Isolation of Bacillus siamensis B-612, a Strain That Is Resistant to Rice Blast Disease and an Investigation of the Mechanisms Responsible for Suppressing Rice Blast Fungus
Abstract
1. Introduction
2. Results
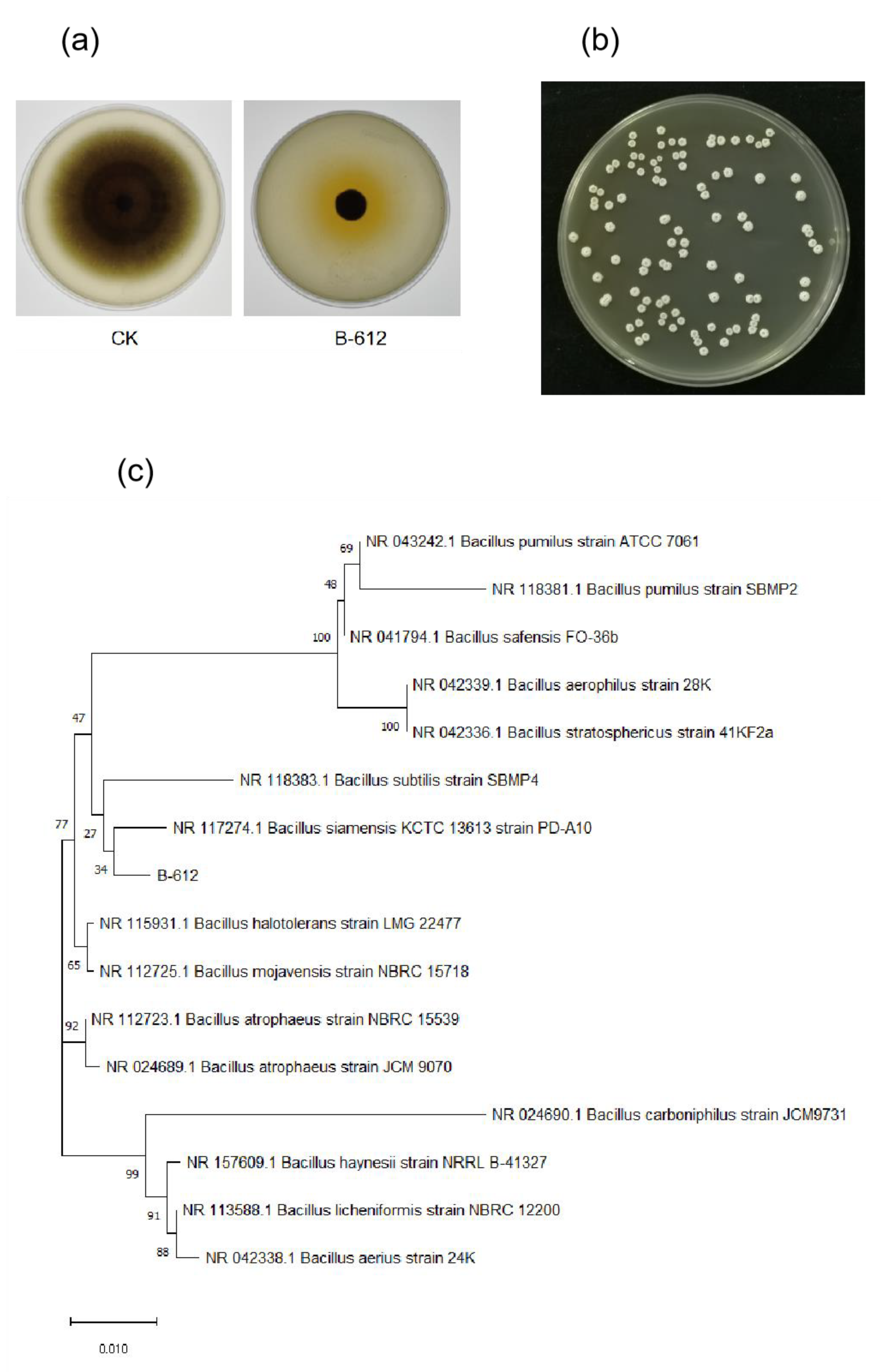
2.1. Isolation and Identification of Endophytes
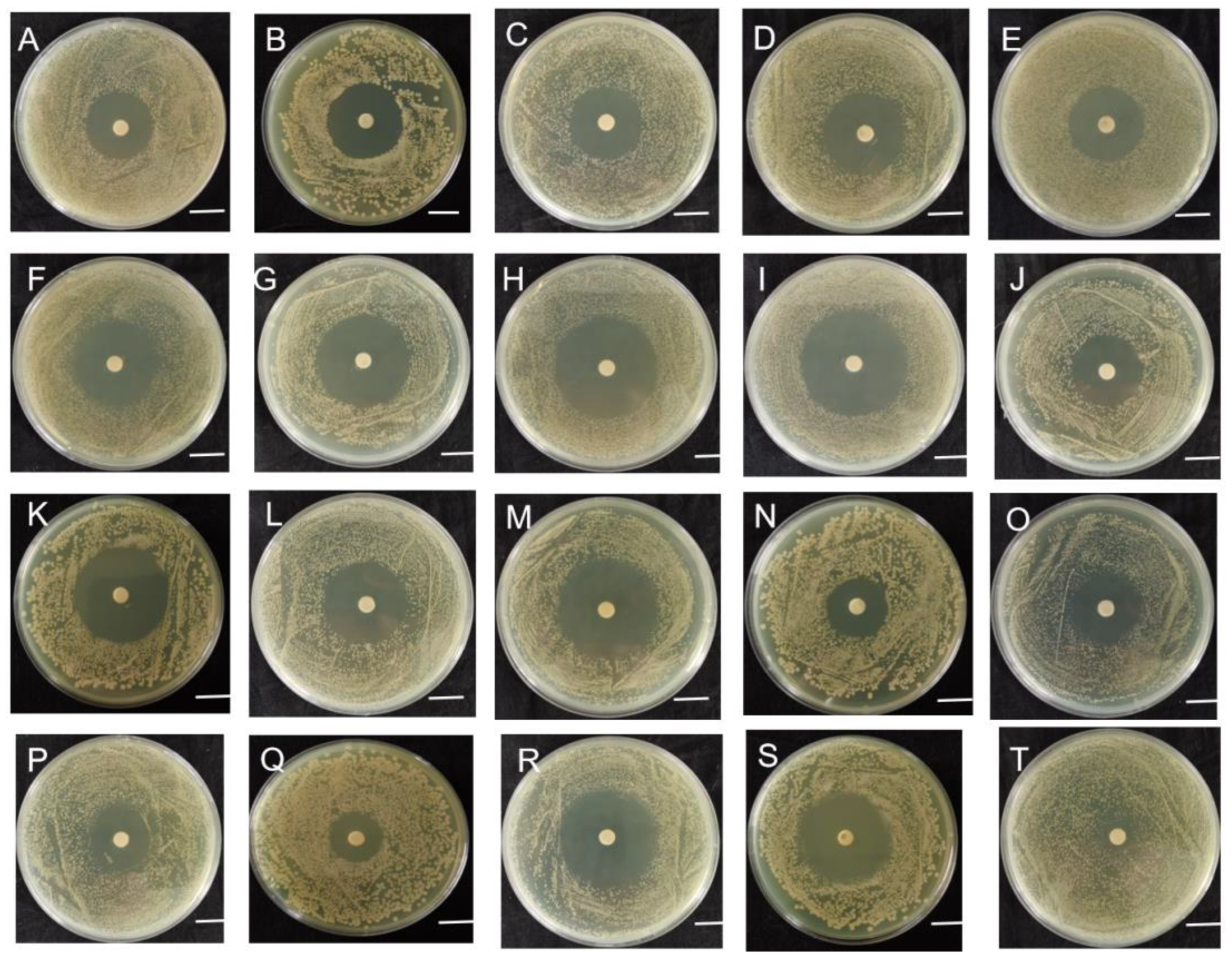
2.2. Sensitivity of Strain B-612 to Various Antibiotics
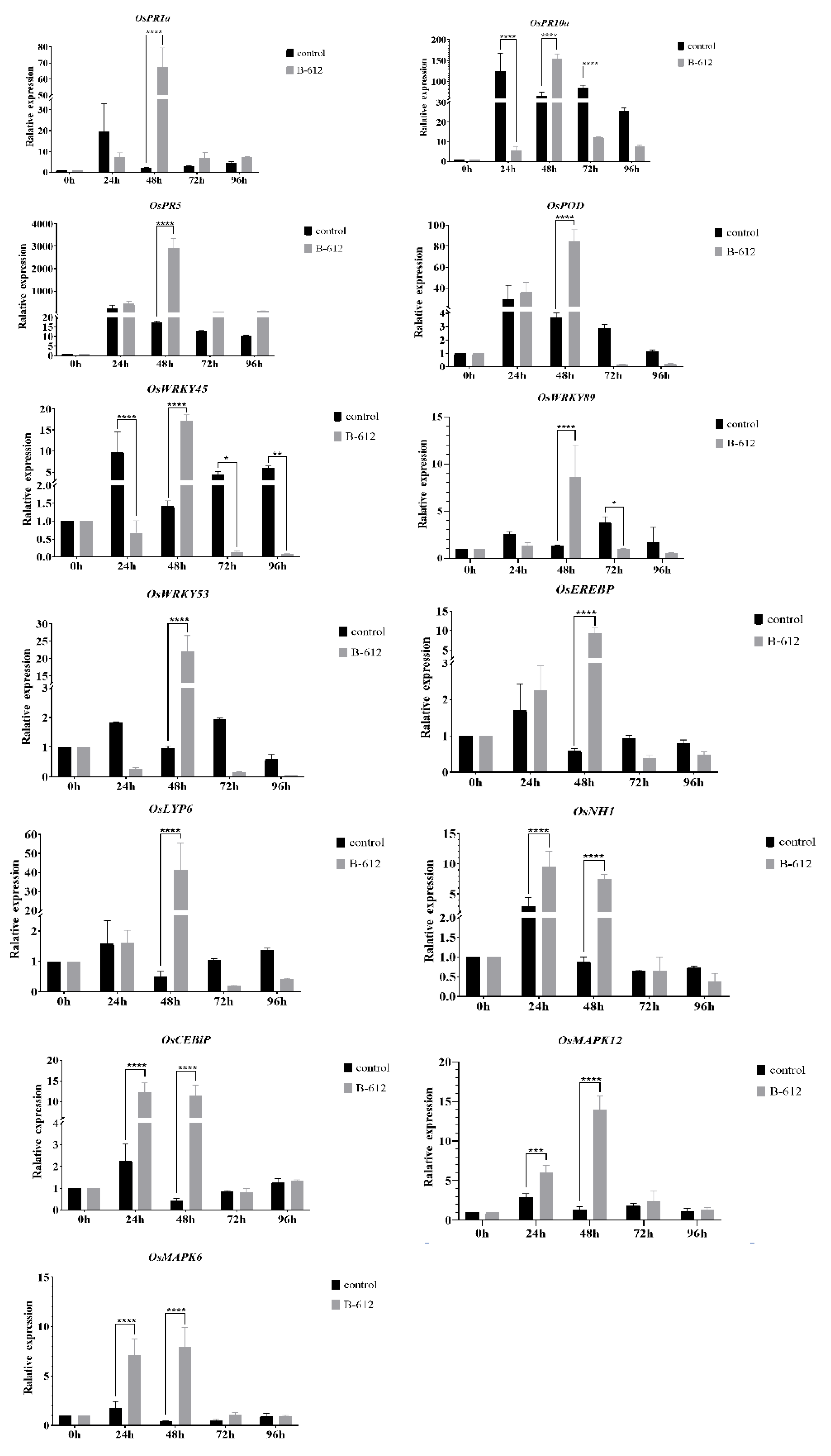
2.3. B-612 Fermentation Solution Enhanced the Resistance of Rice to M. oryzae
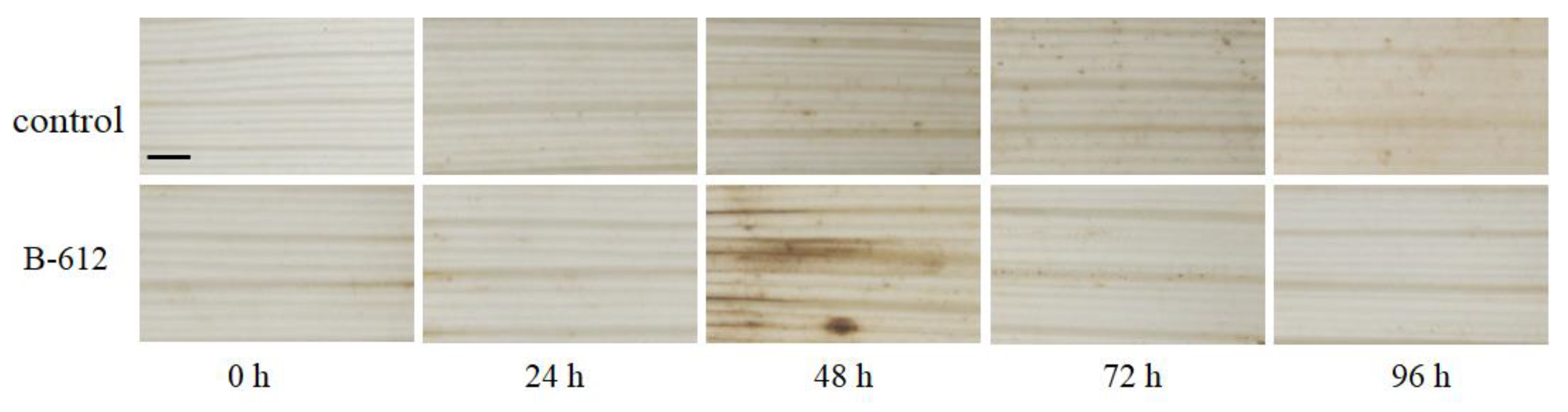
2.4. H2O2 Accumulation
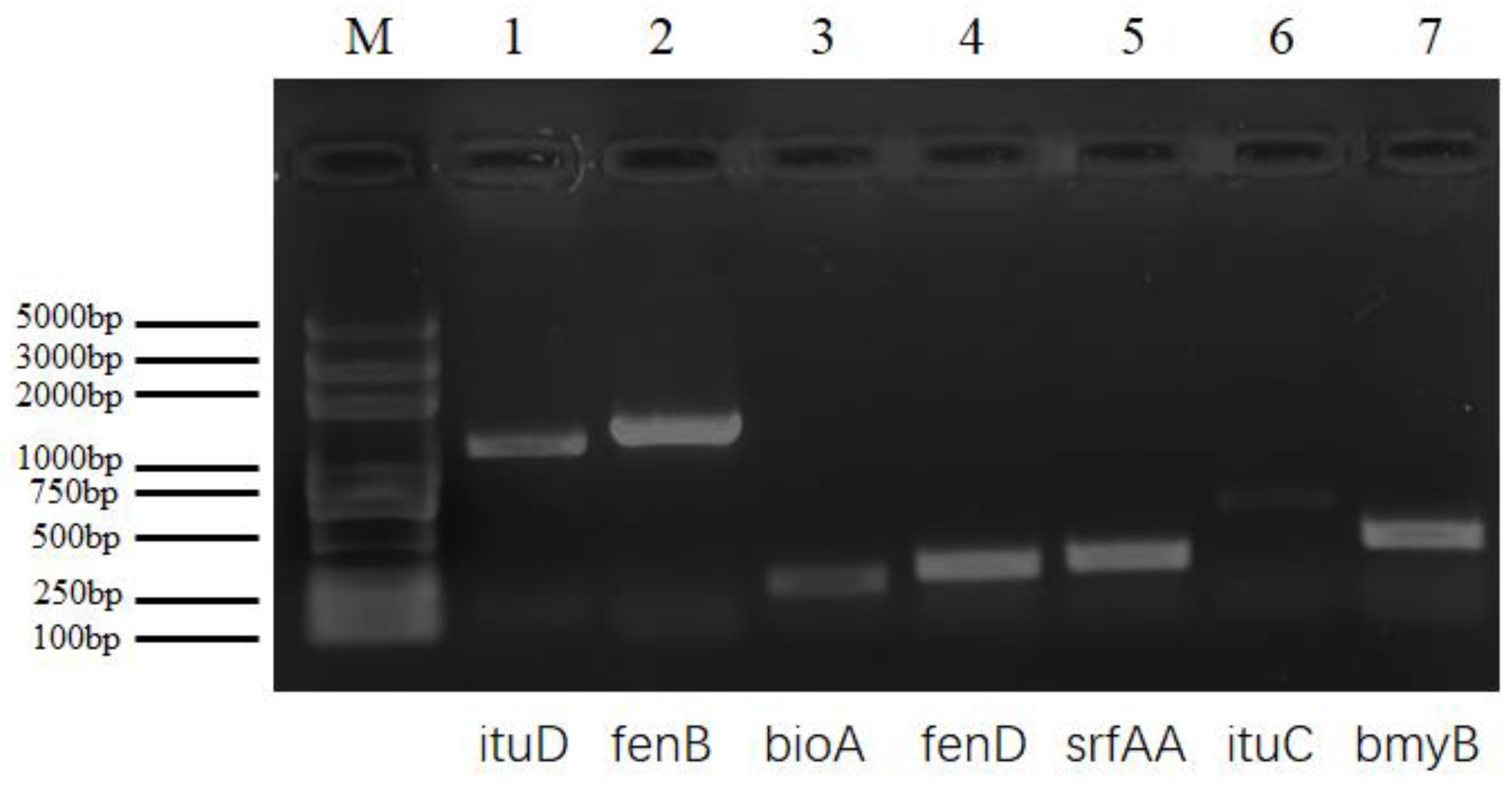
2.5. Detection of Lipopeptide Biosynthetic Genes
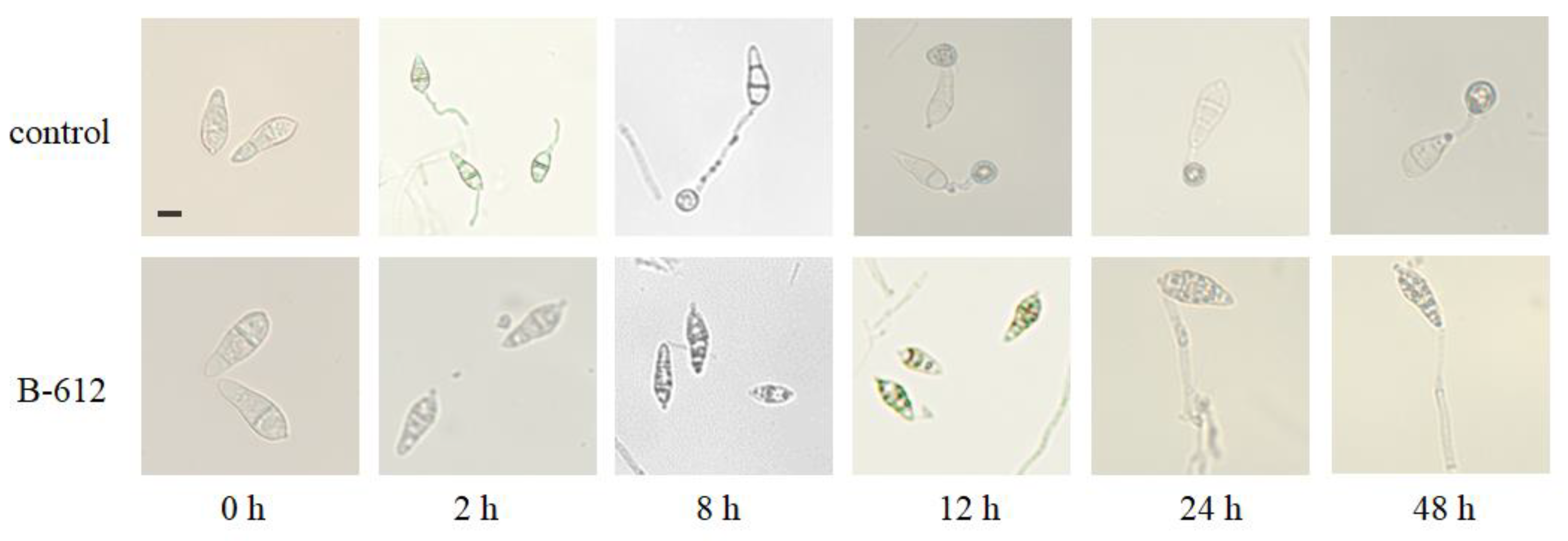
2.6. The Effects of Antifungal Substances Generated by Strain B-612 on M. oryzae Conidia Germination and the Formation of Appressorium
2.7. Biocontrol Efficacy of Strain B-612 and Its Culture Filtrate
3. Discussion
4. Methods
4.1. Isolation and Cultivation of Endophytic Bacteria
4.2. Rice Blast Pathogen (M. oryzae Guy11) and Culture Conditions
4.3. Fungal Antagonism Assay
4.4. Identification of Strain B-612
4.5. Antibiotic Susceptibility Assay
4.6. Defense-Related Gene Expression
4.7. H2O2 Accumulation
4.8. Detection of Lipopeptide Biosynthetic Genes
4.9. B-612 Fermentation Broth Crude Extraction
4.10. Germination Testing of M. oryzae Conidia and the Formation of Appressorium
4.11. In Vivo Experiments with Living Leaves
4.12. Main Instrumentation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, G.S.; Lal, R.; Meena, R.S.; Datta, M.; Babu, S.; Das, A.; Layek, J.; Saha, P. Energy Budgeting for Designing Sustainable and Environmentally Clean/Safer Cropping Systems for Rainfed Rice Fallow Lands in India. J. Clean. Prod. 2017, 158, 29–37. [Google Scholar] [CrossRef]
- Ebbole, D.J. Magnaporthe as a Model for Understanding Host-Pathogen Interactions. Annu. Rev. Phytopathol. 2007, 45, 437–456. [Google Scholar] [CrossRef] [PubMed]
- Saunders, D.G.O.; Aves, S.J.; Talbot, N.J. Cell Cycle-Mediated Regulation of Plant Infection by the Rice Blast Fungus. Plant Cell 2010, 22, 497–507. [Google Scholar] [CrossRef]
- Jasrotia, S.; Salgotra, R.K.; Sharma, M. Efficacy of Bioinoculants to Control of Bacterial and Fungal Diseases of Rice (Oryza sativa L.) in Northwestern Himalaya. Braz. J. Microbiol. 2021, 52, 687–704. [Google Scholar] [CrossRef] [PubMed]
- Shoda, M. Bacterial Control of Plant Diseases. J. Biosci. Bioeng. 2000, 89, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B. Mutualistic Interactions with Fungal Root Endophytes. In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2006; Volume 9, pp. 261–279. ISBN 978-3-540-33525-2. [Google Scholar]
- Huang, W.; Liu, X.; Zhou, X.; Wang, X.; Liu, X.; Liu, H. Calcium Signaling Is Suppressed in Magnaporthe oryzae Conidia by Bacillus Cereus HS24. Phytopathology 2020, 110, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Xu, H.; Li, L.; Chen, R.; Gao, X.; Xu, Z. Antifungal Activity of Endophytic Bacillus Safensis B21 and Its Potential Application as a Biopesticide to Control Rice Blast. Pestic. Biochem. Physiol. 2020, 162, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. The Structure and Diversity of Human, Animal and Environmental Resistomes. Microbiome 2016, 4, 54. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.-G.; Singer, A.C.; Zhu, Y.-G. Review of Antibiotic Resistance in China and Its Environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora Brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168. [Google Scholar]
- Kouzai, Y.; Nakajima, K.; Hayafune, M.; Ozawa, K.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. CEBiP Is the Major Chitin Oligomer-Binding Protein in Rice and Plays a Main Role in the Perception of Chitin Oligomers. Plant Mol. Biol. 2014, 84, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Guo, W.; Lu, J.; Yu, H.; Qu, C.; Tang, Z.; Li, J.; Chai, Y.; Liang, Y. Genome-Wide Survey and Expression Profile Analysis of the Mitogen-Activated Protein Kinase (MAPK) Gene Family in Brassica Rapa. PLoS ONE 2015, 10, e0132051. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.-F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin Motif-Containing Proteins LYP4 and LYP6 Play Dual Roles in Peptidoglycan and Chitin Perception in Rice Innate Immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Slusarenko, A.J. Production of Salicylic Acid Precursors Is a Major Function of Phenylalanine Ammonia-Lyase in the Resistance of Arabidopsis to Peronospora Parasitica. Plant Cell 1996, 8, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.-J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 Plays a Crucial Role in Benzothiadiazole-Inducible Blast Resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Kim, Y.-H.; Kim, M.-D.; Kwon, S.-Y.; Cho, K.; Lee, H.-S.; Kwak, S.-S. Simultaneous Expression of Choline Oxidase, Superoxide Dismutase and Ascorbate Peroxidase in Potato Plant Chloroplasts Provides Synergistically Enhanced Protection against Various Abiotic Stresses. Physiol. Plant 2010, 138, 520–533. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y.; Ikehara, N. Flavonoid-Peroxidase Reaction as a Detoxification Mechanism of Plant Cells against H2O2. Plant Physiol. 1997, 115, 1405–1412. [Google Scholar] [CrossRef]
- Ancheeva, E.; Daletos, G.; Proksch, P. Bioactive Secondary Metabolites from Endophytic Fungi. Curr. Med. Chem. 2020, 27, 1836–1854. [Google Scholar] [CrossRef]
- Lopes, R.; Tsui, S.; Gonçalves, P.J.R.O.; de Queiroz, M.V. A Look into a Multifunctional Toolbox: Endophytic Bacillus Species Provide Broad and Underexploited Benefits for Plants. World J. Microbiol. Biotechnol. 2018, 34, 94. [Google Scholar] [CrossRef]
- Chung, E.J.; Hossain, M.T.; Khan, A.; Kim, K.H.; Jeon, C.O.; Chung, Y.R. Bacillus oryzicola Sp. Nov., an Endophytic Bacterium Isolated from the Roots of Rice with Antimicrobial, Plant Growth Promoting, and Systemic Resistance Inducing Activities in Rice. Plant Pathol. J. 2015, 31, 152–164. [Google Scholar] [CrossRef]
- Hussain, T.; Khan, A.A. Biocontrol Prospective of Bacillus Siamensis-AMU03 against Soil-Borne Fungal Pathogens of Potato Tubers. Indian Phytopathol. 2022, 75, 179–189. [Google Scholar] [CrossRef]
- Ibrahim, E.; Fouad, H.; Zhang, M.; Zhang, Y.; Qiu, W.; Yan, C.; Li, B.; Mo, J.; Chen, J. Biosynthesis of Silver Nanoparticles Using Endophytic Bacteria and Their Role in Inhibition of Rice Pathogenic Bacteria and Plant Growth Promotion. RSC Adv. 2019, 9, 29293–29299. [Google Scholar] [CrossRef]
- Xie, Z.; Li, M.; Wang, D.; Wang, F.; Shen, H.; Sun, G.; Feng, C.; Wang, X.; Chen, D.; Sun, X. Biocontrol Efficacy of Bacillus Siamensis LZ88 against Brown Spot Disease of Tobacco Caused by Alternaria Alternata. Biol. Control 2021, 154, 104508. [Google Scholar] [CrossRef]
- Howard, R.J.; Valent, B. Breaking and Entering: Host Penetration by the Fungal Rice Blast Pathogen Magnaporthe Grisea. Annu. Rev. Microbiol. 1996, 50, 491–512. [Google Scholar] [CrossRef]
- Wilson, R.A.; Talbot, N.J. Under Pressure: Investigating the Biology of Plant Infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Galhano, R.; Talbot, N.J. The Biology of Blast: Understanding How Magnaporthe oryzae Invades Rice Plants. Fungal Biol. Rev. 2011, 25, 61–67. [Google Scholar] [CrossRef]
- Chakraborty, M.; Mahmud, N.U.; Gupta, D.R.; Tareq, F.S.; Shin, H.J.; Islam, T. Inhibitory Effects of Linear Lipopeptides From a Marine Bacillus Subtilis on the Wheat Blast Fungus Magnaporthe oryzae Triticum. Front. Microbiol. 2020, 11, 665. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Zhao, M.; Chen, D.; Cheng, J.; Li, J.; Feng, Z.; Ma, Z.; An, D. Biocontrol of Rice Blast by the Phenaminomethylacetic Acid Producer of Bacillus Methylotrophicus Strain BC79. Crop Prot. 2013, 44, 29–37. [Google Scholar] [CrossRef]
- Li, Y.; Gu, Y.; Li, J.; Xu, M.; Wei, Q.; Wang, Y. Biocontrol Agent Bacillus Amyloliquefaciens LJ02 Induces Systemic Resistance against Cucurbits Powdery Mildew. Front. Microbiol. 2015, 6, 883. [Google Scholar] [CrossRef]
- Lam, V.B.; Meyer, T.; Arias, A.A.; Ongena, M.; Oni, F.E.; Höfte, M. Bacillus Cyclic Lipopeptides Iturin and Fengycin Control Rice Blast Caused by Pyricularia oryzae in Potting and Acid Sulfate Soils by Direct Antagonism and Induced Systemic Resistance. Microorganisms 2021, 9, 1441. [Google Scholar] [CrossRef]
- Joshi, R.; McSpadden Gardener, B.B. Identification and Characterization of Novel Genetic Markers Associated with Biological Control Activities in Bacillus Subtilis. Phytopathology 2006, 96, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; de Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.-W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The Iturin and Fengycin Families of Lipopeptides Are Key Factors in Antagonism of Bacillus Subtilis Toward Podosphaera Fusca. MPMI 2007, 20, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Parker, J.E. Interplay of Signaling Pathways in Plant Disease Resistance. Trends Genet. 2000, 16, 449–455. [Google Scholar] [CrossRef]
- Awan, S.A.; Ilyas, N.; Khan, I.; Raza, M.A.; Rehman, A.U.; Rizwan, M.; Rastogi, A.; Tariq, R.; Brestic, M. Bacillus Siamensis Reduces Cadmium Accumulation and Improves Growth and Antioxidant Defense System in Two Wheat (Triticum aestivum L.) Varieties. Plants 2020, 9, 878. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, J.; Wu, F.; Yin, C.; Kim, K.H.; Zhang, Y. Termite Nest Associated Bacillus Siamensis YC-9 Mediated Biocontrol of Fusarium Oxysporum f. Sp. Cucumerinum. Front. Microbiol. 2022, 13, 893393. [Google Scholar] [CrossRef] [PubMed]
- Del Sal, G.; Manfioletti, G.; Schneider, C. A One-Tube Plasmid DNA Mini-Preparation Suitable for Sequencing. Nucleic Acids Res. 1988, 16, 9878. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, L.; Dong, Y.; Chen, W.; Li, C.; Gao, X.; Chen, R.; Li, L.; Xu, Z. The Antagonistic Mechanism of Bacillus Velezensis ZW10 against Rice Blast Disease: Evaluation of ZW10 as a Potential Biopesticide. PLoS ONE 2021, 16, e0256807. [Google Scholar] [CrossRef]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-Protocol 2012, 2, e263. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126.e15. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, L.; Li, H.; Dong, Y.; Xu, H.; Guan, Y.; Rong, S.; Gao, X.; Chen, R.; Li, L.; et al. The Isolation of the Antagonistic Strain Bacillus Australimaris CQ07 and the Exploration of the Pathogenic Inhibition Mechanism of Magnaporthe oryzae. PLoS ONE 2019, 14, e0220410. [Google Scholar] [CrossRef] [PubMed]

| Product Name | Product Number |
|---|---|
| penicillin | S1001 |
| qxacillin | S1002 |
| ampicillin | S1003 |
| carbenicillin | S1004 |
| piperacillin | S1005 |
| cephalexin | S1011 |
| cefamezin | S1012 |
| cefradine | S1013 |
| cefuroxim | S1015 |
| ceftazidime | S1019 |
| ceftazidime | S1020 |
| ceftriaxone | S1021 |
| doxycycline | S1037 |
| amikacin | S1027 |
| gentamicin | S1028 |
| kanamycin | S1030 |
| neomycin | S1034 |
| tetracycline | S1036 |
| minocycline | S1038 |
| erythromycin | S1039 |
| Primer Name | Primer Sequences (5′—3′) |
|---|---|
| Actin | F:GAGTATGATGAGTCGGGTCCAG |
| R:ACACCAACAATCCCAAACAGAG | |
| PR1a | F:GCTACGTGTTTATGCATGTATGG |
| R:TCGGATTTATTCTCACCAGCA | |
| PR10a | F:AATGAGAGCCGCAGAAATGT |
| RGGCACATAAACACAACCACAA | |
| PR5 | F:GGTACAACGTCGCCATGAGCT |
| R:TGGGCAGAAGACGACTCGGTAG | |
| CEBiP | F:CATCGCTCATCATACAAACCA |
| R:GGAGATAACAGACATGCTCCAC | |
| LYP6 | F:TGCCCAGGACCACATCAGT |
| R:CCAGGGAAGCCCGGAATAT | |
| NH1 | F:AAGCGGTTCAAATCTCAAA |
| R:GCCTCCATCGGAAACATA | |
| MAPK6 | F:CTCGTACCACCTCAGAAAC |
| R:AAATACAGCCCACAGACC | |
| MAPK12 | F:ATCGCTTCAAACGACAGT |
| R:GTGACATTGGAGGGCTTA | |
| POD | F:GGCCTTGGCAAATACCGACC |
| R:TCGTGTGTGCTCCTGAGAGA | |
| WRKY45 | F:GCAGCAATCGTCCGGGAATT |
| R:GCCTTTGGGTGCTTGGAGTTT | |
| WRKY53 | F:ACGGGCAGAAGCAGGTGAAG |
| R:CCCTTGTAGACGATCTGGGTGA | |
| WRKY89 | F:GCACCTCACAATGATGGA |
| R:GGACAGCCTTGCACTTTA | |
| EREBP | F:GTGTTCGTGTCTGGCTTGG |
| R:CACTTGACTTGGGTGCTTTA |
| Primer Sequences(5′—3′) | |
|---|---|
| bioA | F:TTCCACGGCCATTCCTATAC |
| R:TTTGTCCCCTTATCCTGCAC | |
| SrfA | F:GAAAGAGCGGCTGCTGAAAC |
| R:CCCAATATTGCCGCAATGAC | |
| FenD | F:CCTGCAGAAGGAGAAGTGAAG |
| R:TGCTCATCGTCTTCCGTTTC | |
| FenB | F:CTATAGTTTGTTGACGGCTC |
| R:CAGCACTGGTTCTTGTCGCA | |
| ItuC | F:TTCACTTTTGATCTGGCGAT |
| R:CGTCCGGTACATTTTCAC | |
| ItuD | F:ATGAACAATCTTGCCTTTTTA |
| R:TTATTTTAAAATCCGCAATT | |
| bmyB | F:TGAAACAAAGGCATATGCTC |
| R:AAAAATGCATCTGCCGTTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Zhang, Y.; Zhang, L.; Zhou, Z.; Zhang, J.; Yang, J.; Gao, X.; Chen, R.; Huang, Z.; Xu, Z.; et al. Isolation of Bacillus siamensis B-612, a Strain That Is Resistant to Rice Blast Disease and an Investigation of the Mechanisms Responsible for Suppressing Rice Blast Fungus. Int. J. Mol. Sci. 2023, 24, 8513. https://doi.org/10.3390/ijms24108513
Yang Y, Zhang Y, Zhang L, Zhou Z, Zhang J, Yang J, Gao X, Chen R, Huang Z, Xu Z, et al. Isolation of Bacillus siamensis B-612, a Strain That Is Resistant to Rice Blast Disease and an Investigation of the Mechanisms Responsible for Suppressing Rice Blast Fungus. International Journal of Molecular Sciences. 2023; 24(10):8513. https://doi.org/10.3390/ijms24108513
Chicago/Turabian StyleYang, Yanmei, Yifan Zhang, Luyi Zhang, Zhanmei Zhou, Jia Zhang, Jinchang Yang, Xiaoling Gao, Rongjun Chen, Zhengjian Huang, Zhengjun Xu, and et al. 2023. "Isolation of Bacillus siamensis B-612, a Strain That Is Resistant to Rice Blast Disease and an Investigation of the Mechanisms Responsible for Suppressing Rice Blast Fungus" International Journal of Molecular Sciences 24, no. 10: 8513. https://doi.org/10.3390/ijms24108513
APA StyleYang, Y., Zhang, Y., Zhang, L., Zhou, Z., Zhang, J., Yang, J., Gao, X., Chen, R., Huang, Z., Xu, Z., & Li, L. (2023). Isolation of Bacillus siamensis B-612, a Strain That Is Resistant to Rice Blast Disease and an Investigation of the Mechanisms Responsible for Suppressing Rice Blast Fungus. International Journal of Molecular Sciences, 24(10), 8513. https://doi.org/10.3390/ijms24108513





