Relationship between the Reduced Expression of Zinc Finger Protein 668 in Bladder Cancer and Its Invasiveness
Abstract
1. Introduction
2. Results
2.1. Immunohistochemistry
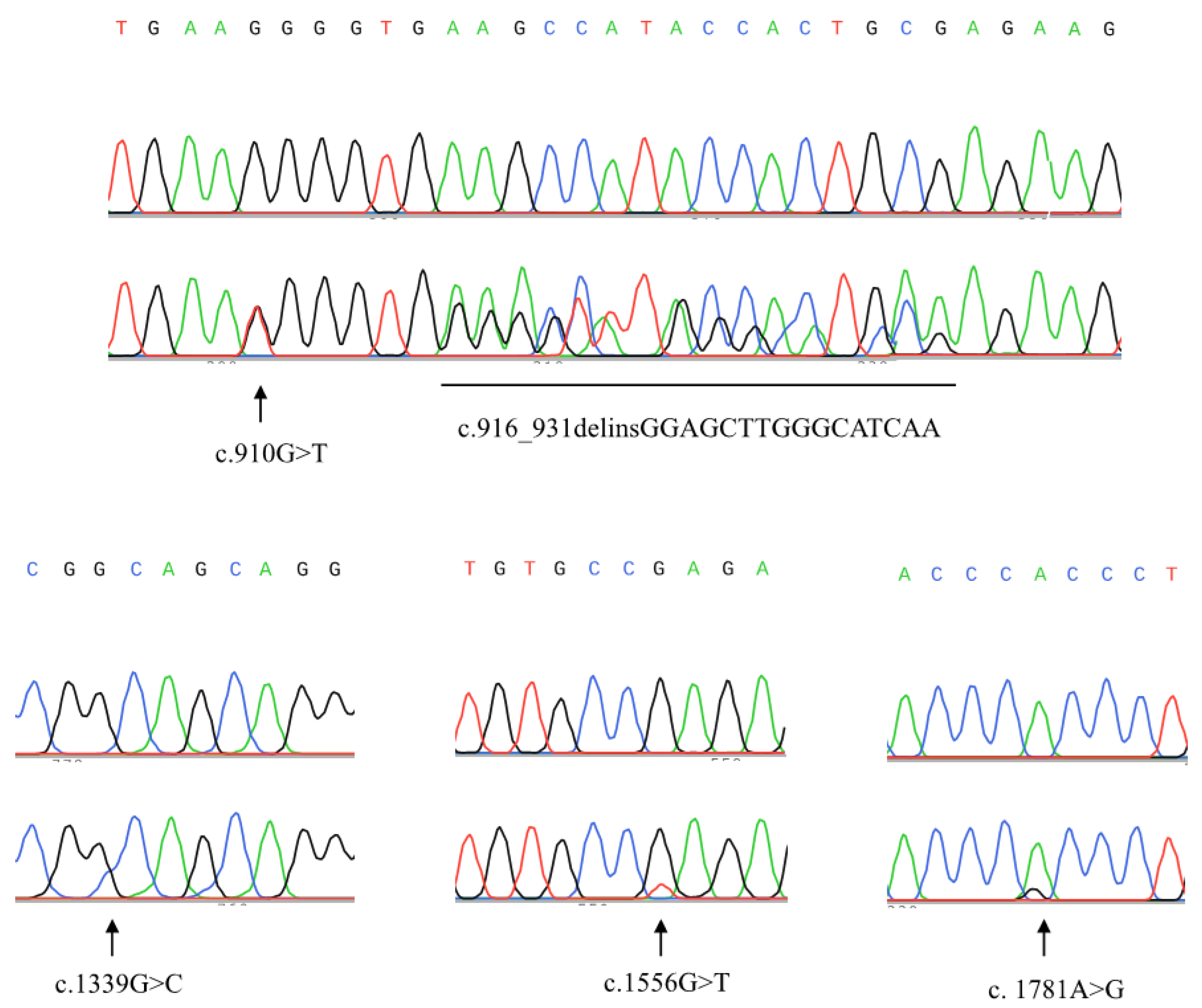
2.2. Structural Analysis of the ZNF668 Gene
3. Discussion
4. Materials and Methods
4.1. Immunohistochemistry
4.2. Structural Analysis of the ZNF668 Gene
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alliance of Genome Resources. Available online: https://www.alliancegenome.org/gene/HGNC:25821 (accessed on 27 May 2022).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- AlphaFold Protein Structure Database. Available online: https://alphafold.ebi.ac.uk/entry/Q96K58 (accessed on 27 May 2022).
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Peng, G.; Dai, H.; Breuer, E.-K.; Stemke-Hale, K.; Li, K.; Gonzalez-Angulo, A.M.; Mills, G.B.; Lin, S.-Y. ZNF668 Functions as a Tumor Suppressor by Regulating P53 Stability and Function in Breast Cancer. Cancer Res. 2011, 71, 6524–6534. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wang, E.; Peng, G.; Dai, H.; Lin, S.-Y. Zinc Finger Protein 668 Interacts with Tip60 to Promote H2AX Acetylation after DNA Damage. Cell Cycle 2013, 12, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Chun-Jen Lin, C.; Mo, W.; Dai, H.; Park, Y.-Y.; Kim, S.M.; Peng, Y.; Mo, Q.; Siwko, S.; Hu, R.; et al. Genome-Wide Transcriptome Profiling of Homologous Recombination DNA Repair. Nat. Commun. 2014, 5, 3361. [Google Scholar] [CrossRef]
- Alsaif, H.S.; Al Ali, H.; Faqeih, E.; Ramadan, S.M.; Barth, M.; Colin, E.; Prouteau, C.; Bonneau, D.; Ziegler, A.; Alkuraya, F.S. ZNF668 Deficiency Causes a Recognizable Disorder of DNA Damage Repair. Hum. Genet. 2021, 140, 1395–1401. [Google Scholar] [CrossRef]
- Cipolla, G.A.; Park, J.K.; de Oliveira, L.A.; Lobo-Alves, S.C.; de Almeida, R.C.; Farias, T.D.J.; Lemos, D.D.S.; Malheiros, D.; Lavker, R.M.; Petzl-Erler, M.L. A 3′UTR Polymorphism Marks Differential KLRG1 MRNA Levels through Disruption of a MiR-584-5p Binding Site and Associates with Pemphigus Foliaceus Susceptibility. Biochim. Biophys. Acta-Gene Regul. Mech. 2016, 1859, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Robertson, K.D. DNA Methyltransferases, DNA Damage Repair, and Cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, G.; Wu, J.; Zhou, H.; Zhang, Y.; Miao, Y.; Feng, Y.; Yu, J. Zinc Finger Protein 668 Suppresses Non-Small Cell Lung Cancer Invasion and Migration by Downregulating Snail and Upregulating E-Cadherin and Zonula Occludens-1. Oncol. Lett. 2018, 15, 3806–3813. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-O.; Kim, H.; Jung, G. P53 Inhibits Tumor Cell Invasion via the Degradation of Snail Protein in Hepatocellular Carcinoma. FEBS Lett. 2010, 584, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987, 8, 138–140. [Google Scholar] [PubMed]
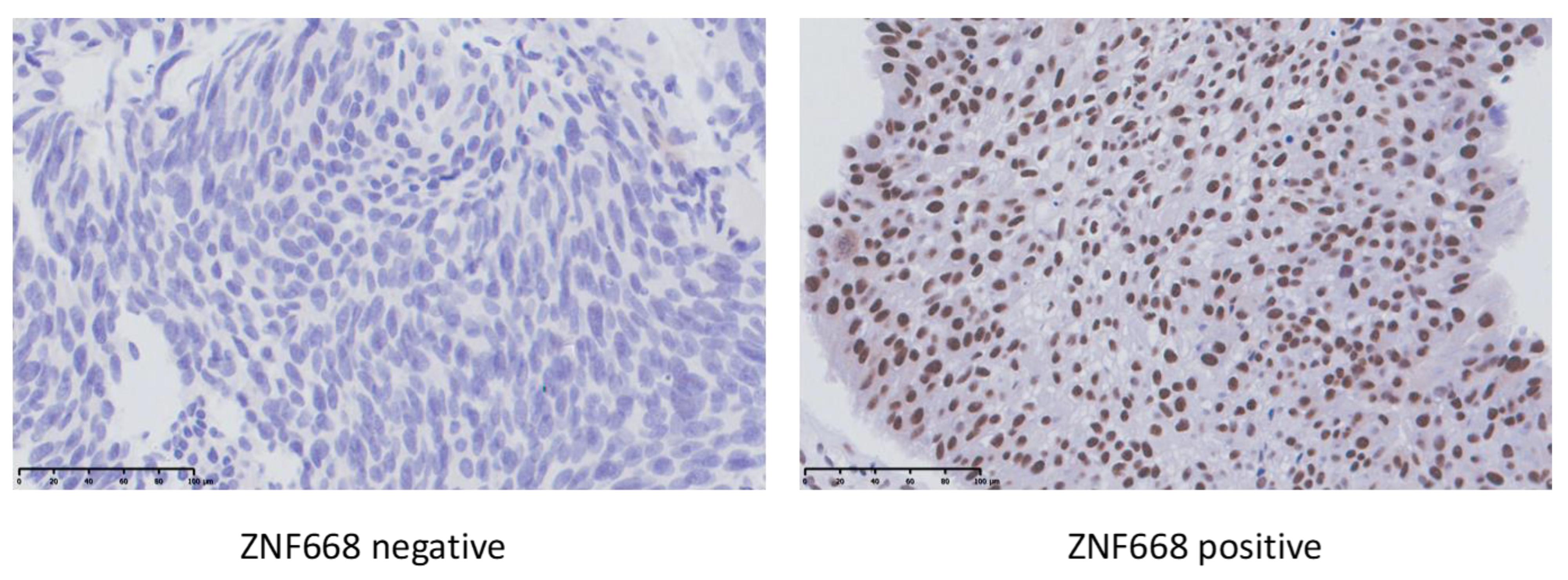
| Submucosal Invasion | |||
|---|---|---|---|
| Plus | Minus | p-Value | |
| IRS | 4.34 ± 2.16 | 6.68 ± 3.23 | 0.0007 |
| muscle invasion | |||
| plus | minus | ||
| IRS | 3.46 ± 2.51 | 6.20 ± 2.92 | 0.0026 |
| rs2303222 (Exon1) | rs2303223 (Exon2) | Somatic Mutation (Exon3) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| G/G (n = 60) | G/A (n = 4) | p-Value | A/A (n = 61) | A/G (n = 5) | p-Value | No (n = 61) | Yes (n = 4) | p-Value | |
| IRS | 5.88 ± 2.73 | 5.50 ± 4.81 | 0.8860 | 5.89 ± 2.80 | 6.40 ± 4.63 | 0.8202 | 5.96 ± 2.85 | 3.88 ± 1.65 | 0.0830 |
| submucosal invasion | 44.6% | 25.0% | 0.6261 | 42.1% | 20.0% | 0.6398 | 42.1% | 50.0% | 1.0000 |
| muscle invasion | 21.4% | 25.0% | 1.0000 | 21.1% | 20.0% | 1.0000 | 19.3% | 50.0% | 0.1963 |
| DNA Changes | Protein Changes | Mean_IRS | |
|---|---|---|---|
| pT1≤ | pT2≤ | ||
| Case A | c.825C>T + | synonymous − | 10.5 |
| Case B | c.[910G>T(;)916_931delinsGGAGCTTGGGCATCAA] − | p.[G298T(;)G304_E311delinsWVGAWASK] − | 4.0 |
| Case C | c.1339G>C + | p.A447P + | 1.5 |
| Case D | c.1556G>T − | p.Q515H − | 5.0 |
| Case E | c.[1781A>G(;)1974C>T(;)2028C>G] + | p.H594R + | 5.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okuno, Y.; Hattori-Kato, M.; Tanaka, H.; Tonooka, A.; Takeuchi, T. Relationship between the Reduced Expression of Zinc Finger Protein 668 in Bladder Cancer and Its Invasiveness. Int. J. Mol. Sci. 2023, 24, 8668. https://doi.org/10.3390/ijms24108668
Okuno Y, Hattori-Kato M, Tanaka H, Tonooka A, Takeuchi T. Relationship between the Reduced Expression of Zinc Finger Protein 668 in Bladder Cancer and Its Invasiveness. International Journal of Molecular Sciences. 2023; 24(10):8668. https://doi.org/10.3390/ijms24108668
Chicago/Turabian StyleOkuno, Yumiko, Mami Hattori-Kato, Hiroki Tanaka, Akiko Tonooka, and Takumi Takeuchi. 2023. "Relationship between the Reduced Expression of Zinc Finger Protein 668 in Bladder Cancer and Its Invasiveness" International Journal of Molecular Sciences 24, no. 10: 8668. https://doi.org/10.3390/ijms24108668
APA StyleOkuno, Y., Hattori-Kato, M., Tanaka, H., Tonooka, A., & Takeuchi, T. (2023). Relationship between the Reduced Expression of Zinc Finger Protein 668 in Bladder Cancer and Its Invasiveness. International Journal of Molecular Sciences, 24(10), 8668. https://doi.org/10.3390/ijms24108668






