Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency
Abstract
1. Introduction
2. Results
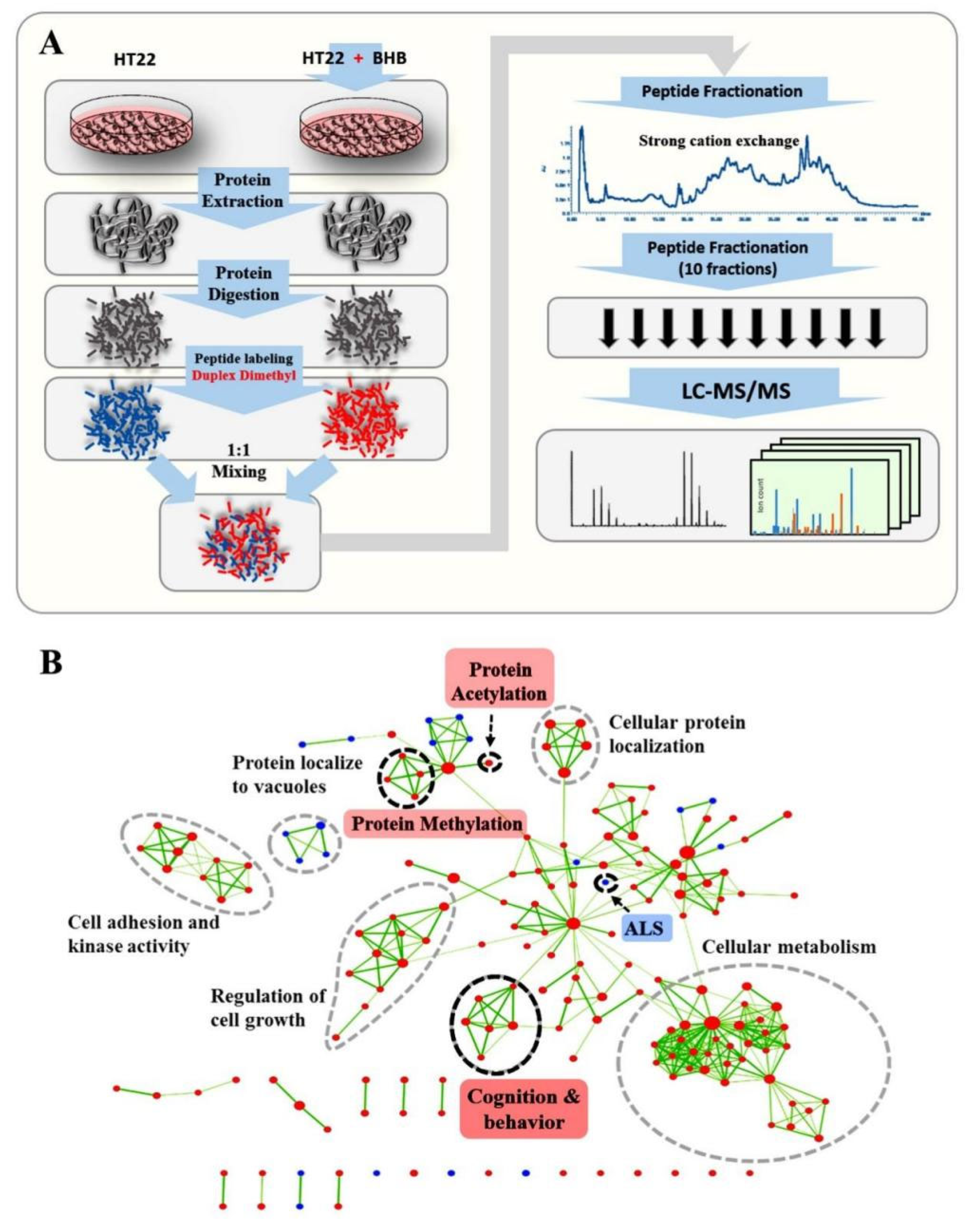
2.1. Proteomics Assessment of the Effects of 3OHB Treatment in Neuronal Cells
2.2. Unbiased Gene Set Enrichment Analysis Identified Pathways That Were Highly Enriched amongst Proteins Altered in Abundance after 3OHB Treatment
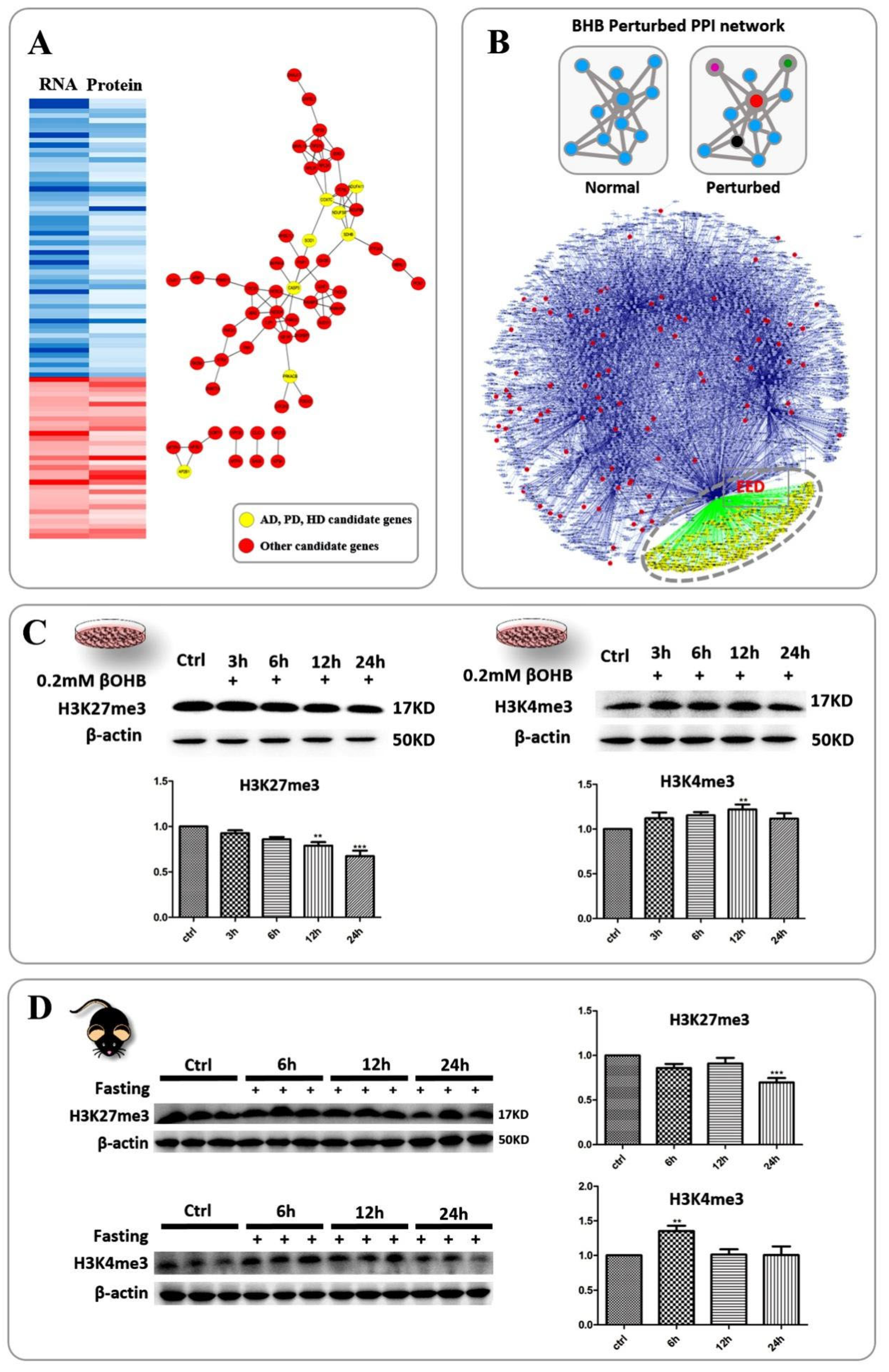
2.3. An Analysis of the Disease-Related Protein-Protein Interaction Network Indicated That H3K27me3 Is a Hub Marker of the Neuronal Response to 3OHB
2.4. The Correlation between Chromatin Bivalency and the Influence of 3OHB on Gene Expression Highlighted the 3OHB-Responsive Transcriptional Regulatory Network as an Intermediary between Disease-Related Genes and 3OHB
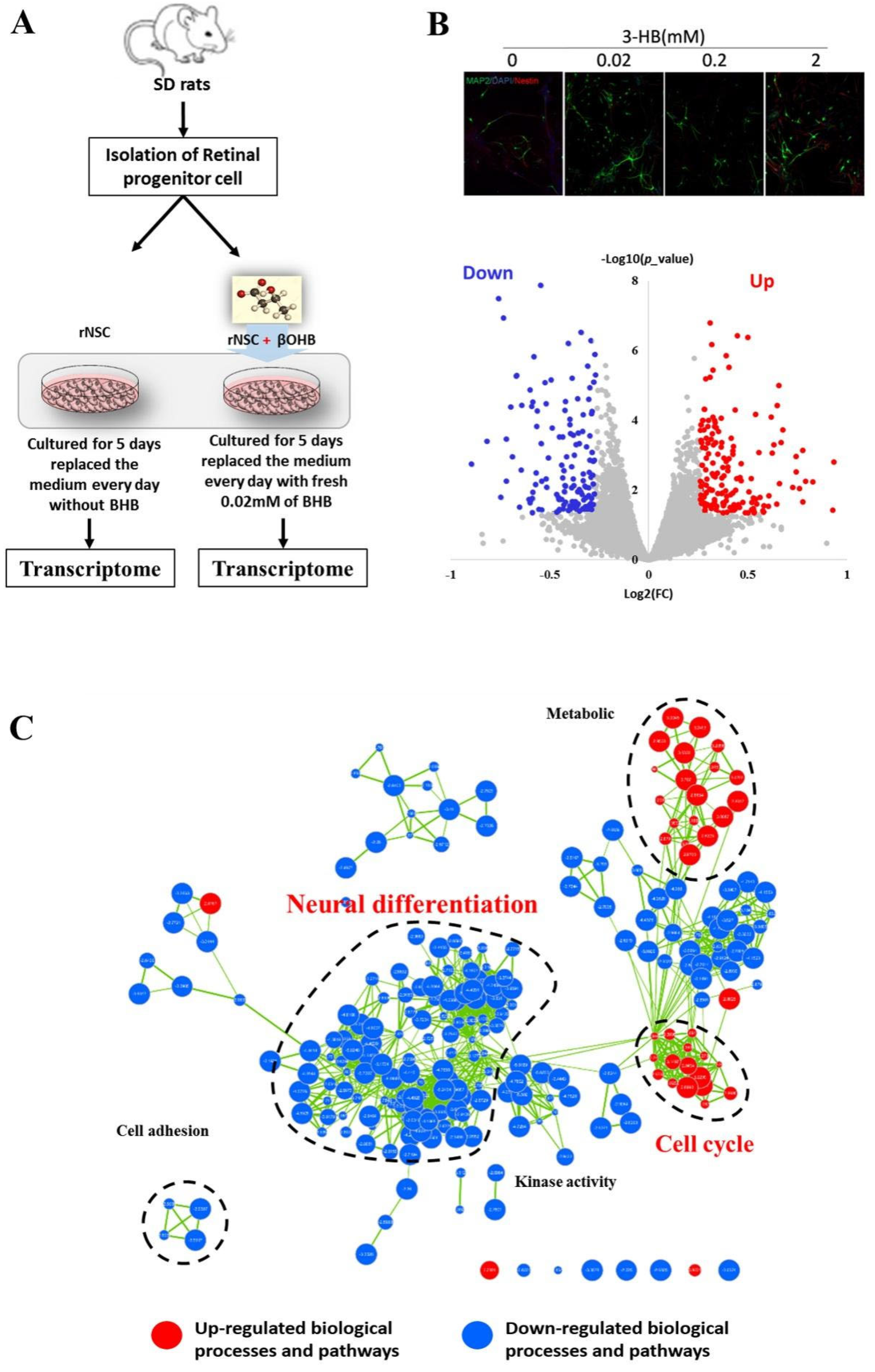
2.5. OHB Perturbed Chromatin Bivalency and Resulted in the Alteration of Neural Differentiation Processes
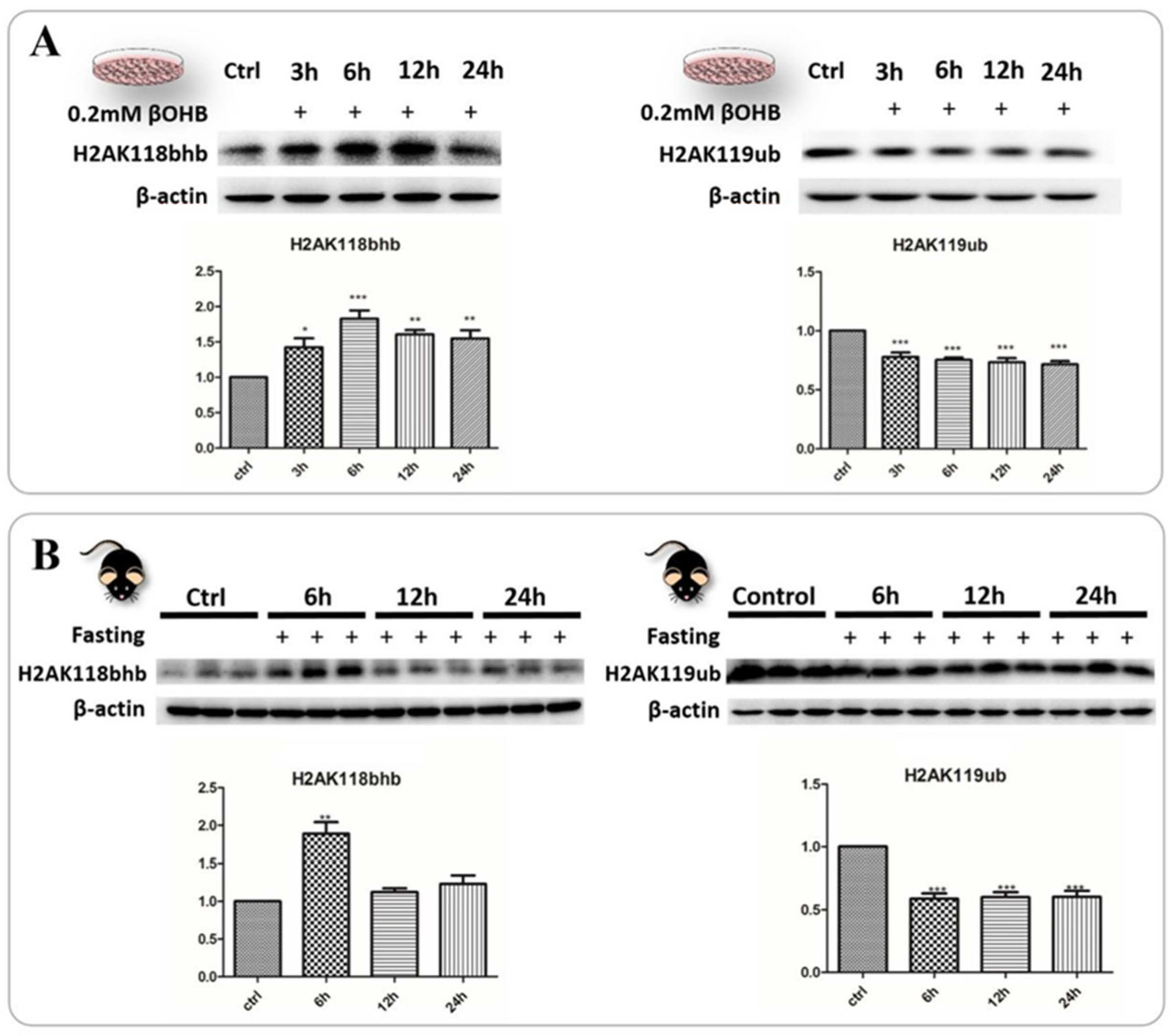
2.6. Identification and Validation of Abundant Histone Lysine Hydroxybutyrylation (Kbhb) Sites
3. Discussion
4. Materials and Methods
4.1. HT22 Cell Culture and 3OHB Treatment
4.2. Culture of NSCs and Transcriptome Sequencing
4.3. Animal Maintenance and Treatment
4.4. Mass Spectrometry of Proteins
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maalouf, M.; Rho, J.M.; Mattson, M.P. The neuroprotective properties of calorie restriction, the ketogenic diet, and ketone bodies. Brain Res. Rev. 2009, 59, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Marosi, K.; Kim, S.W.; Moehl, K.; Scheibye-Knudsen, M.; Cheng, A.; Cutler, R.; Camandola, S.; Mattson, M.P. 3-Hydroxybutyrate regulates energy metabolism and induces BDNF expression in cerebral cortical neurons. J. Neurochem. 2016, 139, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Silver, I.A.; Erecinska, M. Extracellular glucose concentration in mammalian brain: Continuous monitoring of changes during increased neuronal activity and upon limitation in oxygen supply in normo-, hypo-, and hyperglycemic animals. J. Neurosci. 1994, 14, 5068–5076. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Mattson, M.P. Fasting: Molecular mechanisms and clinical applications. Cell Metab. 2014, 19, 181–192. [Google Scholar] [CrossRef]
- Kashiwaya, Y.; Takeshima, T.; Mori, N.; Nakashima, K.; Clarke, K.; Veech, R.L. d-β-Hydroxybutyrate protects neurons in models of Alzheimer’s and Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2000, 97, 5440–5444. [Google Scholar] [CrossRef]
- Henderson, S.T. Ketone bodies as a therapeutic for Alzheimer’s disease. Neurotherapeutics 2008, 5, 470–480. [Google Scholar] [CrossRef]
- Tieu, K.; Perier, C.; Caspersen, C.; Teismann, P.; Wu, D.-C.; Yan, S.-D.; Naini, A.; Vila, M.; Jackson-Lewis, V.; Ramasamy, R. D-β-Hydroxybutyrate rescues mitochondrial respiration and mitigates features of Parkinson disease. J. Clin. Investig. 2003, 112, 892–901. [Google Scholar] [CrossRef]
- Yin, J.X.; Maalouf, M.; Han, P.; Zhao, M.; Gao, M.; Dharshaun, T.; Ryan, C.; Whitelegge, J.; Wu, J.; Eisenberg, D. Ketones block amyloid entry and improve cognition in an Alzheimer’s model. Neurobiol. Aging 2016, 39, 25–37. [Google Scholar] [CrossRef]
- Ruskin, D.N.; Ross, J.L.; Kawamura, M.; Ruiz, T.L.; Geiger, J.D.; Masino, S.A. A ketogenic diet delays weight loss and does not impair working memory or motor function in the R6/2 1J mouse model of Huntington’s disease. Physiol. Behav. 2011, 103, 501–507. [Google Scholar] [CrossRef]
- Zhao, Z.; Lange, D.J.; Voustianiouk, A.; MacGrogan, D.; Ho, L.; Suh, J.; Humala, N.; Thiyagarajan, M.; Wang, J.; Pasinetti, G.M. A ketogenic diet as a potential novel therapeutic intervention in amyotrophic lateral sclerosis. BMC Neurosci. 2006, 7, 29. [Google Scholar] [CrossRef]
- Cotter, D.G.; Schugar, R.C.; Crawford, P.A.; Physiology, C. Ketone body metabolism and cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H1060–H1076. [Google Scholar] [CrossRef] [PubMed]
- Camberos-Luna, L.; Massieu, L. Therapeutic strategies for ketosis induction and their potential efficacy for the treatment of acute brain injury and neurodegenerative diseases. Neurochem. Int. 2020, 133, 104614. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Achanta, L.B.; Rae, C.D. β-Hydroxybutyrate in the brain: One molecule, multiple mechanisms. Neurochem. Res. 2016, 45, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Lund, T.M.; Ploug, K.B.; Iversen, A.; Jensen, A.A.; Jansen-Olesen, I. The metabolic impact of β-hydroxybutyrate on neurotransmission: Reduced glycolysis mediates changes in calcium responses and KATP channel receptor sensitivity. J. Neurochem. 2015, 132, 520–531. [Google Scholar] [CrossRef]
- Benjamin, J.S.; Pilarowski, G.O.; Carosso, G.A.; Zhang, L.; Huso, D.L.; Goff, L.A.; Vernon, H.J.; Hansen, K.D.; Bjornsson, H.T. A ketogenic diet rescues hippocampal memory defects in a mouse model of Kabuki syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, 125–130. [Google Scholar] [CrossRef]
- von Schimmelmann, M.; Feinberg, P.A.; Sullivan, J.M.; Ku, S.M.; Badimon, A.; Duff, M.K.; Wang, Z.; Lachmann, A.; Dewell, S.; Ma’ayan, A. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 2016, 19, 1321–1330. [Google Scholar] [CrossRef]
- Chestnut, B.A.; Chang, Q.; Price, A.; Lesuisse, C.; Wong, M.; Martin, L.J. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011, 31, 16619–16636. [Google Scholar] [CrossRef]
- Chen, S.; Li, J.; Wang, D.-L.; Sun, F.-L. Histone H2B lysine 120 monoubiquitination is required for embryonic stem cell differentiation. Cell Res. 2012, 22, 1402. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Müller, C.W.; Vermeulen, M.; Müller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Barañano, K.W.; Hartman, A.L. The ketogenic diet: Uses in epilepsy and other neurologic illnesses. Curr. Treat. Options Neurol. 2008, 10, 410. [Google Scholar] [CrossRef]
- Ruskin, D.N.; Svedova, J.; Cote, J.L.; Sandau, U.; Rho, J.M.; Kawamura, M., Jr.; Boison, D.; Masino, S.A. Ketogenic diet improves core symptoms of autism in BTBR mice. PLoS ONE 2013, 8, e65021. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, K.; Sandoval, H.; Yamamoto, S.; Jaiswal, M.; Sanz, E.; Li, Z.; Hui, J.; Graham, B.H.; Quintana, A. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 2015, 160, 177–190. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Fang, J.; Yu, F.; Heng, D.; Fan, Y.; Xu, J.; Peng, B.; Liu, W.; Han, S. Decreased Methylation Level of H3K27me3 Increases Seizure Susceptibility. Mol. Neurobiol. 2016, 54, 7343–7352. [Google Scholar] [CrossRef]
- Li, J.; Hart, R.P.; Mallimo, E.M.; Swerdel, M.R.; Kusnecov, A.W.; Herrup, K. EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat. Neurosci. 2013, 16, 1745–1753. [Google Scholar] [CrossRef]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef]
- Schwartz, Y.B.; Pirrotta, V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef]
- Cooper, S.; Dienstbier, M.; Hassan, R.; Schermelleh, L.; Sharif, J.; Blackledge, N.P.; De Marco, V.; Elderkin, S.; Koseki, H.; Klose, R. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014, 7, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Sanulli, S.; Justin, N.; Teissandier, A.; Ancelin, K.; Portoso, M.; Caron, M.; Michaud, A.; Lombard, B.; Da Rocha, S.T.; Offer, J. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell 2015, 57, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zee, B.M.; Wang, Y.; Garcia, B.A.; Dou, Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol. Cell 2011, 43, 132–144. [Google Scholar] [CrossRef]
- Wu, L.; Lee, S.Y.; Zhou, B.; Nguyen, U.T.; Muir, T.W.; Tan, S.; Dou, Y. ASH2L regulates ubiquitylation signaling to MLL: Trans-regulation of H3 K4 methylation in higher eukaryotes. Mol. Cell 2013, 49, 1108–1120. [Google Scholar] [CrossRef]
- Vlaming, H.; van Welsem, T.; de Graaf, E.L.; Ontoso, D.; Altelaar, A.M.; San-Segundo, P.A.; Heck, A.J.; van Leeuwen, F. Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo. EMBO Rep. 2014, 15, 1077–1084. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Shilatifard, A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 2010, 339, 240–249. [Google Scholar] [CrossRef]
- Tahiliani, M.; Mei, P.; Fang, R.; Leonor, T.; Rutenberg, M.; Shimizu, F.; Li, J.; Rao, A.; Shi, Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007, 447, 601–605. [Google Scholar] [CrossRef]
- Shulha, H.P.; Cheung, I.; Whittle, C.; Wang, J.; Virgil, D.; Lin, C.L.; Guo, Y.; Lessard, A.; Akbarian, S.; Weng, Z. Epigenetic signatures of autism: Trimethylated H3K4 landscapes in prefrontal neurons. Arch. Gen. Psychiatry 2012, 69, 314–324. [Google Scholar] [CrossRef]
- Shulha, H.P.; Crisci, J.L.; Reshetov, D.; Tushir, J.S.; Cheung, I.; Bharadwaj, R.; Chou, H.-J.; Houston, I.B.; Peter, C.J.; Mitchell, A.C. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012, 10, e1001427. [Google Scholar] [CrossRef]
- Cheung, I.; Shulha, H.P.; Jiang, Y.; Matevossian, A.; Wang, J.; Weng, Z.; Akbarian, S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 8824–8829. [Google Scholar] [CrossRef] [PubMed]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.-H.; Kellis, M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 2015, 518, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Tsuji, J.; Labadorf, A.; Roussos, P.; Chen, J.-F.; Myers, R.H.; Akbarian, S.; Weng, Z. The role of H3K4me3 in transcriptional regulation is altered in Huntington’s disease. PLoS ONE 2015, 10, e0144398. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.-K.; Koche, R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef]
- Shen, E.; Shulha, H.; Weng, Z.; Akbarian, S. Regulation of histone H3K4 methylation in brain development and disease. Philos. Trans. R. Soc. B 2014, 369, 20130514. [Google Scholar] [CrossRef]
- Fuchs, G.; Shema, E.; Vesterman, R.; Kotler, E.; Wolchinsky, Z.; Wilder, S.; Golomb, L.; Pribluda, A.; Zhang, F.; Haj-Yahya, M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitination. Mol. Cell 2012, 46, 662–673. [Google Scholar] [CrossRef]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484–494. [Google Scholar] [CrossRef]
- Wang, W.; Meng, M.; Zhang, Y.; Wei, C.; Xie, Y.; Jiang, L.; Wang, C.; Yang, F.; Tang, W.; Jin, X. Global transcriptome-wide analysis of CIK cells identify distinct roles of IL-2 and IL-15 in acquisition of cytotoxic capacity against tumor. BMC Med. Genom. 2014, 7, 49. [Google Scholar] [CrossRef]
- Jiao, Q.; Li, X.; An, J.; Zhang, Z.; Chen, X.; Tan, J.; Zhang, P.; Lu, H.; Liu, Y. Cell-Cell Connection Enhances Proliferation and Neuronal Differentiation of Rat Embryonic Neural Stem/Progenitor Cells. Front. Cell. Neurosci. 2017, 11, 200. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Ochagavia, M.E.; Rabasa, L.C.; Miranda, J.; Fernandez-de-Cossio, J.; Bringas, R. BisoGenet: A new tool for gene network building, visualization and analysis. BMC Bioinform. 2010, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Niere, F.; Namjoshi, S.; Song, E.; Dilly, G.A.; Schoenhard, G.; Zemelman, B.V.; Mechref, Y.; Raab-Graham, K.F. Analysis of proteins that rapidly change upon mechanistic/mammalian target of rapamycin complex 1 (mTORC1) repression identifies parkinson protein 7 (PARK7) as a novel protein aberrantly expressed in tuberous sclerosis complex (TSC). Mol. Cell. Proteom. 2016, 15, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Abel, O.; Powell, J.F.; Andersen, P.M.; Al-Chalabi, A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012, 33, 1345–1351. [Google Scholar] [CrossRef]
- Mao, Y.; Kuo, S.-W.; Chen, L.; Heckman, C.; Jiang, M. The essential and downstream common proteins of amyotrophic lateral sclerosis: A protein-protein interaction network analysis. PLoS ONE 2017, 12, e0172246. [Google Scholar] [CrossRef]
- Fujita, K.A.; Ostaszewski, M.; Matsuoka, Y.; Ghosh, S.; Glaab, E.; Trefois, C.; Crespo, I.; Perumal, T.M.; Jurkowski, W.; Antony, P.M. Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol. Neurobiol. 2014, 49, 88–102. [Google Scholar] [CrossRef]
- Foulger, R.E.; Denny, P.; Hardy, J.; Martin, M.J.; Sawford, T.; Lovering, R.C. Using the gene ontology to annotate key players in Parkinson’s disease. Neuroinformatics 2016, 14, 297–304. [Google Scholar] [CrossRef]
- Stroedicke, M.; Bounab, Y.; Strempel, N.; Klockmeier, K.; Yigit, S.; Friedrich, R.P.; Chaurasia, G.; Li, S.; Hesse, F.; Riechers, S.-P. Systematic interaction network filtering identifies CRMP1 as a novel suppressor of huntingtin misfolding and neurotoxicity. Genome Res. 2015, 25, 701–713. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.-L.; Du, H.; Wang, L.-L.; Hu, E.-L.; Li, N.; Lu, H.-X.; Chen, G.-Q.; Lu, X.-Y. Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency. Int. J. Mol. Sci. 2023, 24, 868. https://doi.org/10.3390/ijms24010868
Zhu X-L, Du H, Wang L-L, Hu E-L, Li N, Lu H-X, Chen G-Q, Lu X-Y. Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency. International Journal of Molecular Sciences. 2023; 24(1):868. https://doi.org/10.3390/ijms24010868
Chicago/Turabian StyleZhu, Xin-Liang, Huan Du, Lei-Lei Wang, Er-Ling Hu, Ning Li, Hai-Xia Lu, Guo-Qiang Chen, and Xiao-Yun Lu. 2023. "Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency" International Journal of Molecular Sciences 24, no. 1: 868. https://doi.org/10.3390/ijms24010868
APA StyleZhu, X.-L., Du, H., Wang, L.-L., Hu, E.-L., Li, N., Lu, H.-X., Chen, G.-Q., & Lu, X.-Y. (2023). Integrative Proteome Analysis Revels 3-Hydroxybutyrate Exerts Neuroprotective Effect by Influencing Chromatin Bivalency. International Journal of Molecular Sciences, 24(1), 868. https://doi.org/10.3390/ijms24010868







