Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment
Abstract
1. Introduction
2. Results and Discussion
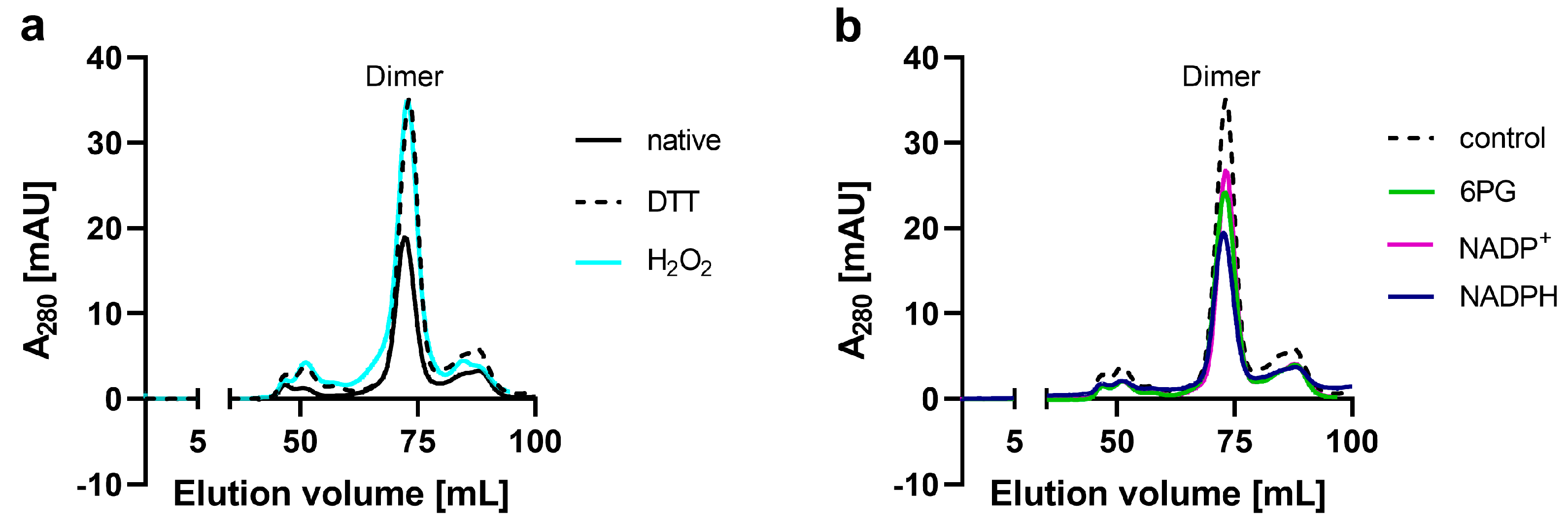
2.1. Production, Oligomerization Behavior and Kinetic Characterization of Ld6PGD wt
2.2. Crystallization and Structure Determination of Ld6PGD
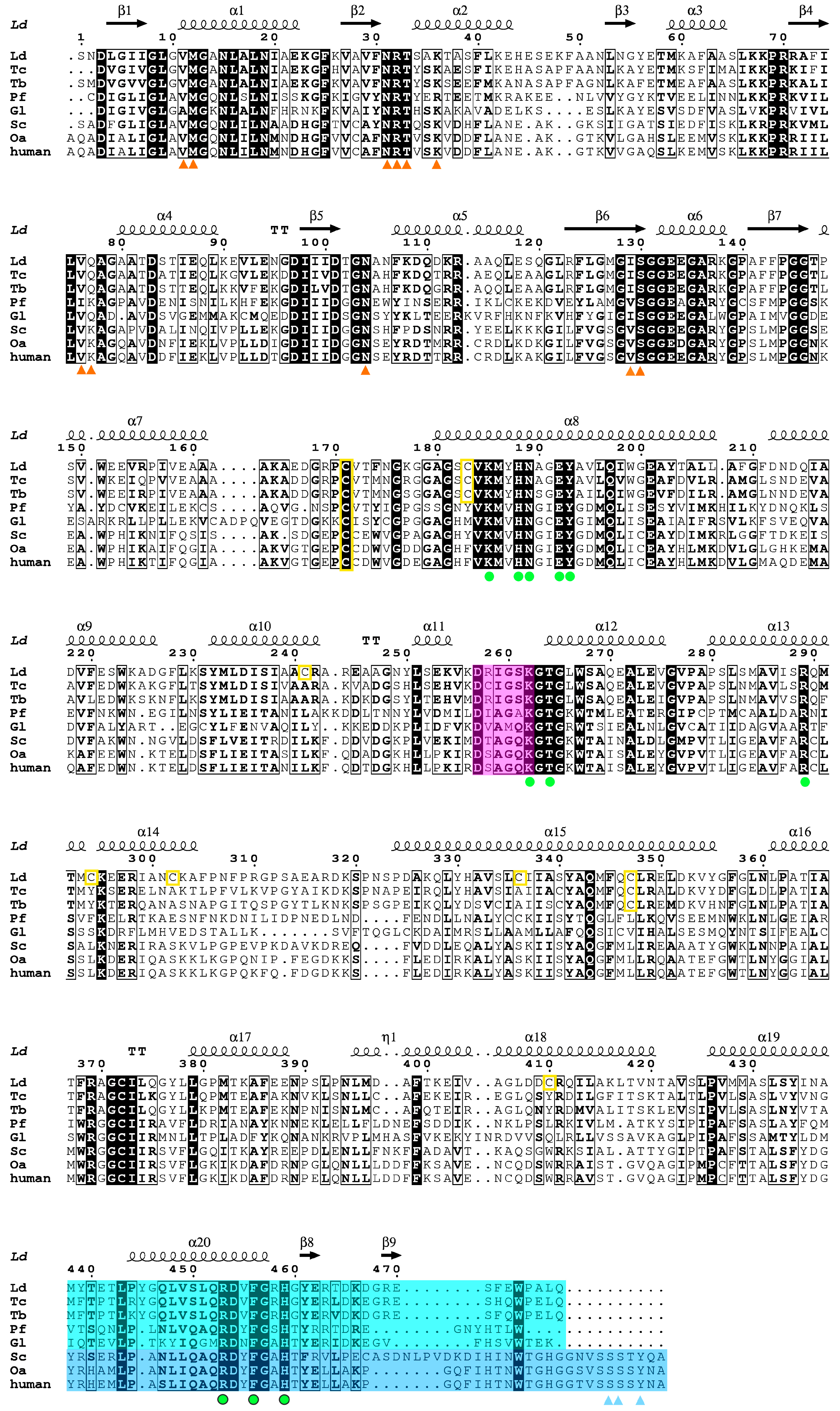
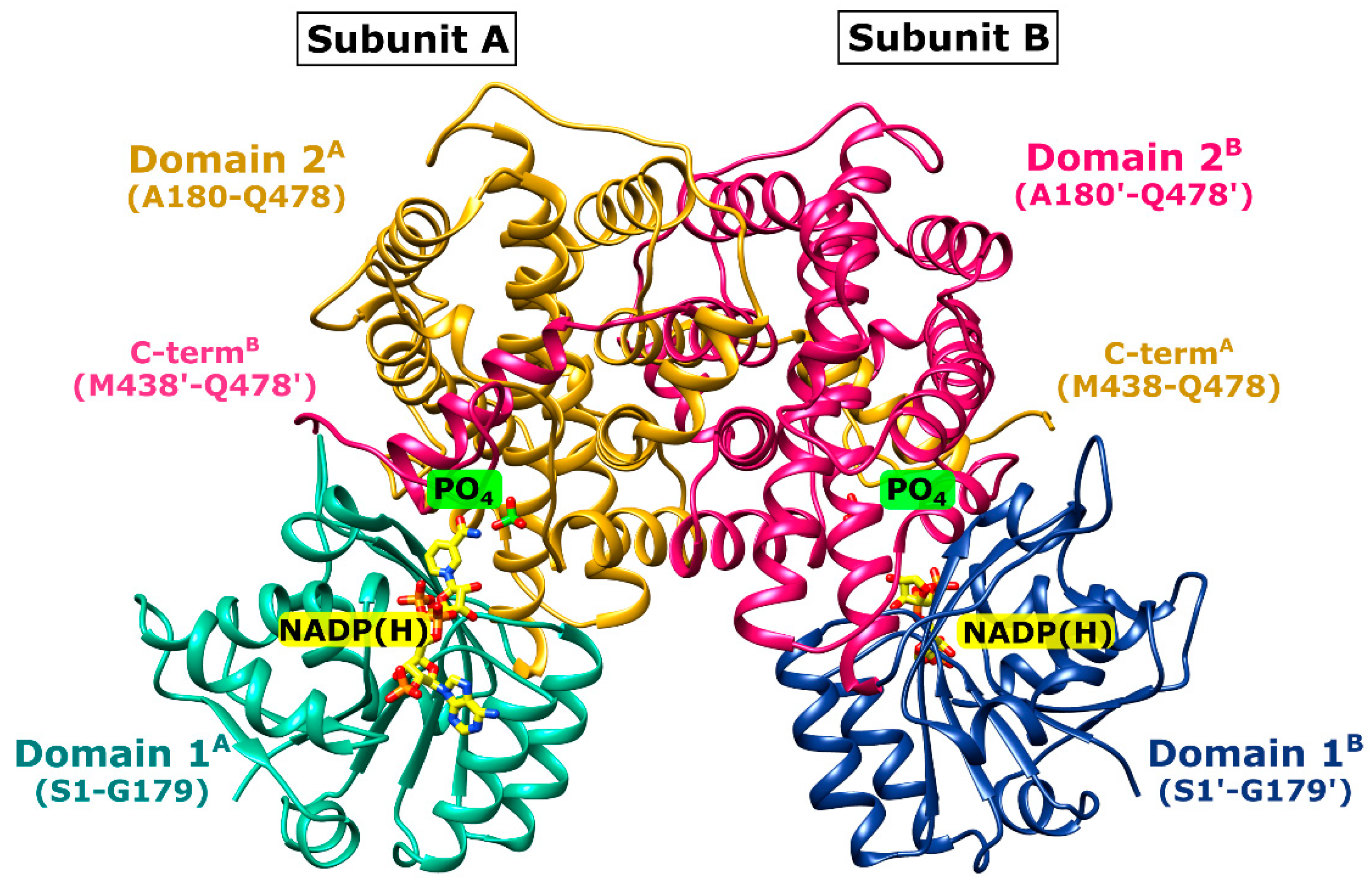
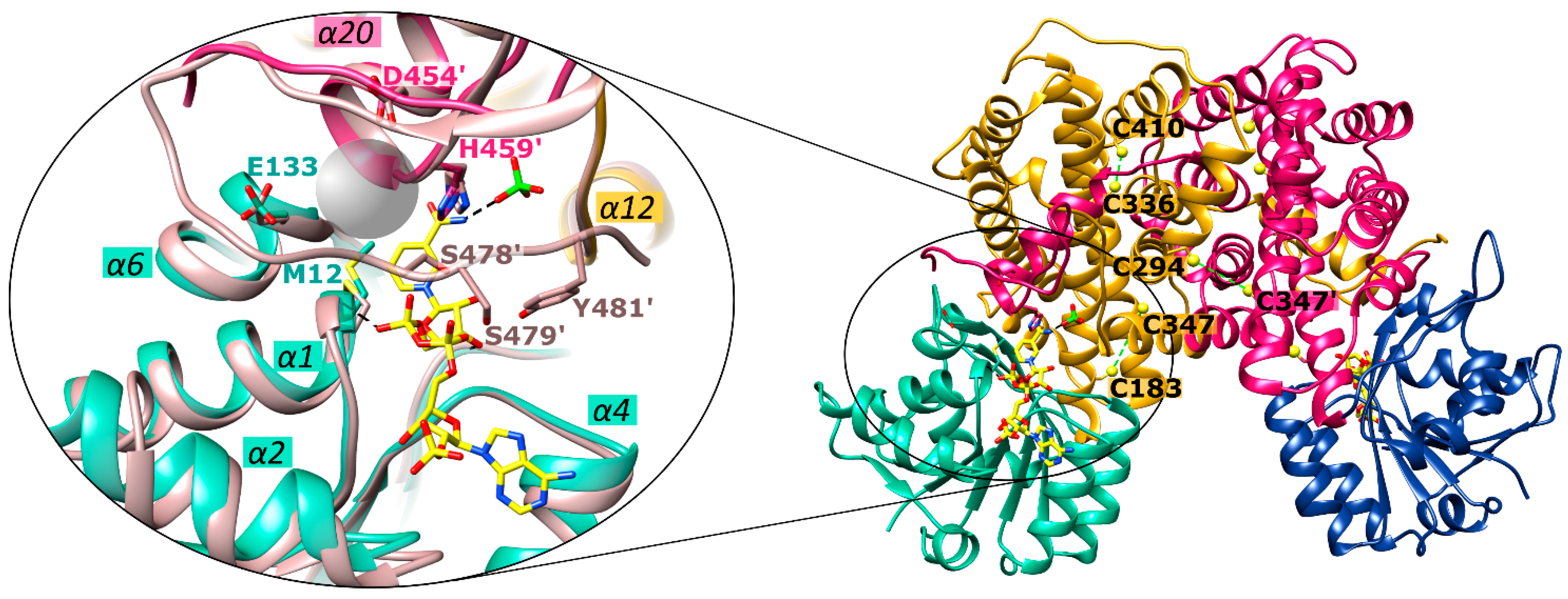
2.3. Overall Structure of Ld6PGD
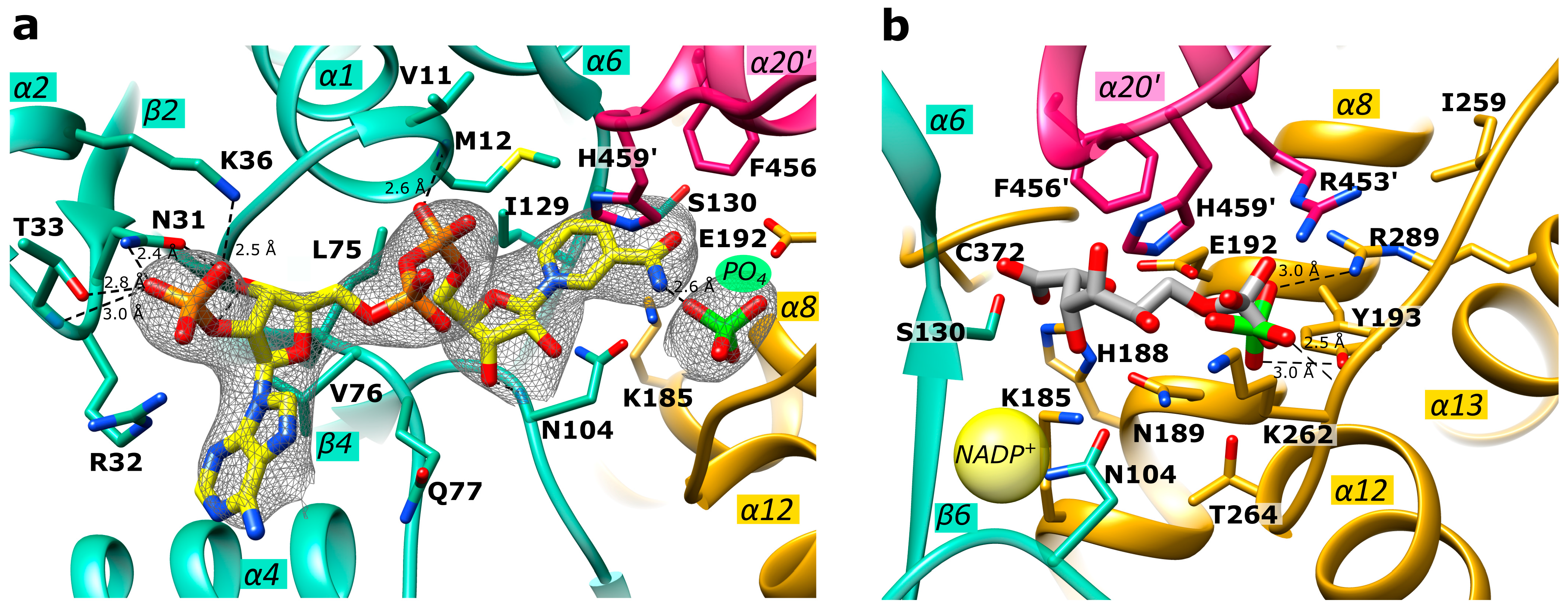
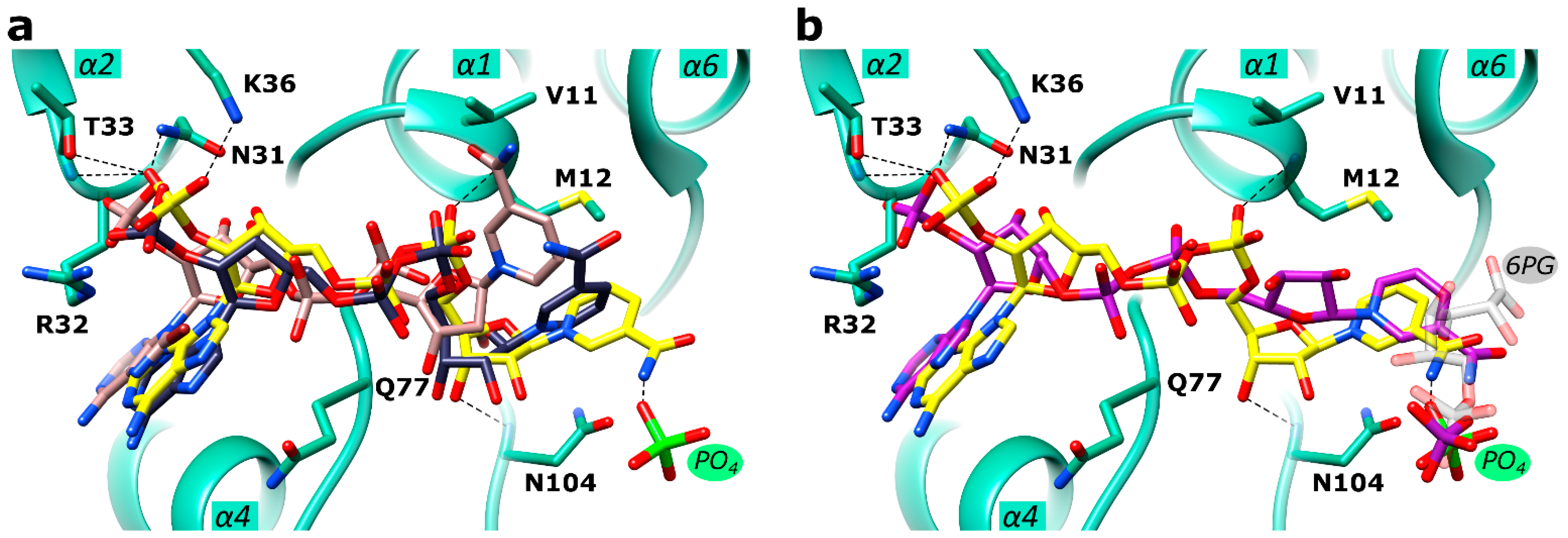
2.4. Active Site in the Ld6PGD Structure
2.5. Mechanistic Considerations
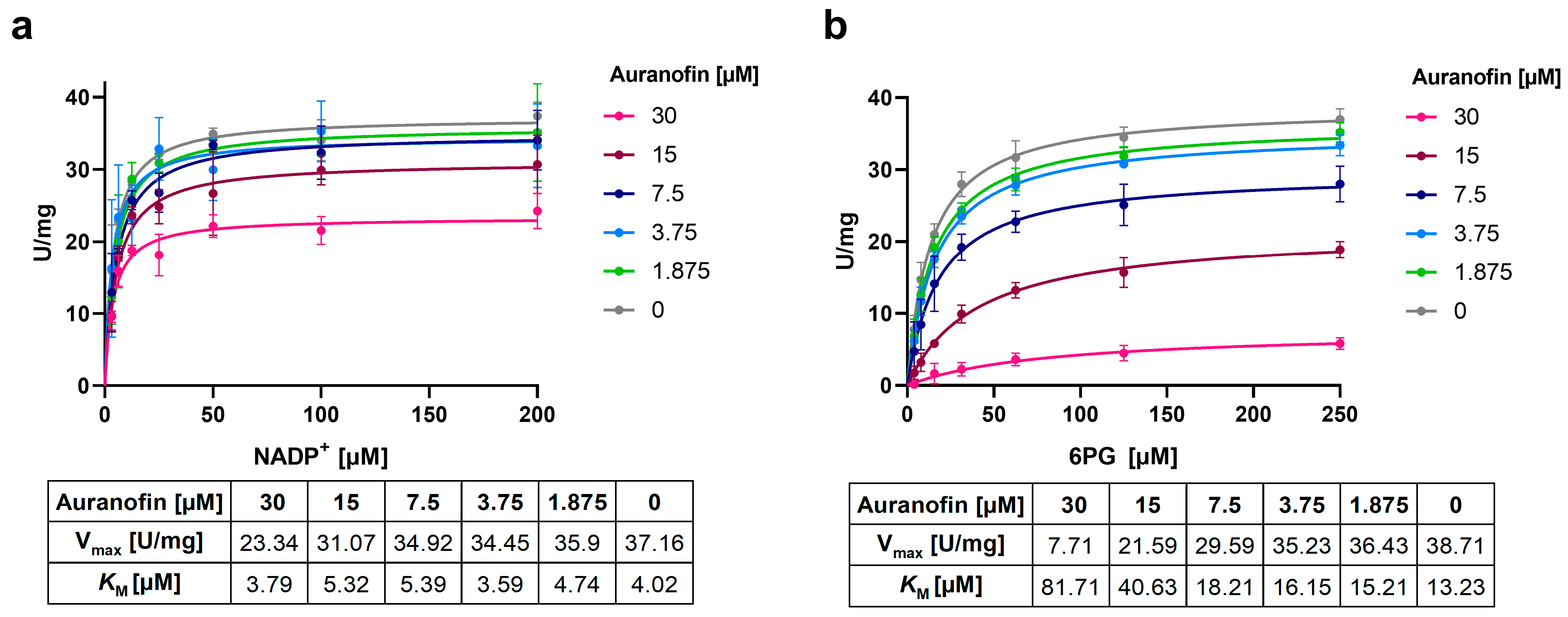
2.6. Ld6PGD Is a Target of the Gold Inhibitor Auranofin
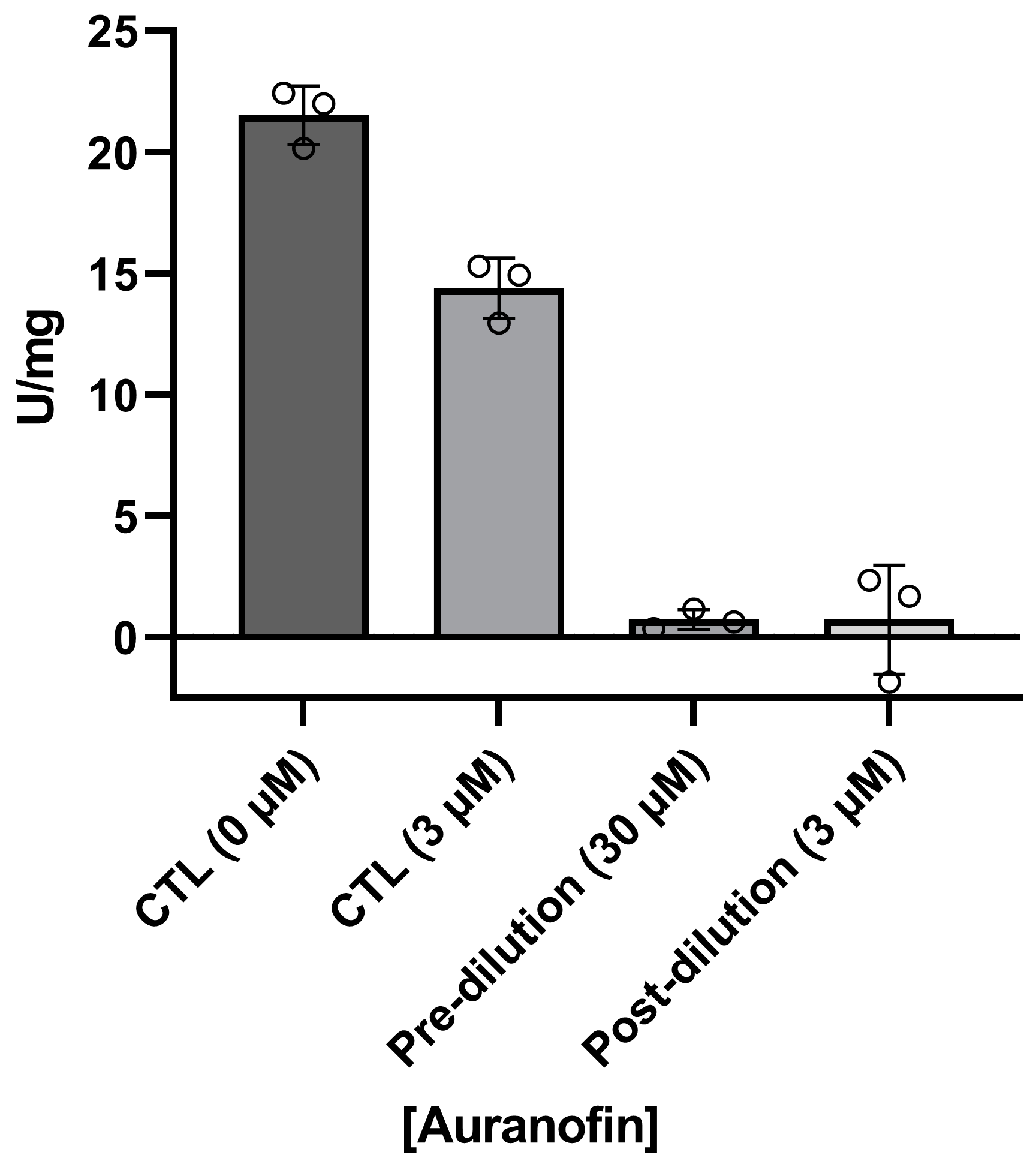
2.7. Auranofin Competes with the 6PG Binding Site and Binds Irreversibly to Ld6PGD
3. Materials and Methods
3.1. Reagents
3.2. Expression and Purification of His-Tagged Ld6PGD wt
3.3. Enzymatic Characterization of Ld6PGD wt
3.4. Enzymatic Characterization of Gold(I)-Based Compounds
3.5. Protein Crystallization
3.6. Data Collections and Processing
3.7. Structure Determination and Refinement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Leishmaniasis. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 27 May 2022).
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, I.; Faria, J.; Santarem, N.; Smith, T.K.; Tavares, J.; Cordeiro-da-Silva, A. Potential Drug Targets in the Pentose Phosphate Pathway of Trypanosomatids. Curr. Med. Chem. 2018, 25, 5239–5265. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, J.; Sundar, S. Current and emerging medications for the treatment of leishmaniasis. Expert Opin. Pharmacother. 2019, 20, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Roatt, B.M.; de Oliveira Cardoso, J.M.; De Brito, R.C.F.; Coura-Vital, W.; de Oliveira Aguiar-Soares, R.D.; Reis, A.B. Recent advances and new strategies on leishmaniasis treatment. Appl. Microbiol. Biotechnol. 2020, 104, 8965–8977. [Google Scholar] [CrossRef] [PubMed]
- Solbach, W.; Laskay, T. The Host Response to Leishmania Infection. In Advances in Immunology, 1st, ed.; Dixon, F.J., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 275–317. ISBN 9780120224746. [Google Scholar]
- Vonlaufen, N.; Kanzok, S.M.; Wek, R.C.; Sullivan, W.J. Stress response pathways in protozoan parasites. Cell. Microbiol. 2008, 10, 2387–2399. [Google Scholar] [CrossRef]
- Moradin, N.; Descoteaux, A. Leishmania promastigotes: Building a safe niche within macrophages. Front. Cell. Infect. Microbiol. 2012, 2, 121. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Sardar, A.H.; Mandal, A.; Saini, S.; Abhishek, K.; Kumar, A.; Purkait, B.; Singh, R.; Das, S.; Mukhopadhyay, R.; et al. Metabolic reconfiguration of the central glucose metabolism: A crucial strategy of Leishmania donovani for its survival during oxidative stress. FASEB J. 2015, 29, 2081–2098. [Google Scholar] [CrossRef]
- Krauth-Siegel, R.L.; Comini, M.A. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta 2008, 1780, 1236–1248. [Google Scholar] [CrossRef]
- Kovářová, J.; Barrett, M.P. The Pentose Phosphate Pathway in Parasitic Trypanosomatids. Trends Parasitol. 2016, 32, 622–634. [Google Scholar] [CrossRef]
- Comini, M.A.; Ortíz, C.; Cazzulo, J.J. Drug Targets in Trypanosomal and Leishmanial Pentose Phosphate Pathway. In Trypanosomatid Diseases; Jäger, T., Koch, O., Flohé, L., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; pp. 297–313. ISBN 9783527670383. [Google Scholar]
- Gupta, S.; Igoillo-Esteve, M.; Michels, P.A.M.; Cordeiro, A.T. Glucose-6-phosphate dehydrogenase of trypanosomatids: Characterization, target validation, and drug discovery. Mol. Biol. Int. 2011, 2011, 135701. [Google Scholar] [CrossRef]
- Allen, S.M.; Lim, E.E.; Jortzik, E.; Preuss, J.; Chua, H.H.; MacRae, J.I.; Rahlfs, S.; Haeussler, K.; Downton, M.T.; McConville, M.J.; et al. Plasmodium falciparum glucose-6-phosphate dehydrogenase 6-phosphogluconolactonase is a potential drug target. FEBS J. 2015, 282, 3808–3823. [Google Scholar] [CrossRef]
- Berneburg, I.; Peddibhotla, S.; Heimsch, K.C.; Haeussler, K.; Maloney, P.; Gosalia, P.; Preuss, J.; Rahbari, M.; Skorokhod, O.; Valente, E.; et al. An Optimized Dihydrodibenzothiazepine Lead Compound (SBI-0797750) as a Potent and Selective Inhibitor of Plasmodium falciparum and P. vivax Glucose 6-Phosphate Dehydrogenase 6-Phosphogluconolactonase. Antimicrob. Agents. Chemother. 2022, 66, e0210921. [Google Scholar] [CrossRef]
- Hannaert, V.; Bringaud, F.; Opperdoes, F.R.; Michels, P.A. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid. Biol. Dis. 2003, 2, 11. [Google Scholar] [CrossRef]
- Michels, P.A.M.; Bringaud, F.; Herman, M.; Hannaert, V. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 2006, 1763, 1463–1477. [Google Scholar] [CrossRef]
- Haanstra, J.R.; van Tuijl, A.; Kessler, P.; Reijnders, W.; Michels, P.A.M.; Westerhoff, H.V.; Parsons, M.; Bakker, B.M. Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl. Acad. Sci. USA 2008, 105, 17718–17723. [Google Scholar] [CrossRef]
- Marchand, M.; Kooystra, U.; Wierenga, R.K.; Lambeir, A.M.; van Beeumen, J.; Opperdoes, F.R.; Michels, P.A. Glucosephosphate isomerase from Trypanosoma brucei. Cloning and characterization of the gene and analysis of the enzyme. Eur. J. Biochem. 1989, 184, 455–464. [Google Scholar] [CrossRef]
- Hanau, S.; Rinaldi, E.; Dallocchio, F.; Gilbert, I.H.; Dardonville, C.; Adams, M.J.; Gover, S.; Barrett, M.P. 6-phosphogluconate dehydrogenase: A target for drugs in African trypanosomes. Curr. Med. Chem. 2004, 11, 2639–2650. [Google Scholar] [CrossRef]
- Hanau, S.; Helliwell, J.R. 6-Phosphogluconate dehydrogenase and its crystal structures. Acta Crystallogr. F Struct. Biol. Commun. 2022, 78, 96–112. [Google Scholar] [CrossRef]
- Kerkhoven, E.J.; Achcar, F.; Alibu, V.P.; Burchmore, R.J.; Gilbert, I.H.; Trybiło, M.; Driessen, N.N.; Gilbert, D.; Breitling, R.; Bakker, B.M.; et al. Handling uncertainty in dynamic models: The pentose phosphate pathway in Trypanosoma brucei. PLoS Comput. Biol. 2013, 9, e1003371. [Google Scholar] [CrossRef]
- Ruda, G.F.; Campbell, G.; Alibu, V.P.; Barrett, M.P.; Brenk, R.; Gilbert, I.H. Virtual fragment screening for novel inhibitors of 6-phosphogluconate dehydrogenase. Bioorg. Med. Chem. 2010, 18, 5056–5062. [Google Scholar] [CrossRef]
- González, D.; Pérez, J.L.; Serrano, M.L.; Igoillo-Esteve, M.; Cazzulo, J.J.; Barrett, M.P.; Bubis, J.; Mendoza-León, A. The 6-phosphogluconate dehydrogenase of Leishmania (Leishmania) mexicana: Gene characterization and protein structure prediction. J. Mol. Microbiol. Biotechnol. 2010, 19, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Mishra, J.; Singh, S. Miltefosine resistance in Leishmania donovani involves suppression of oxidative stress-induced programmed cell death. Exp. Parasitol. 2013, 135, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Purkait, B.; Kumar, A.; Nandi, N.; Sardar, A.H.; Das, S.; Kumar, S.; Pandey, K.; Ravidas, V.; Kumar, M.; De, T.; et al. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob. Agents. Chemother. 2012, 56, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Sudhandiran, G.; Shaha, C. Antimonial-induced increase in intracellular Ca2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular Leishmania donovani amastigotes. J. Biol. Chem. 2003, 278, 25120–25132. [Google Scholar] [CrossRef]
- Finkelstein, A.E.; Walz, D.T.; Batista, V.; Mizraji, M.; Roisman, F.; Misher, A. Auranofin. New oral gold compound for treatment of rheumatoid arthritis. Ann. Rheum. Dis. 1976, 35, 251–257. [Google Scholar] [CrossRef]
- Katz, W.A.; Alexander, S.; Bland, J.H.; Blechman, W.; Bluhm, G.B.; Bonebrake, R.A.; Falbo, A.; Greenwald, R.A.; Hartman, S.; Hobbs, T.; et al. The efficacy and safety of auranofin compared to placebo in rheumatoid arthritis. J. Rheumatol. Suppl. 1982, 8, 173–178. [Google Scholar]
- Kuntz, A.N.; Davioud-Charvet, E.; Sayed, A.A.; Califf, L.L.; Dessolin, J.; Arnér, E.S.J.; Williams, D.L. Thioredoxin glutathione reductase from Schistosoma mansoni: An essential parasite enzyme and a key drug target. PLoS Med. 2007, 4, e206. [Google Scholar] [CrossRef]
- Caroli, A.; Simeoni, S.; Lepore, R.; Tramontano, A.; Via, A. Investigation of a potential mechanism for the inhibition of SmTGR by Auranofin and its implications for Plasmodium falciparum inhibition. Biochem. Biophys. Res. Commun. 2012, 417, 576–581. [Google Scholar] [CrossRef]
- Bonilla, M.; Denicola, A.; Novoselov, S.V.; Turanov, A.A.; Protasio, A.; Izmendi, D.; Gladyshev, V.N.; Salinas, G. Platyhelminth mitochondrial and cytosolic redox homeostasis is controlled by a single thioredoxin glutathione reductase and dependent on selenium and glutathione. J. Biol. Chem. 2008, 283, 17898–17907. [Google Scholar] [CrossRef]
- Tejman-Yarden, N.; Miyamoto, Y.; Leitsch, D.; Santini, J.; Debnath, A.; Gut, J.; McKerrow, J.H.; Reed, S.L.; Eckmann, L. A reprofiled drug, auranofin, is effective against metronidazole-resistant Giardia lamblia. Antimicrob. Agents Chemother. 2013, 57, 2029–2035. [Google Scholar] [CrossRef]
- Ma, C.I.; Tirtorahardjo, J.A.; Jan, S.; Schweizer, S.S.; Rosario, S.A.C.; Du, Y.; Zhang, J.J.; Morrissette, N.S.; Andrade, R.M. Auranofin Resistance in Toxoplasma gondii Decreases the Accumulation of Reactive Oxygen Species but Does Not Target Parasite Thioredoxin Reductase. Front. Cell. Infect. Microbiol. 2021, 11, 618994. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Gromer, S.; Salinas, G.; Gladyshev, V.N. Selenium metabolism in Trypanosoma: Characterization of selenoproteomes and identification of a Kinetoplastida-specific selenoprotein. Nucleic Acids Res. 2006, 34, 4012–4024. [Google Scholar] [CrossRef]
- Da Silva, M.T.A.; Silva-Jardim, I.; Portapilla, G.B.; de Lima, G.M.A.; Costa, F.C.; Anibal, F.d.F.; Thiemann, O.H. In vivo and in vitro auranofin activity against Trypanosoma cruzi: Possible new uses for an old drug. Exp. Parasitol. 2016, 166, 189–193. [Google Scholar] [CrossRef]
- Ilari, A.; Baiocco, P.; Messori, L.; Fiorillo, A.; Boffi, A.; Gramiccia, M.; Di Muccio, T.; Colotti, G. A gold-containing drug against parasitic polyamine metabolism: The X-ray structure of trypanothione reductase from Leishmania infantum in complex with auranofin reveals a dual mechanism of enzyme inhibition. Amino Acids 2012, 42, 803–811. [Google Scholar] [CrossRef]
- Sharlow, E.R.; Leimgruber, S.; Murray, S.; Lira, A.; Sciotti, R.J.; Hickman, M.; Hudson, T.; Leed, S.; Caridha, D.; Barrios, A.M.; et al. Auranofin is an apoptosis-simulating agent with in vitro and in vivo anti-leishmanial activity. ACS Chem. Biol. 2014, 9, 663–672. [Google Scholar] [CrossRef]
- Abou-El-Naga, I.F.; Mady, R.F.; Fawzy Hussien Mogahed, N.M. Efectividad in vitro de tres fármacos aprobados y su interacción sinérgica contra Leishmania infantum. Biomedica 2020, 40, 89–101. [Google Scholar] [CrossRef]
- Gromer, S.; Arscott, L.D.; Williams, C.H.; Schirmer, R.H.; Becker, K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J. Biol. Chem. 1998, 273, 20096–20101. [Google Scholar] [CrossRef]
- Urig, S.; Fritz-Wolf, K.; Réau, R.; Herold-Mende, C.; Tóth, K.; Davioud-Charvet, E.; Becker, K. Undressing of phosphine gold(I) complexes as irreversible inhibitors of human disulfide reductases. Angew. Chem. Int. Ed Engl. 2006, 45, 1881–1886. [Google Scholar] [CrossRef]
- Shoeib, T.; Atkinson, D.W.; Sharp, B.L. Structural analysis of the anti-arthritic drug Auranofin: Its complexes with cysteine, selenocysteine and their fragmentation products. Inorganica Chimica Acta 2010, 363, 184–192. [Google Scholar] [CrossRef]
- Debnath, A.; Parsonage, D.; Andrade, R.M.; He, C.; Cobo, E.R.; Hirata, K.; Chen, S.; García-Rivera, G.; Orozco, E.; Martínez, M.B.; et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat. Med. 2012, 18, 956–960. [Google Scholar] [CrossRef]
- Feng, L.; Pomel, S.; Latre de Late, P.; Taravaud, A.; Loiseau, P.M.; Maes, L.; Cho-Ngwa, F.; Bulman, C.A.; Fischer, C.; Sakanari, J.A.; et al. Repurposing Auranofin and Evaluation of a New Gold(I) Compound for the Search of Treatment of Human and Cattle Parasitic Diseases: From Protozoa to Helminth Infections. Molecules 2020, 25, 5075. [Google Scholar] [CrossRef] [PubMed]
- Shaulov, Y.; Sarid, L.; Trebicz-Geffen, M.; Ankri, S. Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization. Antioxidants 2021, 10, 1240. [Google Scholar] [CrossRef] [PubMed]
- Leitsch, D.; Müller, J.; Müller, N. Evaluation of Giardia lamblia thioredoxin reductase as drug activating enzyme and as drug target. Int. J. Parasitol. Drugs Drug Resist. 2016, 6, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Jakkula, P.; Narsimulu, B.; Qureshi, I.A. Biochemical and structural insights into 6-phosphogluconate dehydrogenase from Leishmania donovani. Appl. Microbiol. Biotechnol. 2021, 105, 5471–5489. [Google Scholar] [CrossRef]
- Pearse, B.M.; Rosemeyer, M.A. Human 6-phosphogluconate dehydrogenase. Purification of the erythrocyte enzyme and the influence of ions and NADPH on its activity. Eur. J. Biochem. 1974, 42, 213–223. [Google Scholar] [CrossRef]
- Haeussler, K.; Fritz-Wolf, K.; Reichmann, M.; Rahlfs, S.; Becker, K. Characterization of Plasmodium falciparum 6-Phosphogluconate Dehydrogenase as an Antimalarial Drug Target. J. Mol. Biol. 2018, 430, 4049–4067. [Google Scholar] [CrossRef]
- Sun, Y.; Bandi, M.; Lofton, T.; Smith, M.; Bristow, C.A.; Carugo, A.; Rogers, N.; Leonard, P.; Chang, Q.; Mullinax, R.; et al. Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation. Cell Rep. 2019, 26, 469–482.e5. [Google Scholar] [CrossRef]
- Adams, M.J.; Ellis, G.H.; Gover, S.; Naylor, C.E.; Phillips, C. Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: Implications for NADP specificity and the enzyme mechanism. Structure 1994, 2, 651–668. [Google Scholar] [CrossRef]
- He, W.; Wang, Y.; Liu, W.; Zhou, C.-Z. Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1. BMC Struct. Biol. 2007, 7, 38. [Google Scholar] [CrossRef]
- Sundaramoorthy, R.; Iulek, J.; Barrett, M.P.; Bidet, O.; Ruda, G.F.; Gilbert, I.H.; Hunter, W.N. Crystal structures of a bacterial 6-phosphogluconate dehydrogenase reveal aspects of specificity, mechanism and mode of inhibition by analogues of high-energy reaction intermediates. FEBS J. 2007, 274, 275–286. [Google Scholar] [CrossRef]
- Cameron, S.; Martini, V.P.; Iulek, J.; Hunter, W.N. Geobacillus stearothermophilus 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 450–454. [Google Scholar] [CrossRef]
- Phillips, C.; Dohnalek, J.; Gover, S.; Barrett, M.P.; Adams, M.J. A 2.8 A resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: Comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues. J. Mol. Biol. 1998, 282, 667–681. [Google Scholar] [CrossRef]
- Hanau, S.; d’Empaire, L.P.; Capone, I.; Alberighi, S.; Montioli, R.; Dallocchio, F. Evidence for dimer/tetramer equilibrium in Trypanosoma brucei 6-phosphogluconate dehydrogenase. Biochim. Biophys. Acta 2013, 1834, 2647–2652. [Google Scholar] [CrossRef]
- Dyson, J.E.; D’Orazio, R.E.; Hanson, W.H. Sheep liver 6-phosphogluconate dehydrogenase: Isolation procedure and effect of pH, ionic strength, and metal ions on the kinetic parameters. Arch. Biochem. Biophys. 1973, 154, 623–635. [Google Scholar] [CrossRef]
- Morales-Luna, L.; Hernández-Ochoa, B.; Martínez-Rosas, V.; González-Valdez, A.; Cárdenas-Rodríguez, N.; Enríquez-Flores, S.; Marcial-Quino, J.; Gómez-Manzo, S. Cloning, purification, and characterization of the 6-phosphogluconate dehydrogenase (6 PGDH) from Giardia lamblia. Mol. Biochem. Parasitol. 2021, 244, 111383. [Google Scholar] [CrossRef]
- Hanau, S.; Rippa, M.; Bertelli, M.; Dallocchio, F.; Barrett, M.P. 6-Phosphogluconate dehydrogenase from Trypanosoma brucei. Kinetic analysis and inhibition by trypanocidal drugs. Eur. J. Biochem. 1996, 240, 592–599. [Google Scholar] [CrossRef]
- Esteve, M.I.; Cazzulo, J.J. The 6-phosphogluconate dehydrogenase from Trypanosoma cruzi: The absence of two inter-subunit salt bridges as a reason for enzyme instability. Mol. Biochem. Parasitol. 2004, 133, 197–207. [Google Scholar] [CrossRef]
- Price, N.E.; Cook, P.F. Kinetic and chemical mechanisms of the sheep liver 6-phosphogluconate dehydrogenase. Arch. Biochem. Biophys. 1996, 336, 215–223. [Google Scholar] [CrossRef]
- Rippa, M.; Giovannini, P.P.; Barrett, M.P.; Dallocchio, F.; Hanau, S. 6-Phosphogluconate dehydrogenase: The mechanism of action investigated by a comparison of the enzyme from different species. Biochim. Biophys. Acta 1998, 1429, 83–92. [Google Scholar] [CrossRef]
- Weisz, K.S.; Schofield, P.J.; Edwards, M.R. Human brain 6-phosphogluconate dehydrogenase: Purification and kinetic properties. J. Neurochem. 1985, 44, 510–517. [Google Scholar] [CrossRef]
- Colotti, G.; Ilari, A.; Fiorillo, A.; Baiocco, P.; Cinellu, M.A.; Maiore, L.; Scaletti, F.; Gabbiani, C.; Messori, L. Metal-based compounds as prospective antileishmanial agents: Inhibition of trypanothione reductase by selected gold complexes. ChemMedChem 2013, 8, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Baiocco, P.; Colotti, G.; Franceschini, S.; Ilari, A. Molecular basis of antimony treatment in leishmaniasis. J. Med. Chem. 2009, 52, 2603–2612. [Google Scholar] [CrossRef] [PubMed]
- Ashutosh; Sundar, S.; Goyal, N. Molecular mechanisms of antimony resistance in Leishmania. J. Med. Microbiol. 2007, 56, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, F.; Sayed, A.A.; Williams, D.L.; Boumis, G.; Brunori, M.; Dimastrogiovanni, D.; Miele, A.E.; Pauly, F.; Bellelli, A. Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: Structural and kinetic aspects. J. Biol. Chem. 2009, 284, 28977–28985. [Google Scholar] [CrossRef] [PubMed]
- Jortzik, E.; Farhadi, M.; Ahmadi, R.; Tóth, K.; Lohr, J.; Helmke, B.M.; Kehr, S.; Unterberg, A.; Ott, I.; Gust, R.; et al. Antiglioma activity of GoPI-sugar, a novel gold(I)-phosphole inhibitor: Chemical synthesis, mechanistic studies, and effectiveness in vivo. Biochim. Biophys. Acta 2014, 1844, 1415–1426. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, Q.; Wang, R.; Wang, H.; Wong, Y.-T.; Wang, M.; Hao, Q.; Yan, A.; Kao, R.Y.-T.; Ho, P.-L.; et al. Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin. Nat. Commun. 2020, 11, 5263. [Google Scholar] [CrossRef]
- Salinas, G.; Gao, W.; Wang, Y.; Bonilla, M.; Yu, L.; Novikov, A.; Virginio, V.G.; Ferreira, H.B.; Vieites, M.; Gladyshev, V.N.; et al. The Enzymatic and Structural Basis for Inhibition of Echinococcus granulosus Thioredoxin Glutathione Reductase by Gold(I). Antioxid. Redox Signal. 2017, 27, 1491–1504. [Google Scholar] [CrossRef]
- Mazzei, L.; Massai, L.; Cianci, M.; Messori, L.; Ciurli, S. Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: Unveiling the structural basis for enzyme inhibition. Dalton Trans. 2021, 50, 14444–14452. [Google Scholar] [CrossRef]
- Marzo, T.; Cirri, D.; Gabbiani, C.; Gamberi, T.; Magherini, F.; Pratesi, A.; Guerri, A.; Biver, T.; Binacchi, F.; Stefanini, M.; et al. Auranofin, Et3PAuCl, and Et3PAuI Are Highly Cytotoxic on Colorectal Cancer Cells: A Chemical and Biological Study. ACS Med. Chem. Lett. 2017, 8, 997–1001. [Google Scholar] [CrossRef]
- Topham, C.M.; Matthews, B.; Dalziel, K. Kinetic studies of 6-phosphogluconate dehydrogenase from sheep liver. Eur. J. Biochem. 1986, 156, 555–567. [Google Scholar] [CrossRef]
- Moritz, B.; Striegel, K.; de Graaf, A.A.; Sahm, H. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 2000, 267, 3442–3452. [Google Scholar] [CrossRef]
- Beutler, E. Red Cell Metabolism: A Manual of Biochemical Methods, 3rd ed.; Grune & Stratton: Orlando, FL, USA, 1984; ISBN 0808916726. [Google Scholar]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Tronrud, D.E.; Adams, P.D.; Karplus, P.A. A new default restraint library for the protein backbone in Phenix: A conformation-dependent geometry goes mainstream. Acta Crystallogr. Sect. D Biol. Crystallogr. 2016, 72, 176–179. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

| 6PG | NADP+ | |||||||
|---|---|---|---|---|---|---|---|---|
| Vmax (U·mg−1) | KM (µM) | kcat (s−1) | kcat/KM (µM−1·s−1) | Vmax (U·mg−1) | KM (µM) | kcat (s−1) | kcat/KM (µM−1·s−1) | |
| Leishmania donovani | 33.3 ± 5.1 | 20.2 ± 2.9 | 30.8 ± 4.7 | 1.5 ± 0.02 | 33.1 ± 5.2 | 13.7 ± 1.2 | 30.5 ± 4.8 | 2.2 ± 0.2 |
| Leishmania donovani [47] | n/a | 80.2 ± 7.4 | 1.0 ± 0.02 | 0.01 | n/a | 22.4 ± 2.7 | 0.9 ± 0.02 | 0.04 |
| Trypanosoma cruzi [60] | 32 | 22.2 | n/a | n/a | 32 | 5.9 | n/a | n/a |
| Trypanosoma brucei [59,60] | 31.3 | 3.5 | 27 | 7.7 | 31.2 | 1.5 | 27 | 18 |
| Plasmodium falciparum [49] | 8.0 ± 1.8 | 11.3 ± 2.7 | 7.1 ± 2.5 | 0.6 | 8.0 ± 1.8 | 9.0 ± 4.2 | 7.6 ± 2.2 | 0.84 |
| Giardia lamblia [58] | 26.2 | 49.2 | n/a | n/a | 26.2 | 139.9 | n/a | n/a |
| Human [49] | 22.1 ± 1.2 | 33.7 ± 7.1 | 22.2 ± 0.3 | 0.6 | 22.1 ± 1.2 | 6.9 ± 2.0 | 21.4 ± 0.8 | 3.1 |
| Sheep liver [57] | 18.8 ± 0.9 | 16.1 ± 1.3 | n/a | n/a | 18.8 ± 0.9 | 6.76 ± 1.6 | n/a | n/a |
| PDB Code | 8C79 | |
|---|---|---|
| Data collection | ||
| Space group | P 21 21 21 | |
| Cell dimensions | ||
| a, b, c (Å) | 65.87, 117.35, 128.91 | |
| α, β, γ (°) | 90, 90, 90 | |
| Resolution (Å) | 46.1–3.1 (3.2–3.1) | |
| R-merge (%) | 8.7 (184.6) | |
| I/σI | 9.9 (0.9) | |
| Completeness (%) | 99.57 (99.95) | |
| Redundancy | 5.1 (5.2) | |
| Molecules per AU | 2 | |
| Wilson B-factor | 129.0 | |
| CC1/2 (%) | 99.8 (34.8) | |
| Refinement | ||
| Resolution (Å) | 3.1 | |
| No. reflections | 18,687 | |
| Rwork/Rfree (%) | 26.2 (41.3)/31.7 (45.0) | |
| No. atoms | ||
| Proteins | 7278 | |
| Ligands | 87 | |
| Water | 1 | |
| Protein residues | 956 | |
| B-factors | ||
| Proteins | 126.6 | |
| Ligands | 121.0 | |
| Water | 93.8 | |
| Ramachandran plot (%) | ||
| Favored (%) | 97.5 | |
| R. m. s. deviations | ||
| Bonds lengths | 0.011 | |
| Bond angles | 0.94 |
| IC50 [µM] for Recombinant Protein | |||
|---|---|---|---|
| Compound | Ld6PGD | Pf6PGD | Hs6PGD |
| Auranofin | 8.6 ± 1.0 | 20.1 ± 1.5 | >100 |
| GoPi-sugar | 9.4 ± 2.1 | n.d. | n.d. |
| Ph3PAuCl | 2.1 ± 0.3 | n.d. | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berneburg, I.; Stumpf, M.; Velten, A.-S.; Rahlfs, S.; Przyborski, J.; Becker, K.; Fritz-Wolf, K. Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment. Int. J. Mol. Sci. 2023, 24, 8615. https://doi.org/10.3390/ijms24108615
Berneburg I, Stumpf M, Velten A-S, Rahlfs S, Przyborski J, Becker K, Fritz-Wolf K. Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment. International Journal of Molecular Sciences. 2023; 24(10):8615. https://doi.org/10.3390/ijms24108615
Chicago/Turabian StyleBerneburg, Isabell, Michaela Stumpf, Ann-Sophie Velten, Stefan Rahlfs, Jude Przyborski, Katja Becker, and Karin Fritz-Wolf. 2023. "Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment" International Journal of Molecular Sciences 24, no. 10: 8615. https://doi.org/10.3390/ijms24108615
APA StyleBerneburg, I., Stumpf, M., Velten, A.-S., Rahlfs, S., Przyborski, J., Becker, K., & Fritz-Wolf, K. (2023). Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment. International Journal of Molecular Sciences, 24(10), 8615. https://doi.org/10.3390/ijms24108615






