Acacia senegal Budmunchiamines as a Potential Adjuvant for Rejuvenating Phenicol Activities towards Escherichia coli-Resistant Strains
Abstract
1. Introduction
2. Results
2.1. Purification of the HEASG Extract by Flash Chromatography
2.2. Effect of Extract Fractions on Direct and Adjuvant Antibacterial Assays
2.3. NMR Structure Determination
2.4. Structure Annotation of LC-MS/MS
3. Discussion
4. Materials and Methods
4.1. Plant Collection
4.2. Hydroethanolic Extracts (HE)
4.3. Flash Chromatography Purification
4.4. Bacterial Strains
4.5. Bacterial Susceptibility Determinations
4.6. Combination with Antibiotics
4.7. Ultra-Performance Liquid Chromatography Coupled to Mass Spectrometry Analysis
4.8. Molecular Networking
4.9. NMR
Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GA, USA, 2019; p. 150.
- OMS. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022; WHO: Geneva, Switzerland, 2022; p. 82.
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Masi, M.; Refregiers, M.; Pos, K.M.; Pages, J.-M. Mechanisms of Envelope Permeability and Antibiotic Influx and Efflux in Gram-Negative Bacteria. Nat. Microbiol. 2017, 2, 17001. [Google Scholar] [CrossRef] [PubMed]
- OMS. Who Global Report on Traditional and Complementary Medicine 2019; WHO: Geneva, Switzerland, 2019; p. 226.
- Lomartire, S.; Gonçalves, A.M.M. An Overview on Antimicrobial Potential of Edible Terrestrial Plants and Marine Macroalgae Rhodophyta and Chlorophyta Extracts. Mar. Drugs 2023, 21, 163. [Google Scholar] [CrossRef] [PubMed]
- Magnini, R.D.; Hilou, A.; Millogo-Koné, H.; Pagès, J.-M.; Davin-Regli, A. Acacia Senegal Extract Rejuvenates the Activity of Phenicols on Selected Enterobacteriaceae Multi Drug Resistant Strains. Antibiotics 2020, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Tabcheh, J.; Vergalli, J.; Davin-Régli, A.; Ghanem, N.; Pagès, J.-M.; Al-Bayssari, C.; Brunel, J.-M. Rejuvenating the Activity of Usual Antibiotics on Resistant Gram-Negative Bacteria: Recent Issues and Perspectives. Int. J. Mol. Sci. 2023, 24, 1515. [Google Scholar] [CrossRef] [PubMed]
- Magnini, R.D.; Nitiéma, M.; Ouédraogo, G.G.; Ilboudo, S.; Bancé, A.; Millogo-Koné, H.; Giorgio, C.D.; Pagès, J.-M.; Hilou, A.; Davin-Regli, A. Toxicity and Bacterial Anti-Motility Activities of the Hydroethanolic Extract of Acacia senegal (L.) Willd (Fabaceae) Leaves. BMC Complement. Med. Ther. 2021, 21, 178. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Purson, S.; Hamzaoui, M.; Borie, N.; Reynaud, R.; Renault, J.-H. Identification of Natural Metabolites in Mixture: A Pattern Recognition Strategy Based on (13)C NMR. Anal. Chem. 2014, 86, 2955–2962. [Google Scholar] [CrossRef] [PubMed]
- Spraul, M.; Freund, A.S.; Nast, R.E.; Withers, R.S.; Maas, W.E.; Corcoran, O. Advancing NMR Sensitivity for LC-NMR-MS Using a Cryoflow Probe: Application to the Analysis of Acetaminophen Metabolites in Urine. Anal. Chem. 2003, 75, 1536–1541. [Google Scholar] [CrossRef]
- Corcoran, O.; Spraul, M. LC-NMR-MS in Drug Discovery. Drug Discov. Today 2003, 8, 624–631. [Google Scholar] [CrossRef]
- Godejohann, M.; Tseng, L.-H.; Braumann, U.; Fuchser, J.; Spraul, M. Characterization of a Paracetamol Metabolite Using On-Line LC-SPE-NMR-MS and a Cryogenic NMR Probe. J. Chromatogr. A 2004, 1058, 191–196. [Google Scholar] [CrossRef]
- Misra, L.N.; Dixit, A.K.; Wagner, H. N-Demethyl Budmunchiamines from Albizzia Lebbek Seeds. Phytochemistry 1995, 39, 247–249. [Google Scholar] [CrossRef]
- Dixit, A.K.; Misra, L.N. Macrocyclic Budmunchiamine Alkaloids from Albizia Lebbek. J. Nat. Prod. 1997, 60, 1036–1037. [Google Scholar] [CrossRef]
- Ovenden, S.P.B.; Cao, S.; Leong, C.; Flotow, H.; Gupta, M.P.; Buss, A.D.; Butler, M.S. Spermine Alkaloids from Albizia Adinocephala with Activity against Plasmodium Falciparum Plasmepsin II. Phytochemistry 2002, 60, 175–177. [Google Scholar] [CrossRef]
- Pezzuto, J.M.; Che, C.-T.; McPherson, D.D.; Zhu, J.-P.; Topcu, G.; Erdelmeier, C.A.J.; Cordell, G.A. DNA as an Affinity Probe Useful in the Detection and Isolation of Biologically Active Natural Products. J. Nat. Prod. 1991, 54, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, J.M.; Mar, W.; Lin, L.-Z.; Cordell, G.A.; Neszmélyi, A.; Wagner, H. Budmunchiamines D–I from Albizia Amara. Phytochemistry 1992, 31, 1795–1800. [Google Scholar] [CrossRef]
- Rukunga, G.M.; Waterman, P.G. New Macrocyclic Spermine (Budmunchiamine) Alkaloids from Albizia Gummifera: With Some Observations on the Structure—Activity Relationships of the Budmunchiamines. J. Nat. Prod. 1996, 59, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Mar, W.; Tan, G.T.; Cordell, G.A.; Pezzuto, J.M.; Jurcic, K.; Offermann, F.; Redl, K.; Steinke, B.; Wagner, H. Biological Activity of Novel Macrocyclic Alkaloids (Budmunchiamines) from Albizia Amara Detected on the Basis of Interaction with DNA. J. Nat. Prod. 1991, 54, 1531–1542. [Google Scholar] [CrossRef]
- Thippeswamy, S.; Mohana, D.C.; Abhishek, R.U.; Manjunath, K. Evaluation of Antimicrobial and Antioxidant Properties of Pithecolobine Isolated from Albizia Saman. J. Herbs Spices Med. Plants 2015, 21, 438–446. [Google Scholar] [CrossRef]
- Thippeswamy, S.; Mohana, D.C.; Abhishek, R.U.; Manjunath, K. Efficacy of Bioactive Compounds Isolated from Albizia Amara and Albizia Saman as Source of Antifungal and Antiaflatoxigenic Agents. J. Verbr. Lebensm. 2013, 8, 297–305. [Google Scholar] [CrossRef]
- Samoylenko, V.; Jacob, M.R.; Khan, S.I.; Zhao, J.; Tekwani, B.L.; Midiwo, J.O.; Walker, L.A.; Muhammad, I. Antimicrobial, Antiparasitic and Cytotoxic Spermine Alkaloids from Albizia Schimperiana. Nat. Prod. Commun. 2009, 4, 791–796. [Google Scholar] [CrossRef]
- Boes, A.; Brunel, J.M.; Derouaux, A.; Kerff, F.; Bouhss, A.; Touze, T.; Breukink, E.; Terrak, M. Squalamine and Aminosterol Mimics Inhibit the Peptidoglycan Glycosyltransferase Activity of PBP1b. Antibiotics 2020, 9, 373. [Google Scholar] [CrossRef]
- Borselli, D.; Blanchet, M.; Bolla, J.-M.; Muth, A.; Skruber, K.; Phanstiel, O.; Brunel, J.M. Motuporamine Derivatives as Antimicrobial Agents and Antibiotic Enhancers against Resistant Gram-Negative Bacteria. ChemBioChem 2017, 18, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Magoulas, G.E.; Kostopoulou, O.N.; Garnelis, T.; Athanassopoulos, C.M.; Kournoutou, G.G.; Leotsinidis, M.; Dinos, G.P.; Papaioannou, D.; Kalpaxis, D.L. Synthesis and Antimicrobial Activity of Chloramphenicol-Polyamine Conjugates. Bioorg. Med. Chem. 2015, 23, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Vergalli, J.; Atzori, A.; Pajovic, J.; Dumont, E.; Malloci, G.; Masi, M.; Vargiu, A.V.; Winterhalter, M.; Réfrégiers, M.; Ruggerone, P.; et al. The Challenge of Intracellular Antibiotic Accumulation, a Function of Fluoroquinolone Influx versus Bacterial Efflux. Commun. Biol. 2020, 3, 198. [Google Scholar] [CrossRef] [PubMed]
- Vergalli, J.; Dumont, E.; Cinquin, B.; Maigre, L.; Pajovic, J.; Bacqué, E.; Mourez, M.; Réfrégiers, M.; Pagès, J.-M. Fluoroquinolone Structure and Translocation Flux across Bacterial Membrane. Sci. Rep. 2017, 7, 9821. [Google Scholar] [CrossRef] [PubMed]
- Fadli, M.; Saad, A.; Sayadi, S.; Chevalier, J.; Mezrioui, N.-E.; Pagès, J.-M.; Hassani, L. Antibacterial Activity of Thymus Maroccanus and Thymus Broussonetii Essential Oils against Nosocomial Infection–Bacteria and Their Synergistic Potential with Antibiotics. Phytomedicine 2012, 19, 464–471. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Pagès, J.-M.; Pradel, E. Overexpression and Purification of the Three Components of the Enterobacter Aerogenes AcrA–AcrB–TolC Multidrug Efflux Pump. J. Chromatogr. B 2003, 786, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Mallea, M.; Chevalier, J.; Bornet, C.; Eyraud, A.; Davin-Regli, A.; Bollet, C.; Pages, J.-M. Porin Alteration and Active Efflux: Two in Vivo Drug Resistance Strategies Used by Enterobacter Aerogenes. Microbiology 1998, 144, 3003–3009. [Google Scholar] [CrossRef]
- Ghisalberti, D.; Masi, M.; Pagès, J.-M.; Chevalier, J. Chloramphenicol and Expression of Multidrug Efflux Pump in Enterobacter Aerogenes. Biochem. Biophys. Res. Commun. 2005, 328, 1113–1118. [Google Scholar] [CrossRef]
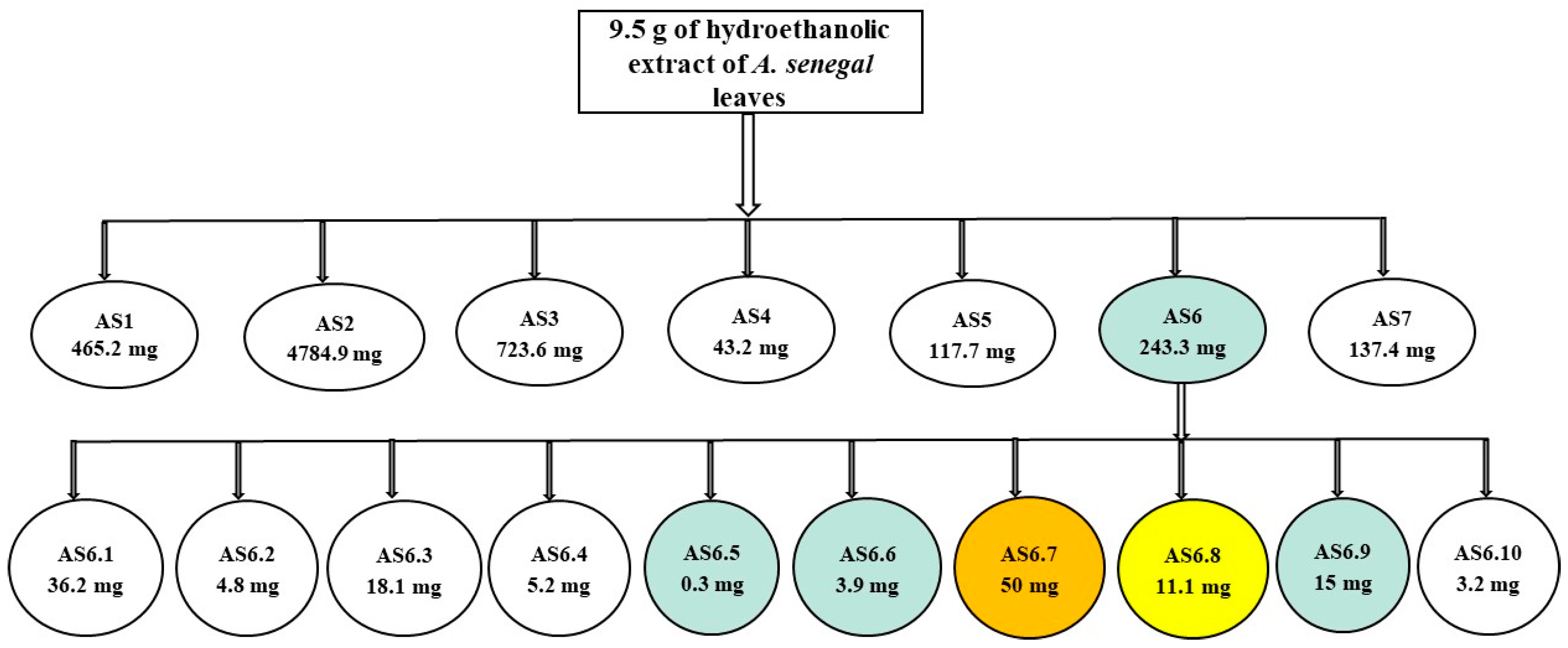
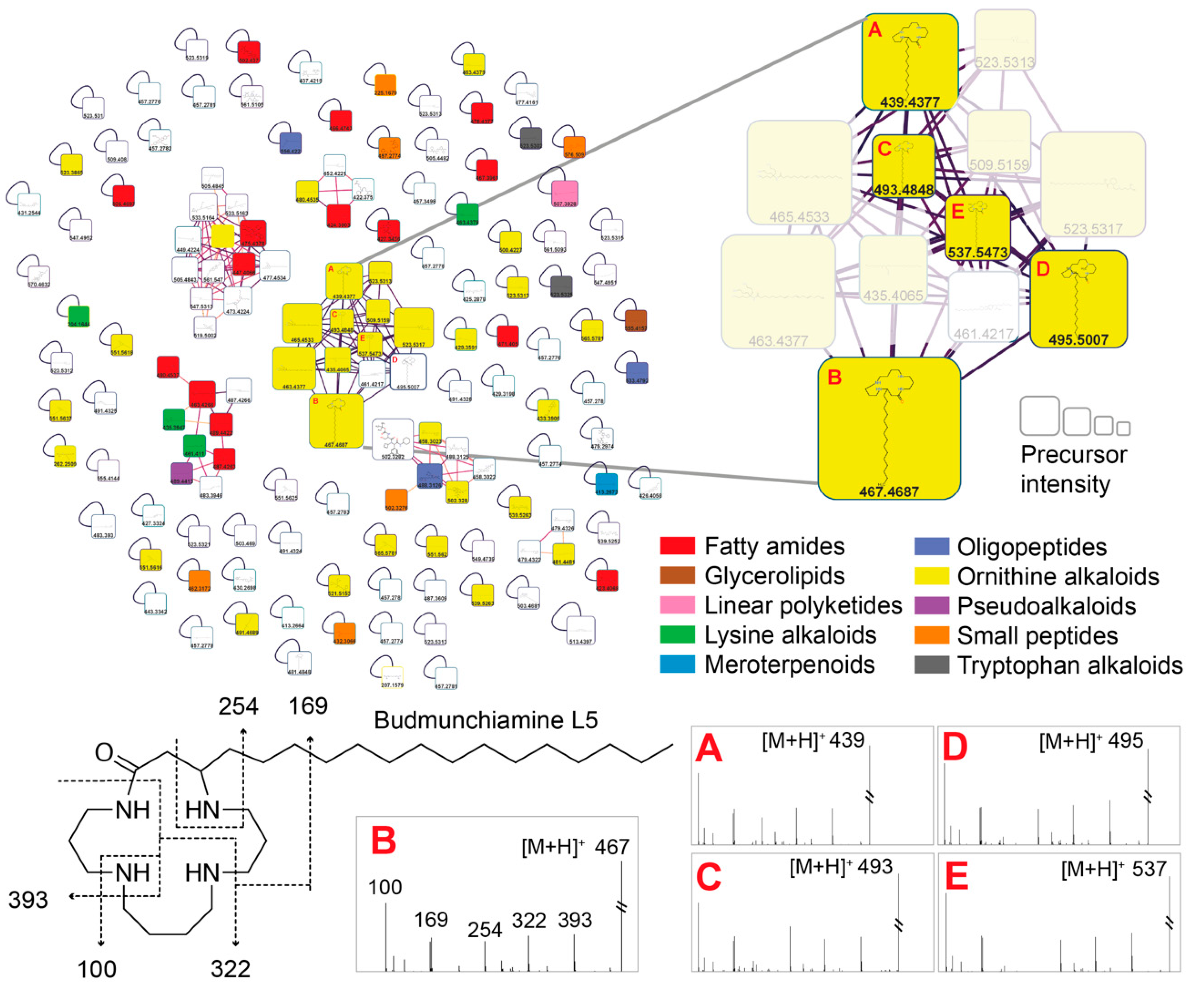
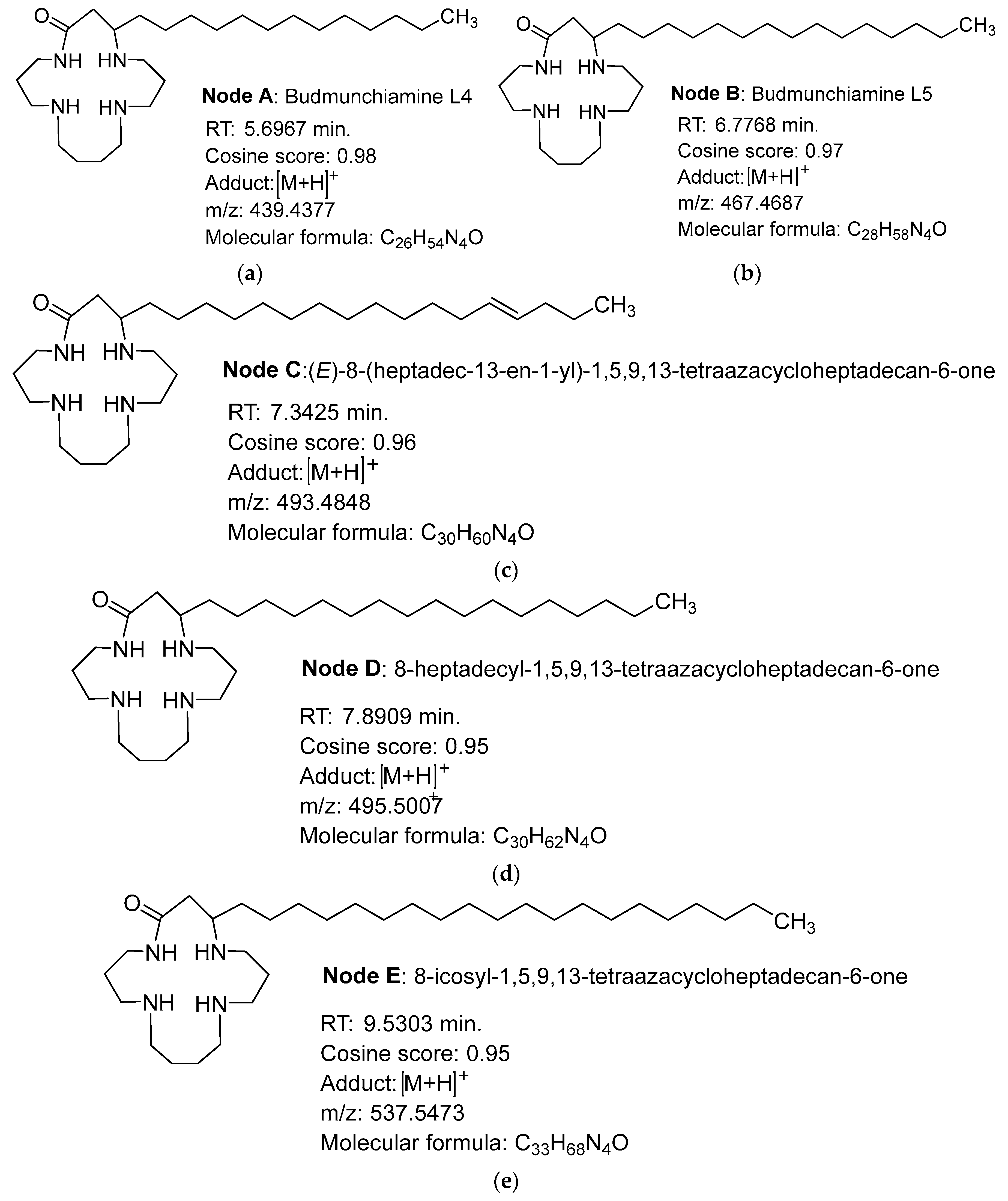
| Fraction | Weight (mg) | Yield (%) |
|---|---|---|
| AS1 (F1–F17) | 465 | 5 |
| AS2 (F18–F33) | 4785 | 50 |
| AS3 (F34–F38) | 724 | 8 |
| AS4 (F39–F45) | 43 | 0.5 |
| AS5 (F46–F50) | 118 | 1 |
| AS6 (F51–F55) | 243 | 3 |
| AS7 (F56–F60) | 137 | 2 |
| Fractions | Weight (mg) | Yield (%) |
|---|---|---|
| AS6.1 (F1–F19) | 36 | 15 |
| AS6.2 (F20–F23) | 5 | 2 |
| AS6.3 (F24–F25) | 18 | 8 |
| AS6.4 (F26–F27) | 5 | 2 |
| AS6.5 (F28–F29) | 1 | 1 |
| AS6.6 (F30–F33) | 4 | 2 |
| AS6.7 (F34–F35) | 50 | 21 |
| AS6.8 (F36–F37) | 11 | 5 |
| AS6.9 (F38–F39) | 15 | 6 |
| AS6.10 (F40) | 3 | 2 |
| E. coli | K. aerogenes | ||||||
|---|---|---|---|---|---|---|---|
| Fractions | AG100 | AG100A | AG102 | Ea 289 | Ea 298 | EaATCC 15038 | CM 64 |
| AS1 | >512 | >512 | >512 | >512 | >512 | >512 | >512 |
| AS2 | >512 | >512 | >512 | >512 | >512 | >512 | >512 |
| AS3 | >512 | >512 | >512 | >512 | >512 | >512 | >512 |
| AS4 | 64 | 64 | 64 | 128 | 32 | 64 | 128 |
| AS5 | 32 | 16 | 64 | 64 | 32 | 32 | 64 |
| AS6 | 16 | 16 | 16 | 32 | 16 | 16 | 64 |
| AS7 | 64 | 32 | 64 | 128 | 64 | 64 | 256 |
| E. coli MIC (mg/L) | |||
|---|---|---|---|
| Fractions | AG100 | AG100A | AG102 |
| AS6.1 | 16 | 16 | 32 |
| AS6.2 | >64 | >64 | >64 |
| AS6.3 | >64 | >64 | >64 |
| AS6.4 | >64 | 64 | >64 |
| AS6.5 | 64 | 64 | >64 |
| AS6.6 | 16 | 16 | 16 |
| AS6.7 | 8 | 16 | 16 |
| AS6.8 | 8 | 16 | 8 |
| AS6.9 | 16 | 16 | 16 |
| AS6.10 | >64 | >64 | >64 |
| Strains | ATBs | Fractions | Combination with AS5 at Conc | ||
|---|---|---|---|---|---|
| E. coli | MIC CHL | MIC AS5 | 16 | 8 | 4 |
| AG100 | 8 | 16 | 2 | 2 | 4 |
| AG100A | 1 | 16 | - | 1 | 1 |
| AG102 | 64 | 32 | 8 | 16 | 32 |
| Combination with AS6 at Conc | |||||
| Strains | MIC CHL | MIC AS6 | 16 | 8 | 4 |
| AG100 | 8 | 16 | - | 1 | 2 (4) |
| AG100A | 1 | 16 | - | 1 | 1 |
| AG102 | 64 | 16 | - | 2 | 4 (16) |
| Combination with AS7 at Conc | |||||
| Strains | MIC CHL | MIC AS7 | 16 | 8 | 4 |
| AG100 | 8 | 32 | 1 | 2 | 4 |
| AG100A | 1 | 32 | 1 | 1 | 1 |
| AG102 | 64 | 32 | 4 | 8 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dofini Magnini, R.; Pedinielli, F.; Vergalli, J.; Ouedraogo, N.; Remy, S.; Hilou, A.; Brunel, J.-M.; Pagès, J.-M.; Davin-Regli, A. Acacia senegal Budmunchiamines as a Potential Adjuvant for Rejuvenating Phenicol Activities towards Escherichia coli-Resistant Strains. Int. J. Mol. Sci. 2023, 24, 8790. https://doi.org/10.3390/ijms24108790
Dofini Magnini R, Pedinielli F, Vergalli J, Ouedraogo N, Remy S, Hilou A, Brunel J-M, Pagès J-M, Davin-Regli A. Acacia senegal Budmunchiamines as a Potential Adjuvant for Rejuvenating Phenicol Activities towards Escherichia coli-Resistant Strains. International Journal of Molecular Sciences. 2023; 24(10):8790. https://doi.org/10.3390/ijms24108790
Chicago/Turabian StyleDofini Magnini, René, François Pedinielli, Julia Vergalli, Noufou Ouedraogo, Simon Remy, Adama Hilou, Jean-Michel Brunel, Jean-Marie Pagès, and Anne Davin-Regli. 2023. "Acacia senegal Budmunchiamines as a Potential Adjuvant for Rejuvenating Phenicol Activities towards Escherichia coli-Resistant Strains" International Journal of Molecular Sciences 24, no. 10: 8790. https://doi.org/10.3390/ijms24108790
APA StyleDofini Magnini, R., Pedinielli, F., Vergalli, J., Ouedraogo, N., Remy, S., Hilou, A., Brunel, J.-M., Pagès, J.-M., & Davin-Regli, A. (2023). Acacia senegal Budmunchiamines as a Potential Adjuvant for Rejuvenating Phenicol Activities towards Escherichia coli-Resistant Strains. International Journal of Molecular Sciences, 24(10), 8790. https://doi.org/10.3390/ijms24108790







