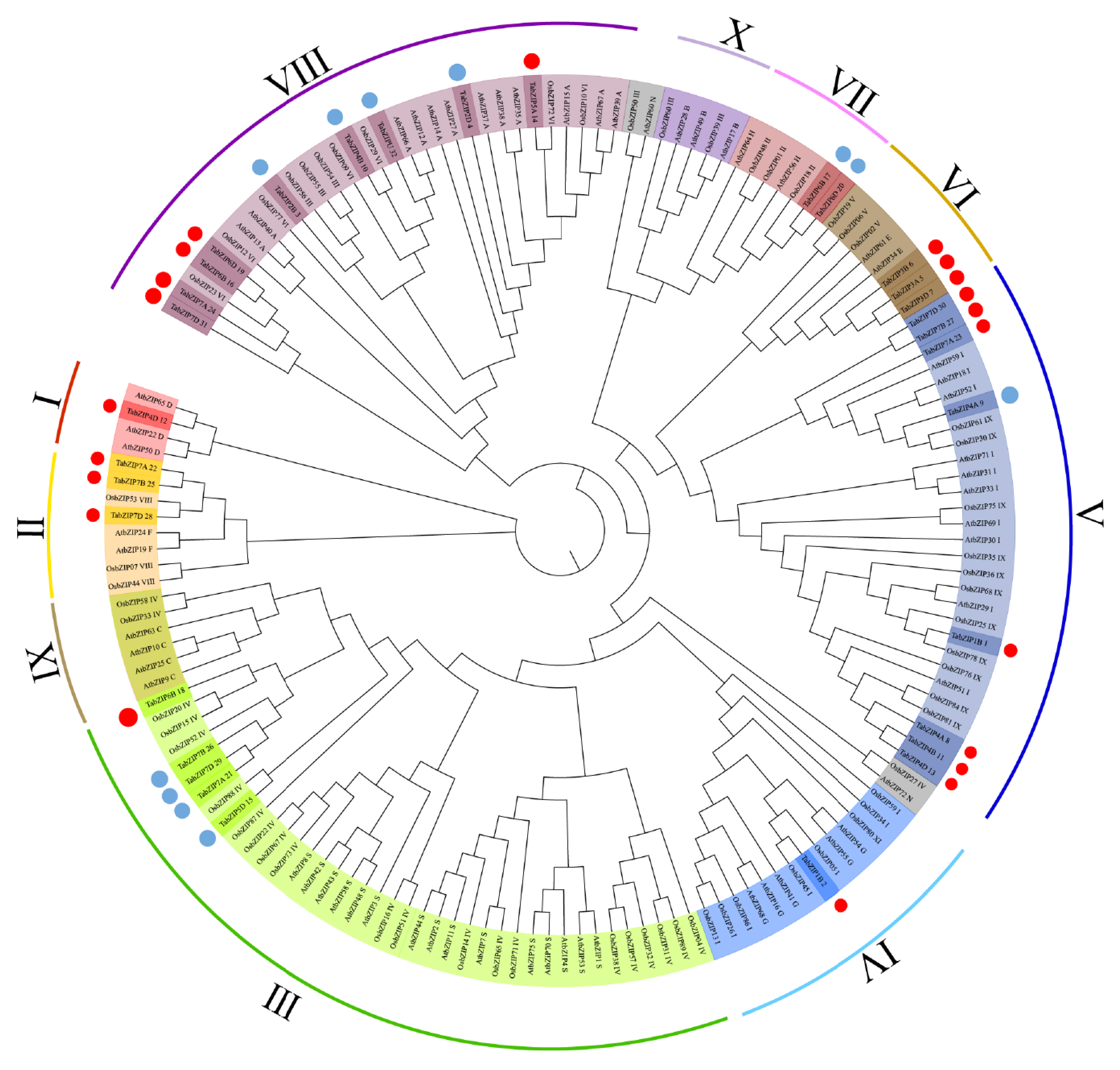
2.3. Phylogenetic Analysis, Conserved Motif, and Gene Structure Analysis of the Auxin-Responsive TabZIP Genes in Wheat Leaves
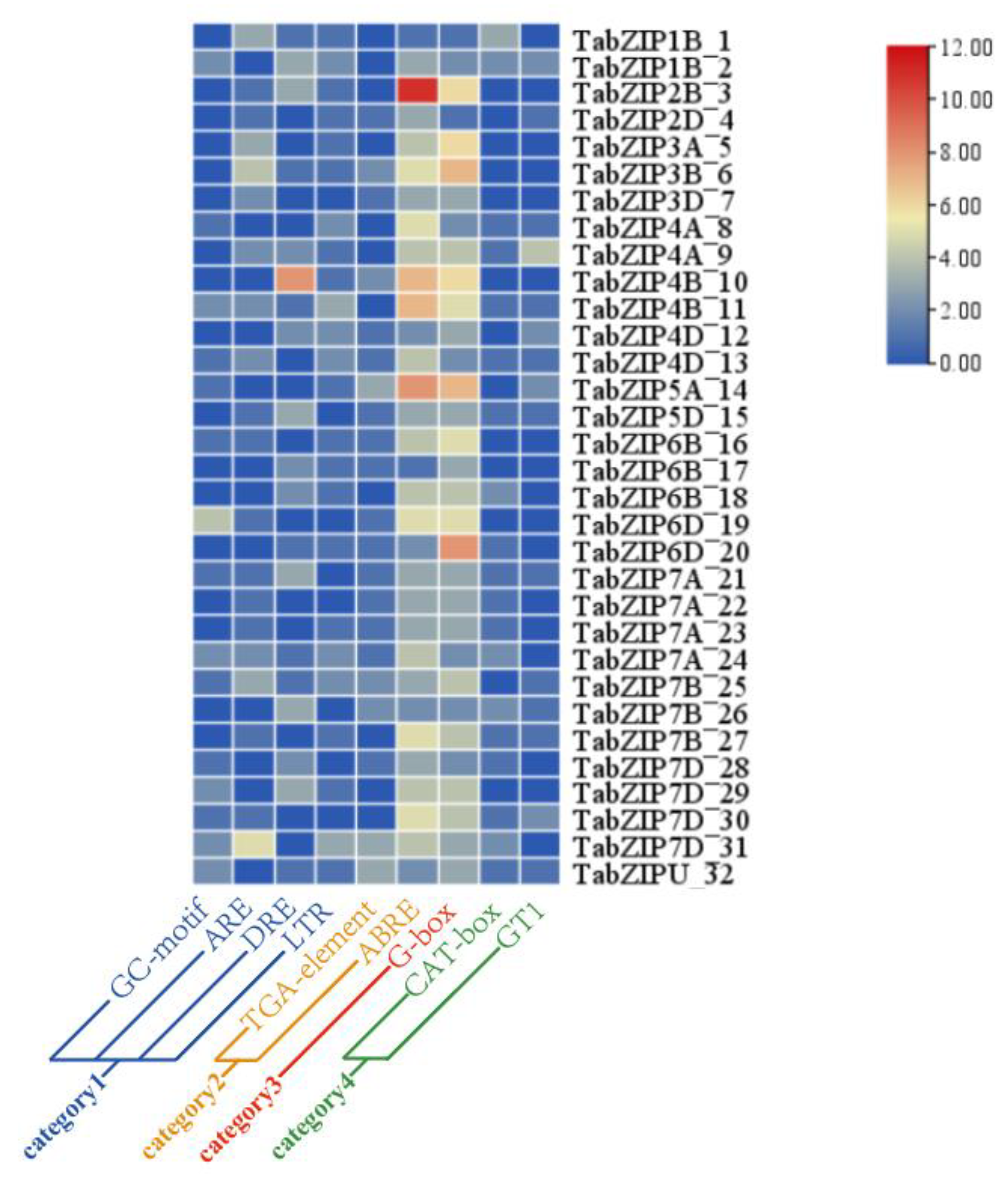
2.4. Cis-Acting Element Analysis of the 32 Auxin-Responsive TabZIP Genes
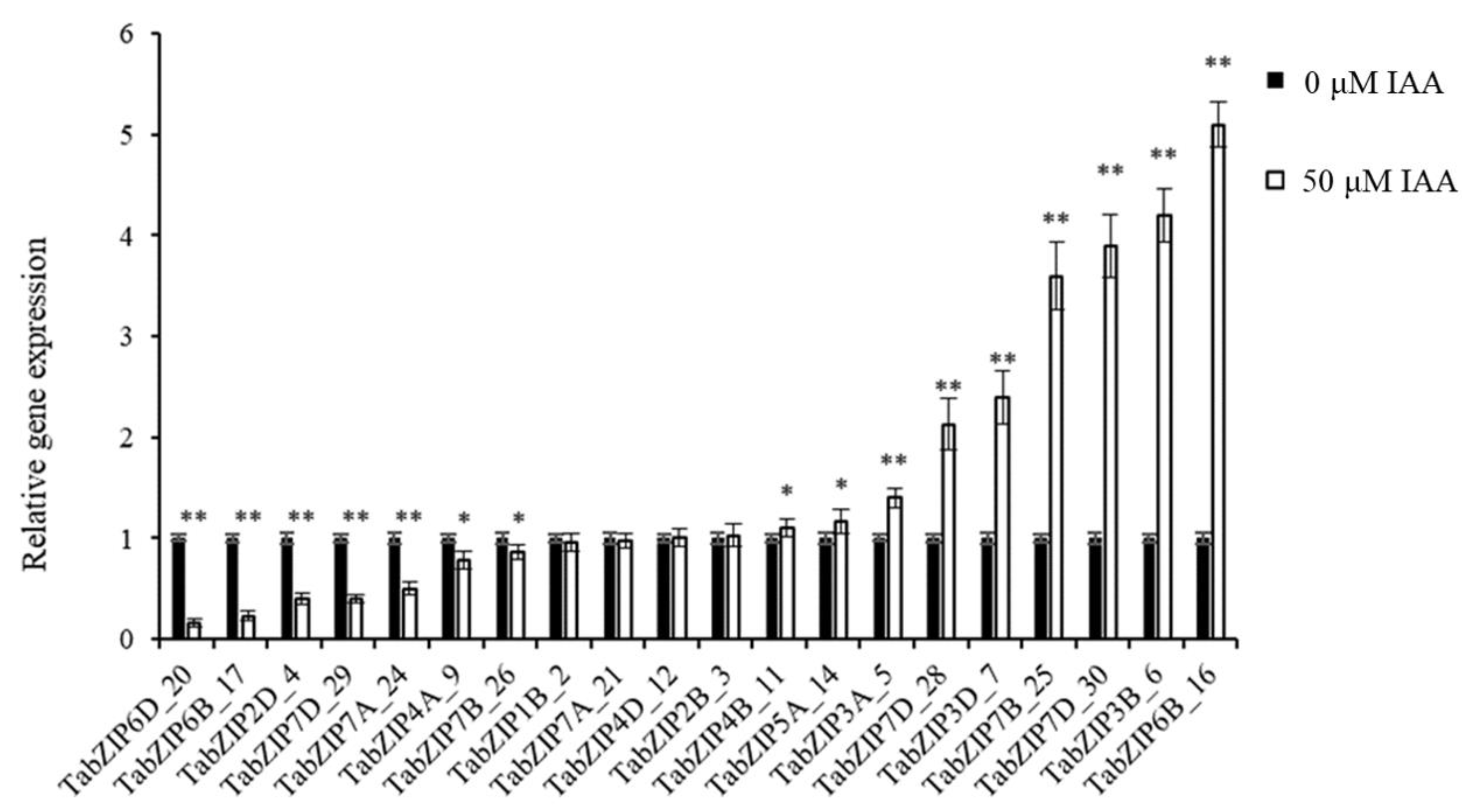
2.5. Gene Expression Analysis of Auxin-Responsive TabZIP Genes in Wheat Leaves by qRT-PCR
2.6. Down-Regulated TabZIP6D_20 Is a Negative Regulator of Auxin Response
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and IAA Treatments
4.2. Extracting RNA, Preparing Library and Sequencing
4.3. RNA-seq Reads Assembly and Quantification of Gene Expression
4.4. GO and KEGG Analysis of DEGs
4.5. Phylogenetic Analysis of Auxin-Responsive TabZIP Genes in Wheat Leaf
4.6. Gene Structure, Conserved Motif and Promoter Cis-Element Analysis of Auxin-Responsive TabZIP Genes in Wheat Leaf
4.7. Heatmap on Expression Level of the Auxin-Responsive TabZIP Genes in Wheat Leaf
4.8. qRT-PCR Analysis of 20 Auxin-Responsive TabZIP Genes in Wheat Leaves
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Blomme, J.; Inzé, D.; Gonzalez, N. The cell-cycle interactome: A source of growth regulators? J. Exp. Bot. 2014, 65, 2715–2730. [Google Scholar] [CrossRef] [PubMed]
- Omidbakhshfard, M.A.; Fujikura, U.; Olas, J.J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genet. 2018, 14, e1007484. [Google Scholar] [CrossRef]
- Bohn-Courseau, I. Auxin: A major regulator of organogenesis. Comptes Rendus Biol. 2010, 333, 290–296. [Google Scholar] [CrossRef]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Negin, A.; Kangarshahi, A.A.; Kazem, A.; Mohsen, B. Calyx Biochemical Changes and Possibility of Reducing Thomson Orange June Drop by Nutrition Elements and Growth Regulators. Int. J. Hortic. Sci. Technol. 2016, 3, 179–186. [Google Scholar] [CrossRef]
- Khodadadi, F.; Tohidfar, M.; Vahdati, K.; Dandekar, A.M.; Leslie, C.A. Functional analysis of walnut polyphenol oxidase gene (JrPPO1) in transgenic tobacco plants and PPO induction in response to walnut bacterial blight Plant Pathology. Plant Pathol. 2020, 69, 756–764. [Google Scholar] [CrossRef]
- Kalve, S.; Sizani, B.L.; Markakis, M.N.; Helsmoortel, C.; Vandeweyer, G.; Laukens, K.; Sommen, M.; Naulaerts, S.; Vissenberg, K.; Prinsen, E.; et al. Osmotic stress inhibits leaf growth of Arabidopsis thaliana by enhancing ARF-mediated auxin responses. New Phytol. 2020, 226, 1766–1780. [Google Scholar] [CrossRef]
- Guan, C.; Wu, B.; Yu, T.; Wang, Q.; Krogan, N.T.; Liu, X.; Jiao, Y. Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis. Curr. Biol. 2017, 27, 2940–2950.e4. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Berendzen, K.W.; Weiste, C.; Wanke, D.; Kilian, J.; Harter, K.; Dröge-Laser, W. Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. BMC Plant Biol. 2012, 12, 125. [Google Scholar] [CrossRef] [PubMed]
- Cherenkov, P.; Novikova, D.; Omelyanchuk, N.; Levitsky, V.; Grosse, I.; Weijers, D.; Mironova, V. Diversity of cis-regulatory elements associated with auxin response in Arabidopsis thaliana. J. Exp. Bot. 2018, 69, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Research Group bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Tian, X.; Jiang, Q.; Jia, Z.; Fang, Y.; Wang, Z.; Wang, J. Identification of TabZIP family members with possible roles in the response to auxin in wheat roots. Phytochemistry 2022, 196, 113103. [Google Scholar] [CrossRef]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide Analysis of bZIP Transcription Factors in wheat and Functional Characterization of a TabZIP under Abiotic Stress. Sci. Rep. 2019, 9, 4608. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Van Leene, J.; Blomme, J.; Kulkarni, S.R.; Cannoot, B.; De Winne, N.; Eeckhout, D.; Persiau, G.; Van De Slijke, E.; Vercruysse, L.; Vanden Bossche, R.; et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J. Exp. Bot. 2016, 67, 5825–5840. [Google Scholar] [CrossRef]
- Vikman, P.; Fadista, J.; Oskolkov, N. RNA sequencing: Current and prospective uses in metabolic research. J. Mol. Endocrinol. 2014, 53, R93–R101. [Google Scholar] [CrossRef] [PubMed]
- Sadat-Hosseini, M.; Bakhtiarizadeh, M.R.; Boroomand, N.; Tohidfar, M.; Vahdati, K. Combining independent de novo assemblies to optimize leaf transcriptome of Persian walnut. PloS ONE 2020, 15, e0232005. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; Neff, M.M.; Hong, S.W.; Zhang, H.; Deng, X.W.; Xiong, L. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 2008, 105, 4495–4500. [Google Scholar] [CrossRef]
- Balcerowicz, M.; Mahjoub, M.; Nguyen, D.; Lan, H.; Stoeckle, D.; Conde, S.; Jaeger, K.E.; Wigge, P.A.; Ezer, D. An early-morning gene network controlled by phytochromes and cryptochromes regulates photomorphogenesis pathways in Arabidopsis. Mol. Plant 2021, 14, 983–996. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Habricot, Y.; Condiff, A.S.; Maldiney, R.; Van der Straeten, D.; Ahmad, M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2007, 49, 428–441. [Google Scholar] [CrossRef]
- Mancinelli, A.L.; Rossi, F.; Moroni, A. Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol. 1991, 96, 1079–1085. [Google Scholar] [CrossRef][Green Version]
- Sharma, P.; Mishra, S.; Burman, N.; Chatterjee, M.; Singh, S.; Pradhan, A.K.; Khurana, P.; Khurana, J.P. Characterization of Cry2 genes (CRY2a and CRY2b) of B. napus and comparative analysis of BnCRY1 and BnCRY2a in regulating seedling photomorphogenesis. Plant Mol. Biol. 2022, 110, 161–186. [Google Scholar] [CrossRef]
- Tsuchida-Mayama, T.; Sakai, T.; Hanada, A.; Uehara, Y.; Asami, T.; Yamaguchi, S. Role of the phytochrome and cryptochrome signaling pathways in hypocotyl phototropism. Plant J. Cell Mol. Biol. 2010, 62, 653–662. [Google Scholar] [CrossRef]
- Xing, Y.; Sun, W.; Sun, Y.; Li, J.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. MPK6-mediated HY5 phosphorylation regulates light-induced anthocyanin accumulation in apple fruit. Plant Biotechnol. J. 2022. advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]




| Sample Name | Raw Reads | Clean Reads | Clean Bases | Clean Reads Q20 (%) | Clean Reads Q30 (%) | GC Content of Clean Reads (%) | Total Mapped Rate (%) |
|---|---|---|---|---|---|---|---|
| 0 µM_1 | 94,625,386 | 92,792,406 | 13.74 G | 95.72 | 88.78 | 55.9 | 93.74% |
| 0 µM_2 | 79,896,388 | 79,079,780 | 11.76 G | 96.72 | 91.6 | 57.22 | 94.52% |
| 0 µM_3 | 78,477,378 | 77,536,150 | 11.53 G | 96.47 | 91.09 | 57.73 | 94.20% |
| 50 µM_1 | 68,767,174 | 68,035,660 | 10.12 G | 96.63 | 91.41 | 55.23 | 94.04% |
| 50 µM_2 | 82,892,820 | 81,892,170 | 12.16 G | 96.75 | 90.96 | 55.9 | 95.03% |
| 50 µM_3 | 70,708,528 | 69,210,504 | 10.23 G | 95.24 | 87.74 | 54.26 | 92.96% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Z.; Zhang, M.; Ma, C.; Wang, Z.; Wang, Z.; Fang, Y.; Wang, J. Identification and Functional Validation of Auxin-Responsive Tabzip Genes from Wheat Leaves in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 756. https://doi.org/10.3390/ijms24010756
Jia Z, Zhang M, Ma C, Wang Z, Wang Z, Fang Y, Wang J. Identification and Functional Validation of Auxin-Responsive Tabzip Genes from Wheat Leaves in Arabidopsis. International Journal of Molecular Sciences. 2023; 24(1):756. https://doi.org/10.3390/ijms24010756
Chicago/Turabian StyleJia, Ziyao, Mengjie Zhang, Can Ma, Zanqiang Wang, Zhonghua Wang, Yan Fang, and Jun Wang. 2023. "Identification and Functional Validation of Auxin-Responsive Tabzip Genes from Wheat Leaves in Arabidopsis" International Journal of Molecular Sciences 24, no. 1: 756. https://doi.org/10.3390/ijms24010756
APA StyleJia, Z., Zhang, M., Ma, C., Wang, Z., Wang, Z., Fang, Y., & Wang, J. (2023). Identification and Functional Validation of Auxin-Responsive Tabzip Genes from Wheat Leaves in Arabidopsis. International Journal of Molecular Sciences, 24(1), 756. https://doi.org/10.3390/ijms24010756





