Characterization and Engineering Studies of a New Endolysin from the Propionibacterium acnes Bacteriophage PAC1 for the Development of a Broad-Spectrum Artilysin with Altered Specificity
Abstract
1. Introduction
2. Results and Discussion
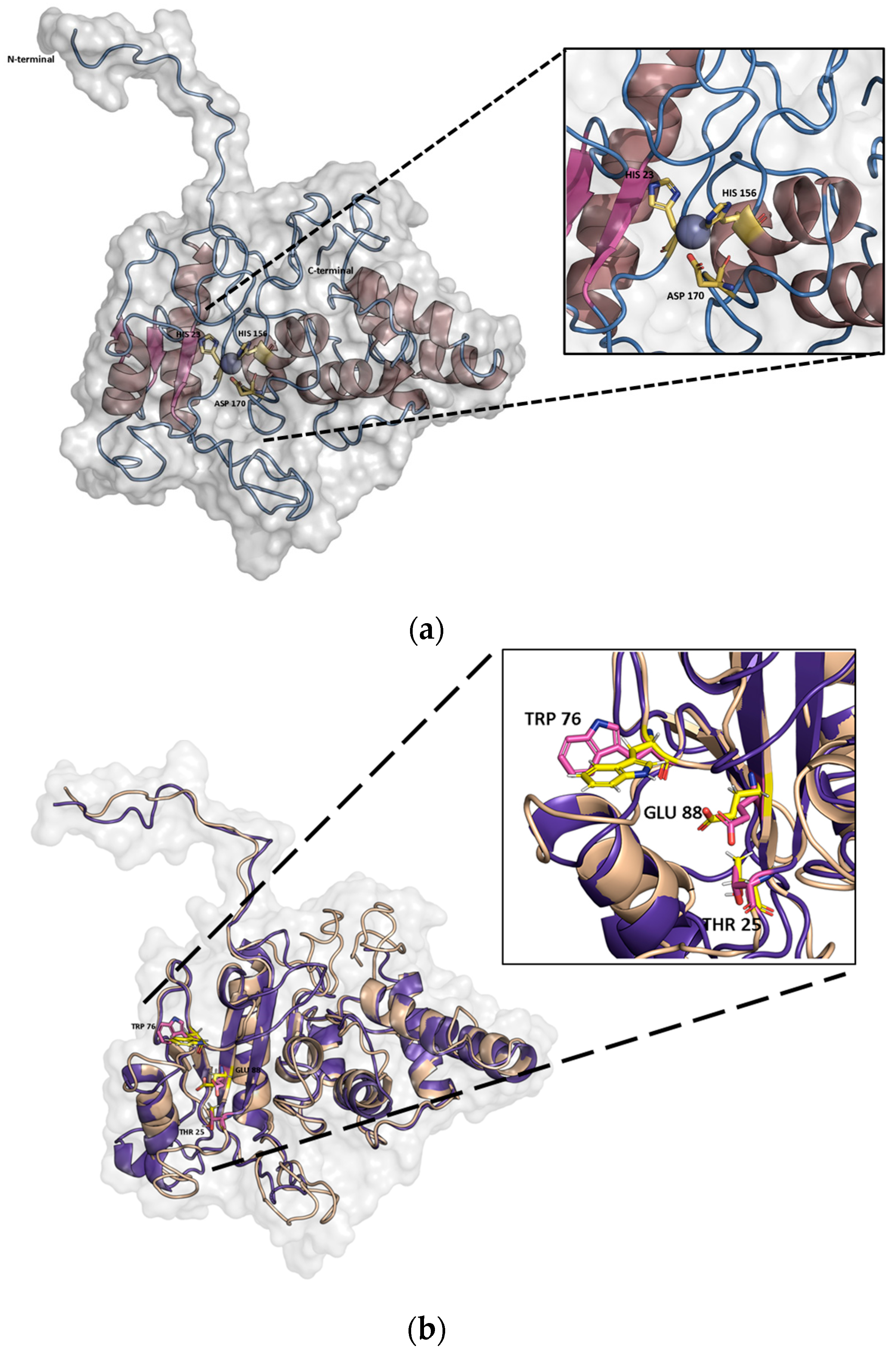
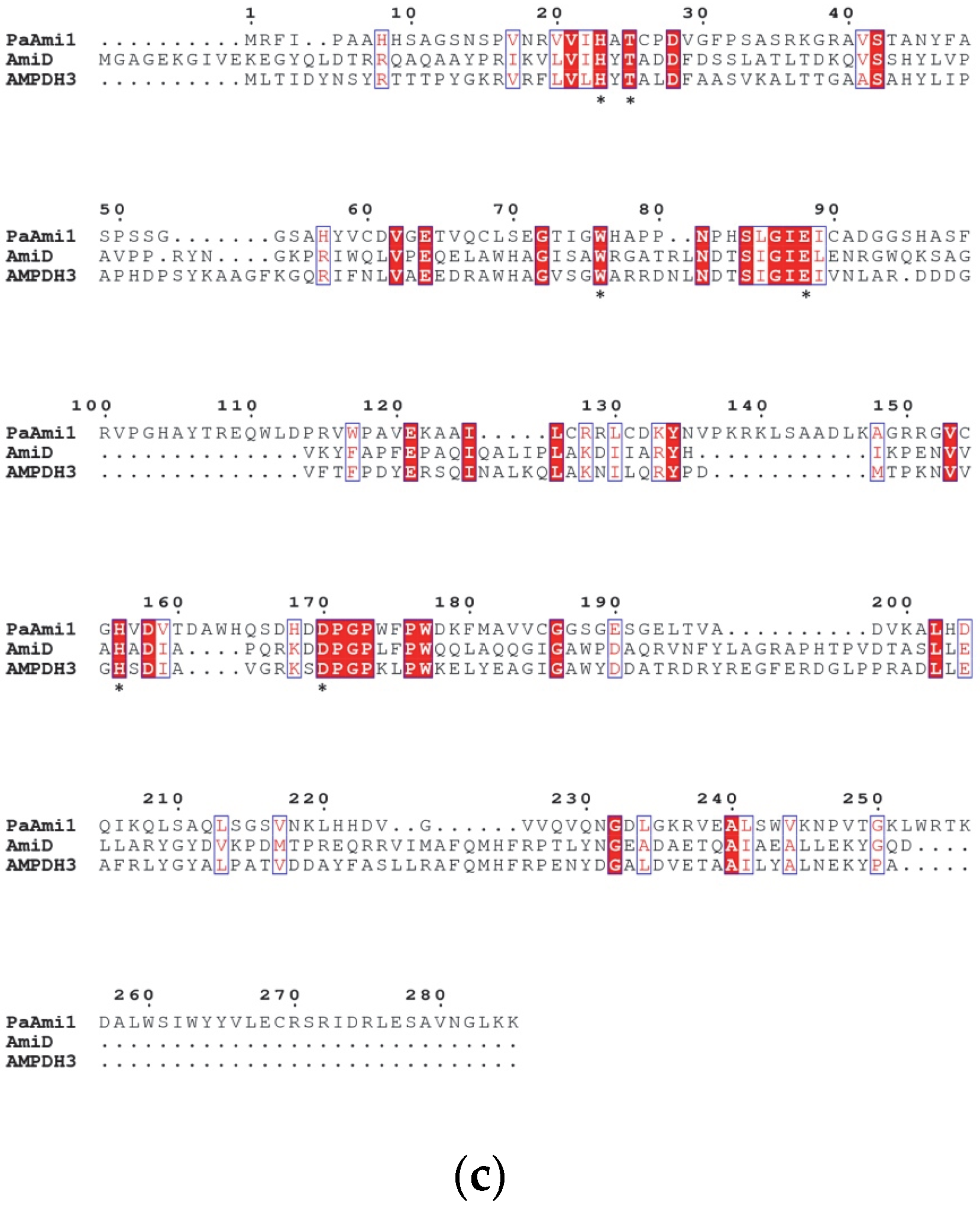
2.1. In Silico Characterization of PaAmi1
2.2. Expression and Purification of PaAmi1
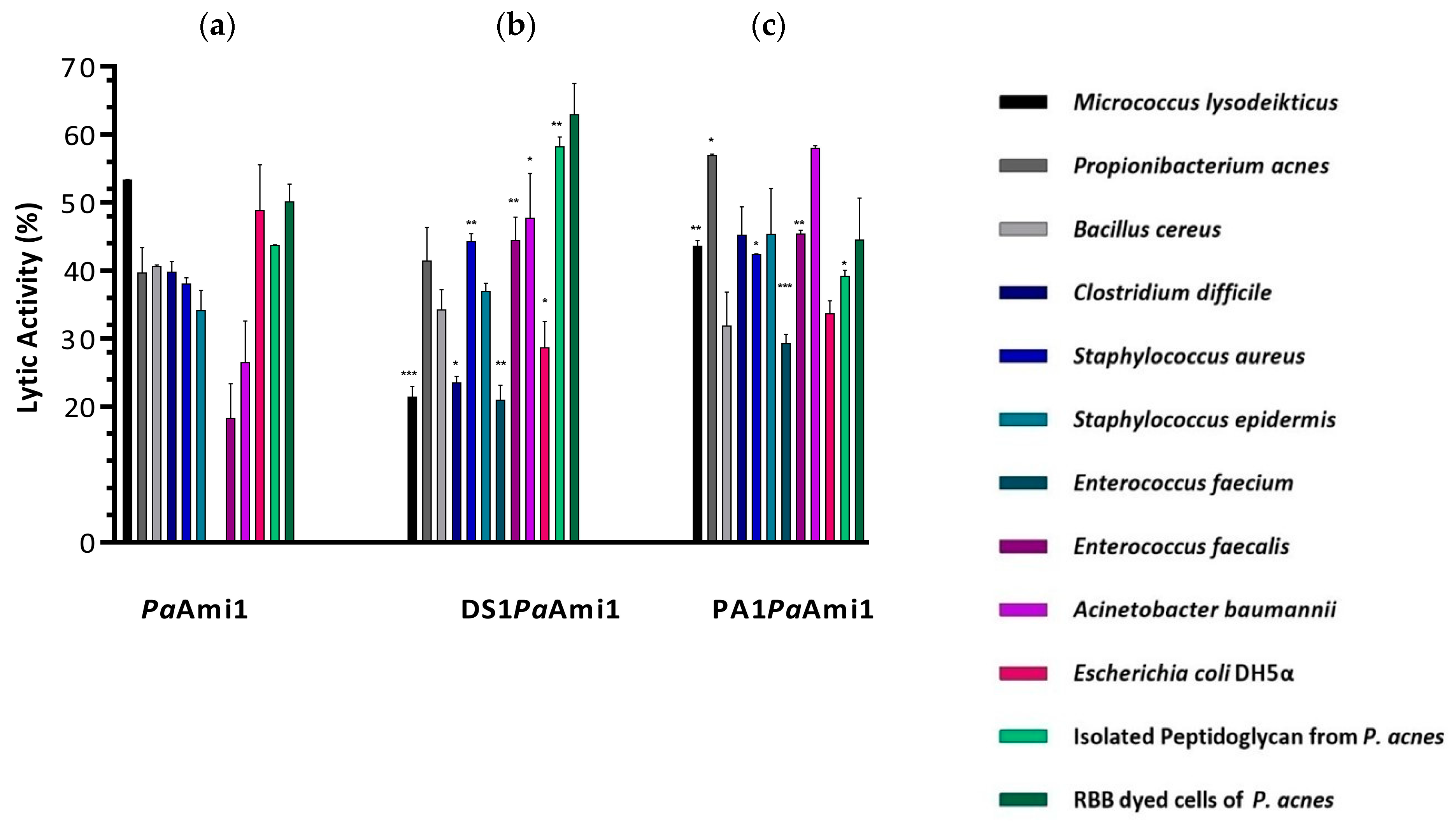
2.3. Demonstration of Lytic Activity and Substrate Specificity of PaAmi1
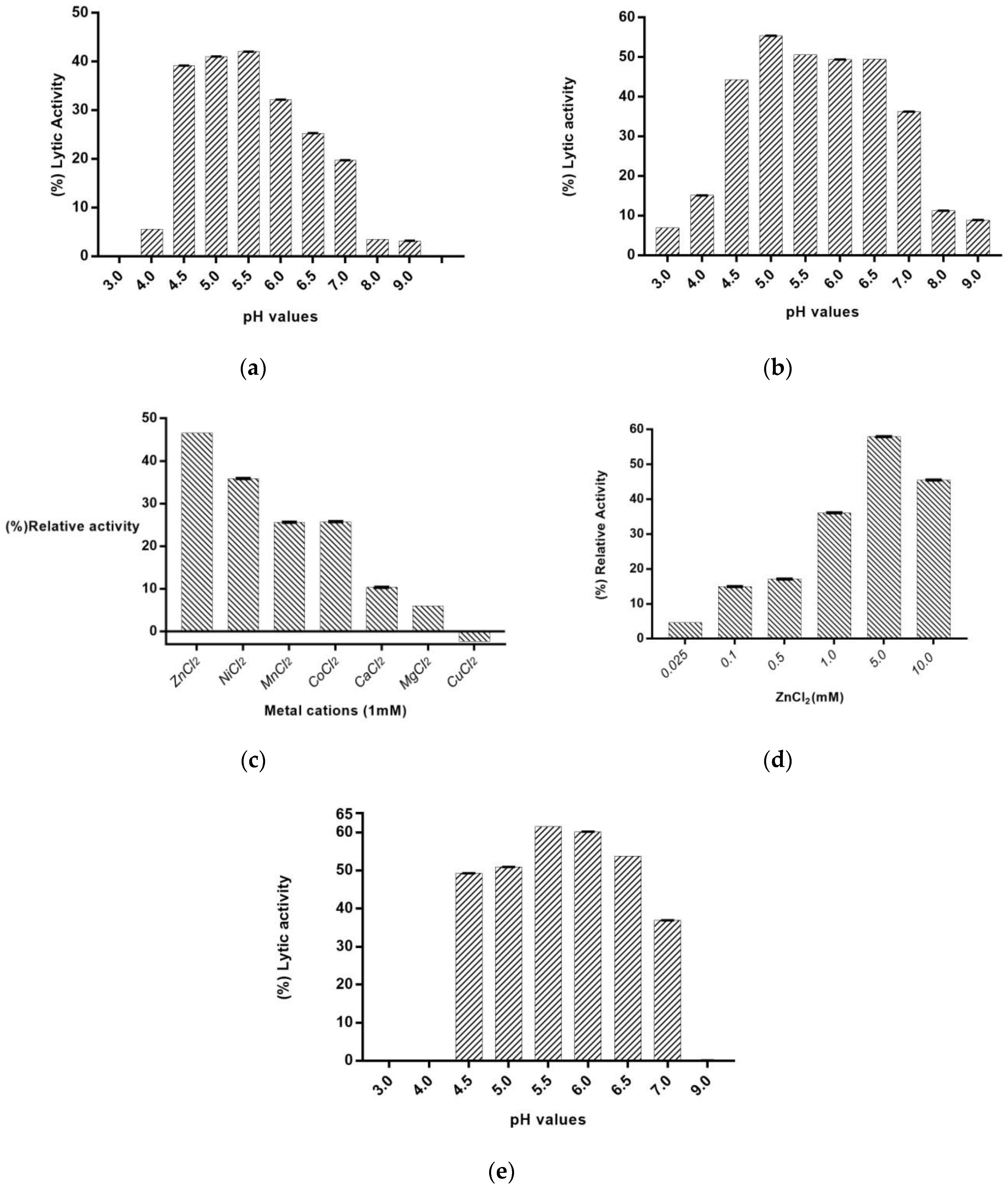
2.4. The Effect of pH and Divalent Metal Ions on PaAmi1 Activity
2.5. Evaluation of the Inhibitory and Bactericidal Activity of PaAmi1 against Live Cultures of P. acnes
2.6. ‘Artilysation’ of the Endolysin PaAmi1
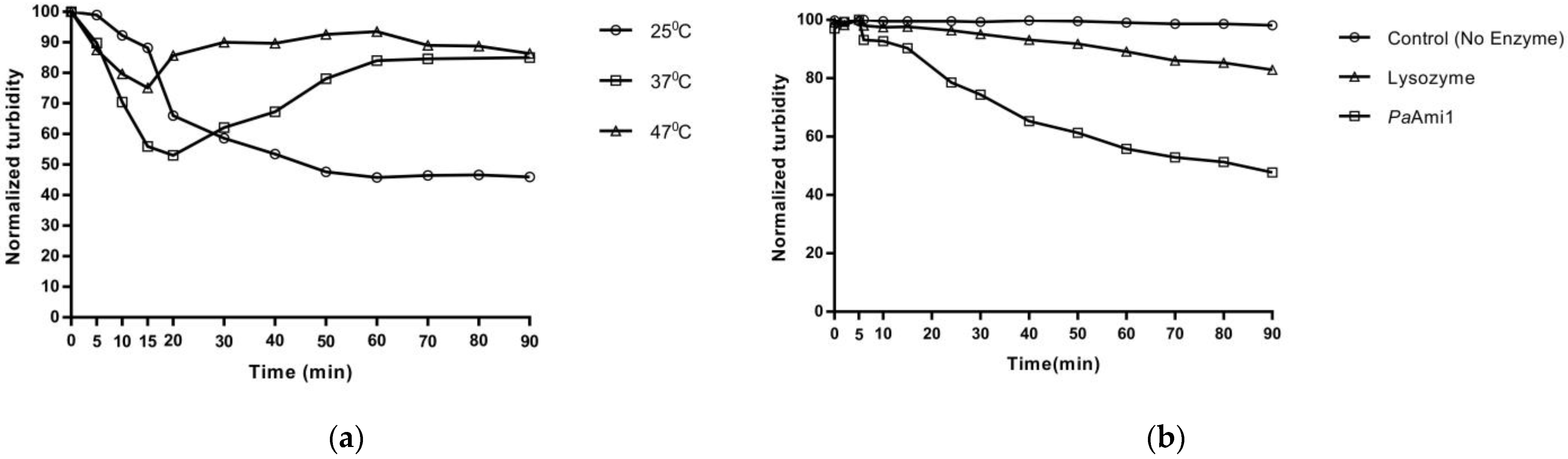
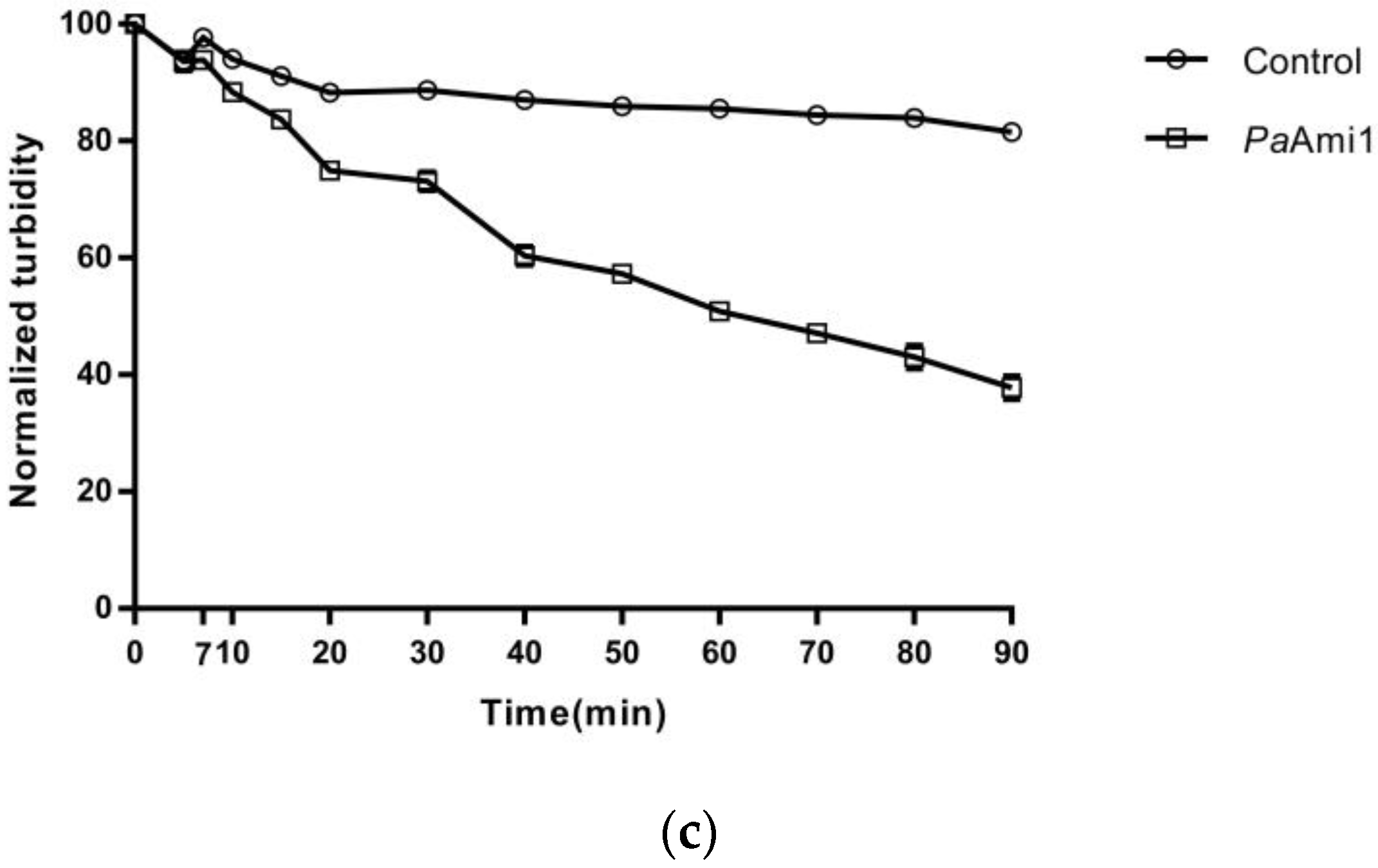
2.7. Thermostability of PaAmi1, DS1PaAmi1, and PA1PaAmi1
3. Materials and Methods
3.1. Biocomputing Analysis
3.2. Synthesis and Cloning of PaAmi1
3.3. Expression and Purification of PaAmi1
3.4. Lytic Activity Assays
3.5. Effect of pH, Divalent Metal Ions, and Temperature
3.6. Turbidity Reduction Assays Using Isolated Peptidoglycan
3.7. Turbidity Reduction Assays Using Remazol Brilliant Blue R-Dyed Cells
3.8. Lytic Activity of PaAmi1 Using Live Propionibacterium Acnes Cells on Agar Plates
3.9. Fusion of PaAmi1 with Antimicrobial Peptides (AMP)
- ds1F(5′-ATGCGCATTCGCTTATTACAGCGCTTCAACAAGCGCCGCTTCATTCCGGCT-3′) and
- pa1F(5′-ATGCGCGTATTCCGCCGCGCAGCACGCATTGCACAGCGCTTCATTCCGGCT-3′).
3.10. Expression, Purification, and Lytic Activity of DS1PaAmi1 and PA1PaAmi1
3.11. Thermostability Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, A.; Rappuoli, R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Front. Immunol. 2018, 9, 1068. [Google Scholar] [CrossRef] [PubMed]
- Premetis, G.E.; Stathi, A.; Papageorgiou, A.C.; Labrou, N.E. Characterization of a glycoside hydrolase endolysin from Acinetobacter baumannii phage AbTZA1 with high antibacterial potency and novel structural features. FEBS J. 2023, 290, 2146–2164. [Google Scholar] [CrossRef] [PubMed]
- Pai, A.; Sudhakar, G.K.; Kamath, V. Enzybiotics—A Review. Int. J. Pharmacol. Res. 2013, 3, 69–71. [Google Scholar]
- Villa, T.G.; Feijoo-Siota, L.; Rama, J.L.R.; Sánchez-Pérez, A.; de Miguel-Bouzas, T. Enzybiotics. Antimicrobial Food Packaging; Elsevier: Amsterdam, The Netherlands, 2016; pp. 491–502. ISBN 978-0-12-800723-5. [Google Scholar]
- Brives, C.; Pourraz, J. Phage Therapy as a Potential Solution in the Fight against AMR: Obstacles and Possible Futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage Therapy: A Potential Solution for the Antibiotic Resistance Crisis. J. Infect. Dev. Ctries 2014, 8, 129–136. [Google Scholar] [CrossRef]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The Advantages and Challenges of Using Endolysins in a Clinical Setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as Emerging Alternative Therapeutic Agents to Counter Drug-Resistant Infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-Based Antibacterials as Therapeutics. In Therapeutic Enzymes: Function and Clinical Implications; Labrou, N., Ed.; Springer Singapore: Singapore, 2019; Volume 1148, pp. 233–253. ISBN 9789811377082. [Google Scholar]
- Love, M.; Bhandari, D.; Dobson, R.; Billington, C. Potential for Bacteriophage Endolysins to Supplement or Replace Antibiotics in Food Production and Clinical Care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef]
- Thummeepak, R.; Kitti, T.; Kunthalert, D.; Sitthisak, S. Enhanced Antibacterial Activity of Acinetobacter Baumannii Bacteriophage ØABP-01 Endolysin (LysABP-01) in Combination with Colistin. Front. Microbiol. 2016, 7, 1402. [Google Scholar] [CrossRef]
- Taati Moghadam, M.; Khoshbayan, A.; Chegini, Z.; Farahani, I.; Shariati, A. Bacteriophages, a New Therapeutic Solution for Inhibiting Multidrug-Resistant Bacteria Causing Wound Infection: Lesson from Animal Models and Clinical Trials. Drug Des. Dev. Ther. 2020, 14, 1867–1883. [Google Scholar] [CrossRef]
- Vázquez, R.; García, E.; García, P. Phage Lysins for Fighting Bacterial Respiratory Infections: A New Generation of Antimicrobials. Front. Immunol. 2018, 9, 2252. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, D.-W.; Jin, J.-S.; Kim, J. Antimicrobial Activity of LysSS, a Novel Phage Endolysin, against Acinetobacter Baumannii and Pseudomonas Aeruginosa. J. Glob. Antimicrob. Resist. 2020, 22, 32–39. [Google Scholar] [CrossRef]
- Son, B.; Yun, J.; Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an Endolysin from the Bacillus Cereus-Infecting Bacteriophage B4. BMC Microbiol. 2012, 12, 33. [Google Scholar] [CrossRef]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R. Role of Net Charge on Catalytic Domain and Influence of Cell Wall Binding Domain on Bactericidal Activity, Specificity, and Host Range of Phage Lysins. J. Biol. Chem. 2011, 286, 34391–34403. [Google Scholar] [CrossRef]
- Dörr, T.; Moynihan, P.J.; Mayer, C. Editorial: Bacterial Cell Wall Structure and Dynamics. Front. Microbiol. 2019, 10, 2051. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan Structure and Architecture. FEMS Microbiol Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef]
- Neuhaus, F.C.; Baddiley, J. A Continuum of Anionic Charge: Structures and Functions of d-Alanyl-Teichoic Acids in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef]
- Kamisango, K.; Saiki, I.; Tanio, Y.; Okumura, H.; Araki, Y.; Sekikawa, I.; Azuma, I.; Yamamura, Y. Structures and Biological Activities of Peptidoglycans of Listeria Monocytogenes and Propionibacterium Acnes12. J. Biochem. 1982, 92, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kandler, O. Peptidoglycan Types of Bacterial Cell Walls and Their Taxonomic Implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Coates, P.; Vyakrnam, S.; Eady, E.A.; Jones, C.E.; Cove, J.H.; Cunliffe, W.J. Prevalence of Antibiotic-Resistant Propionibacteria on the Skin of Acne Patients: 10-Year Surveillance Data and Snapshot Distribution Study. Br. J. Derm. 2002, 146, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Elliott, A.G.; Kavanagh, A.M.; Ramu, S.; Cooper, M.A. In Vitro Antimicrobial Activity of Acne Drugs Against Skin-Associated Bacteria. Sci. Rep. 2019, 9, 14658. [Google Scholar] [CrossRef] [PubMed]
- McDowell, A.; Patrick, S.; Eishi, Y.; Lambert, P.; Eady, A. Propionibacterium Acnes in Human Health and Disease. BioMed Res. Int. 2013, 2013, 493564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Lu, Z.; Ma, Y. Draft Genome Sequences of Three Multidrug-Resistant Cutibacterium (Formerly Propionibacterium) Acnes Strains Isolated from Acne Patients, China. J. Glob. Antimicrob. Resist. 2017, 11, 114–115. [Google Scholar] [CrossRef]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium Acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin Microbiota: A Source of Disease or Defence?: Skin Microbiota. Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Alkhawaja, E.; Hammadi, S.; Abdelmalek, M.; Mahasneh, N.; Alkhawaja, B.; Abdelmalek, S.M. Antibiotic Resistant Cutibacterium Acnes among Acne Patients in Jordan: A Cross Sectional Study. BMC Dermatol. 2020, 20, 17. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P.; et al. Engineered Endolysin-Based “Artilysins” To Combat Multidrug-Resistant Gram-Negative Pathogens. mBio 2014, 5, e01379-14. [Google Scholar] [CrossRef]
- Gerstmans, H.; Rodríguez-Rubio, L.; Lavigne, R.; Briers, Y. From Endolysins to Artilysin®s: Novel Enzyme-Based Approaches to Kill Drug-Resistant Bacteria. Biochem. Soc. Trans. 2016, 44, 123–128. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Chang, W.-L.; Gutiérrez, D.; Lavigne, R.; Martínez, B.; Rodríguez, A.; Govers, S.K.; Aertsen, A.; Hirl, C.; Biebl, M.; et al. ‘Artilysation’ of Endolysin ΛSa2lys Strongly Improves Its Enzymatic and Antibacterial Activity against Streptococci. Sci. Rep. 2016, 6, 35382. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The Formulation of Bacteriophage in a Semi Solid Preparation for Control of Propionibacterium Acnes Growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The Diversity and Host Interactions of Propionibacterium Acnes Bacteriophages on Human Skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Żaczek, M.; Międzybrodzki, R.; Letkiewicz, S.; Łusiak-Szelchowska, M.; Górski, A. Prospects of Phage Application in the Treatment of Acne Caused by Propionibacterium Acnes. Front. Microbiol. 2017, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved Protein Function Prediction by Combining Structure, Sequence and Protein–Protein Interaction Information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Firczuk, M.; Bochtler, M. Folds and Activities of Peptidoglycan Amidases. FEMS Microbiol. Rev. 2007, 31, 676–691. [Google Scholar] [CrossRef] [PubMed]
- Kerff, F.; Petrella, S.; Mercier, F.; Sauvage, E.; Herman, R.; Pennartz, A.; Zervosen, A.; Luxen, A.; Frère, J.-M.; Joris, B.; et al. Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family. J. Mol. Biol. 2010, 397, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Artola-Recolons, C.; Carrasco-López, C.; Martínez-Caballero, S.; Hesek, D.; Spink, E.; Lastochkin, E.; Zhang, W.; Hellman, L.M.; Boggess, B.; et al. Cell-Wall Remodeling by the Zinc-Protease AmpDh3 from Pseudomonas Aeruginosa. J. Am. Chem. Soc. 2013, 135, 12604–12607. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein–Ligand Binding Site Recognition Using Complementary Binding-Specific Substructure Comparison and Sequence Profile Alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- Zhang, Y. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Gouet, P. ESPript/ENDscript: Extracting and Rendering Sequence and 3D Information from Atomic Structures of Proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef]
- Steinmetz, E. Expresso® Cloning and Expression Systems: ExpressioneeringTM Technology Streamlines Recombinant Protein Expression. Nat. Methods 2011, 8, 3–4. [Google Scholar] [CrossRef]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a Novel Escherichia Coli Protein Expression Strain Capable of Correctly Folding Disulfide Bonded Proteins in Its Cytoplasm. Microb. Cell Fact. 2012, 11, 753. [Google Scholar] [CrossRef]
- Shugar, D. The Measurement of Lysozyme Activity and the Ultra-Violet Inactivation of Lysozyme. Biochim. Biophys. Acta 1952, 8, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, D.; De Marco, C.; Duprè, S.; Rotilio, G. The Copper Catalyzed Oxidation of Cysteine to Cystine. Arch. Biochem. Biophys. 1969, 130, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2020, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Saito, K.; Nomoto, T.; Namae, T.; Ochiai, A.; Saitoh, E.; Tanaka, T. Identification and Characterization of Multifunctional Cationic and Amphipathic Peptides from Soybean Proteins: Multifunctional Cationic and Amphipathic Peptides from Soybean Proteins. Biopolymers 2017, 108, e23023. [Google Scholar] [CrossRef] [PubMed]
- Santos-Júnior, C.D.; Pan, S.; Zhao, X.-M.; Coelho, L.P. Macrel: Antimicrobial Peptide Screening in Genomes and Metagenomes. PeerJ 2020, 8, e10555. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMP R3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides: Table 1. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Waghu, F.H.; Idicula-Thomas, S. Collection of Antimicrobial Peptides Database and Its Derivatives: Applications and Beyond. Protein Sci. 2020, 29, 36–42. [Google Scholar] [CrossRef]
- Lin, T.-T.; Yang, L.-Y.; Lu, I.-H.; Cheng, W.-C.; Hsu, Z.-R.; Chen, S.-H.; Lin, C.-Y. AI4AMP: Sequence-Based Antimicrobial Peptides Predictor Using Physicochemical Properties-Based Encoding Method and Deep Learning. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lin, T.-T.; Yang, L.-Y.; Lu, I.-H.; Cheng, W.-C.; Hsu, Z.-R.; Chen, S.-H.; Lin, C.-Y. AI4AMP: An Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning. mSystems 2021, 6, e00299-21. [Google Scholar] [CrossRef]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W.I. AmPEP: Sequence-Based Prediction of Antimicrobial Peptides Using Distribution Patterns of Amino Acid Properties and Random Forest. Sci. Rep. 2018, 8, 1697. [Google Scholar] [CrossRef] [PubMed]
- Timmons, P.B.; Hewage, C.M. HAPPENN Is a Novel Tool for Hemolytic Activity Prediction for Therapeutic Peptides Which Employs Neural Networks. Sci. Rep. 2020, 10, 10869. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the Mechanisms of Acinetobacter Baumannii Virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef]
- Vázquez, R.; Blanco-Gañán, S.; Ruiz, S.; García, P. Mining of Gram-Negative Surface-Active Enzybiotic Candidates by Sequence-Based Calculation of Physicochemical Properties. Front. Microbiol. 2021, 12, 660403. [Google Scholar] [CrossRef]
- Kumar, S.; Tsai, C.-J.; Nussinov, R. Factors Enhancing Protein Thermostability. Protein Eng. Des. Sel. 2000, 13, 179–191. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 50, D20–D26. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A New Bioinformatics Analysis Tools Framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD2: The Updated Antimicrobial Peptide Database and Its Application in Peptide Design. Nucleic Acids Res. 2008, 37, D933–D937. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. APD: The Antimicrobial Peptide Database. Nucleic Acids Res. 2004, 32, 590D–592D. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein Recovery from Inclusion Bodies of Escherichia Coli Using Mild Solubilization Process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef]
- Singh, S.M.; Upadhyay, A.K.; Panda, A.K. Solubilization at High PH Results in Improved Recovery of Proteins from Inclusion Bodies of E. Coli. J. Chem. Technol. Biotechnol. 2008, 83, 1126–1134. [Google Scholar] [CrossRef]
- Fukushima, T.; Sekiguchi, J. Zymographic Techniques for the Analysis of Bacterial Cell Wall in Bacillus. In Bacterial Cell Wall Homeostasis; Hong, H.-J., Ed.; Springer New York: New York, NY, USA, 2016; Volume 1440, pp. 87–98. ISBN 978-1-4939-3674-8. [Google Scholar]
- Zhou, R.; Chen, S.; Recsei, P. A Dye Release Assay for Determination of Lysostaphin Activity. Anal. Biochem. 1988, 171, 141–144. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varotsou, C.; Premetis, G.E.; Labrou, N.E. Characterization and Engineering Studies of a New Endolysin from the Propionibacterium acnes Bacteriophage PAC1 for the Development of a Broad-Spectrum Artilysin with Altered Specificity. Int. J. Mol. Sci. 2023, 24, 8523. https://doi.org/10.3390/ijms24108523
Varotsou C, Premetis GE, Labrou NE. Characterization and Engineering Studies of a New Endolysin from the Propionibacterium acnes Bacteriophage PAC1 for the Development of a Broad-Spectrum Artilysin with Altered Specificity. International Journal of Molecular Sciences. 2023; 24(10):8523. https://doi.org/10.3390/ijms24108523
Chicago/Turabian StyleVarotsou, Christina, Georgios E. Premetis, and Nikolaos E. Labrou. 2023. "Characterization and Engineering Studies of a New Endolysin from the Propionibacterium acnes Bacteriophage PAC1 for the Development of a Broad-Spectrum Artilysin with Altered Specificity" International Journal of Molecular Sciences 24, no. 10: 8523. https://doi.org/10.3390/ijms24108523
APA StyleVarotsou, C., Premetis, G. E., & Labrou, N. E. (2023). Characterization and Engineering Studies of a New Endolysin from the Propionibacterium acnes Bacteriophage PAC1 for the Development of a Broad-Spectrum Artilysin with Altered Specificity. International Journal of Molecular Sciences, 24(10), 8523. https://doi.org/10.3390/ijms24108523






