GNAQ-Regulated ZO-1 and ZO-2 Act as Tumor Suppressors by Modulating EMT Potential and Tumor-Repressive Microenvironment in Lung Cancer
Abstract
:1. Introduction
2. Results
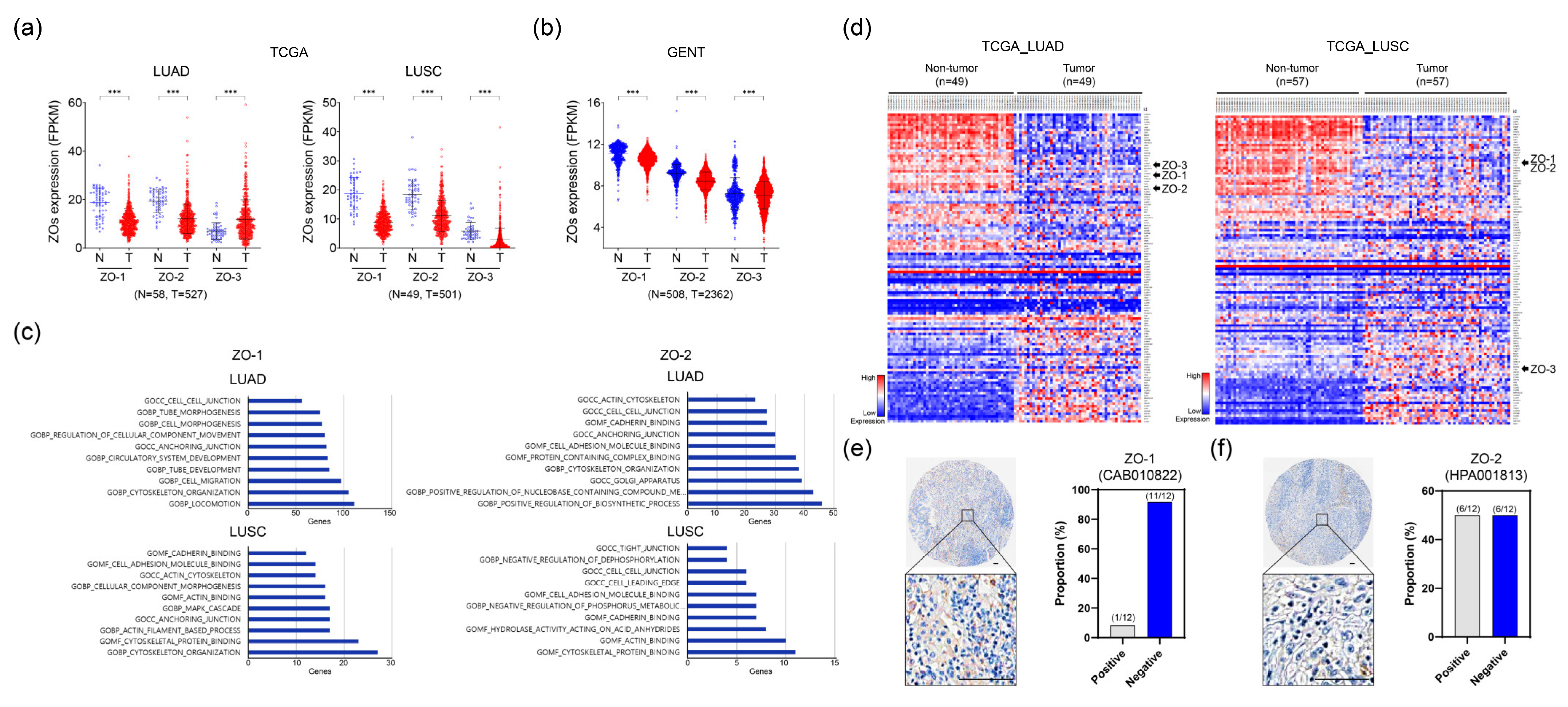
2.1. ZO-1 and ZO-2 Expression Levels Are Downregulated in Lung Cancer Tissues
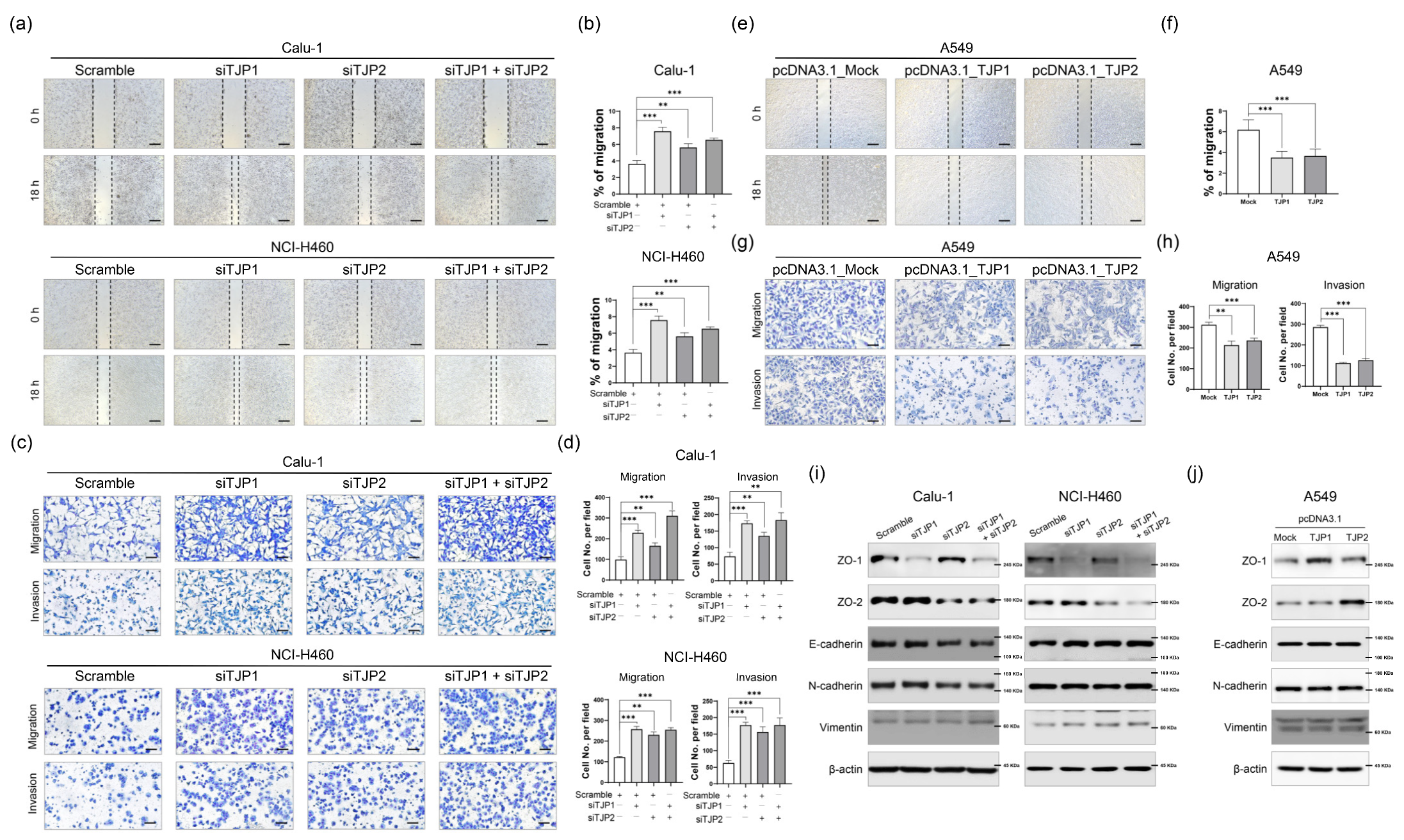
2.2. ZO-1 and ZO-2 Function as Metastatic Suppressors in Lung Cancer
2.3. ZO-1 and ZO-2 Suppresses Distinct M2 Macrophage Polarization Phenotypes
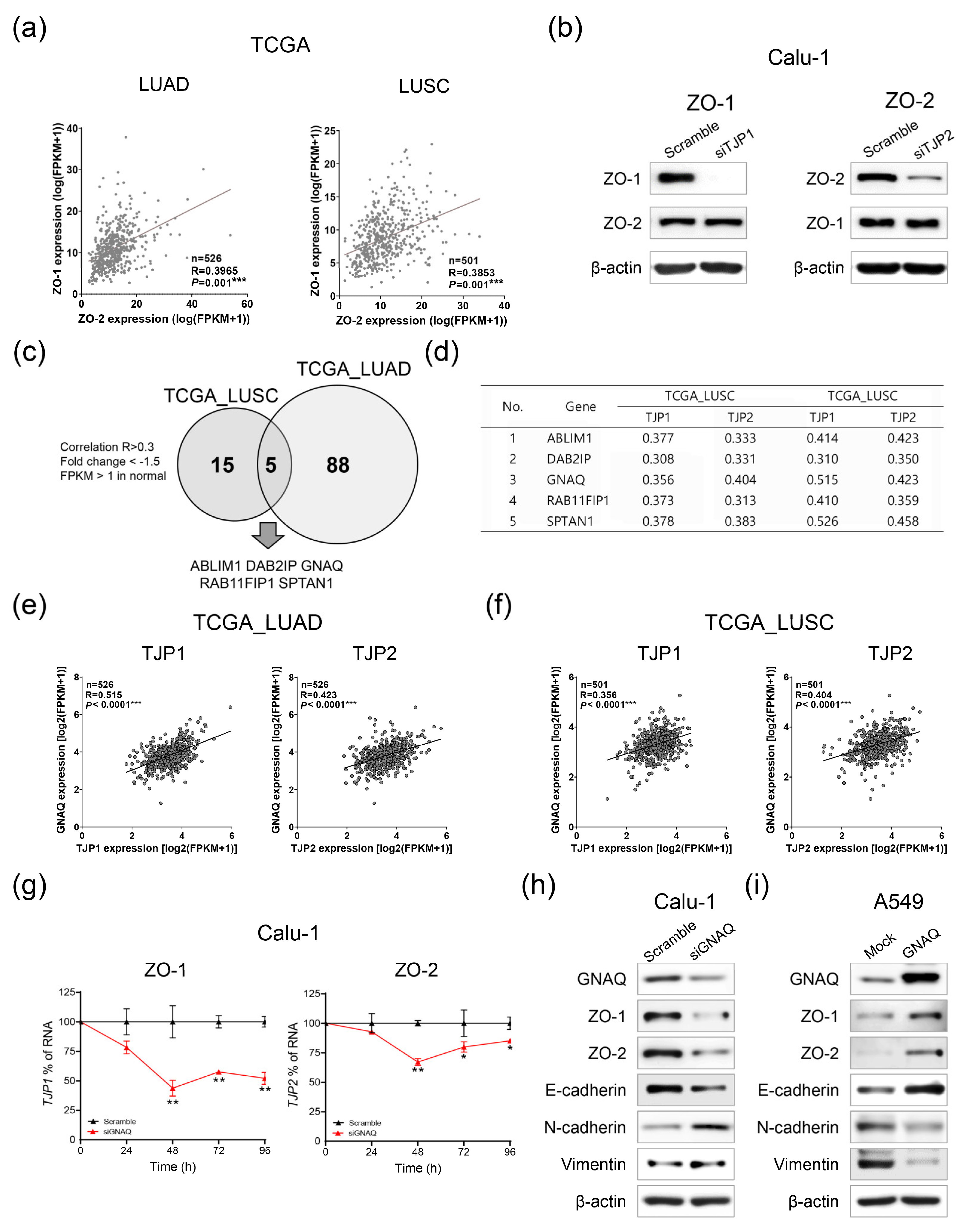
2.4. GNAQ Is an Upstream Regulatory Factor of ZO-1 and ZO-2
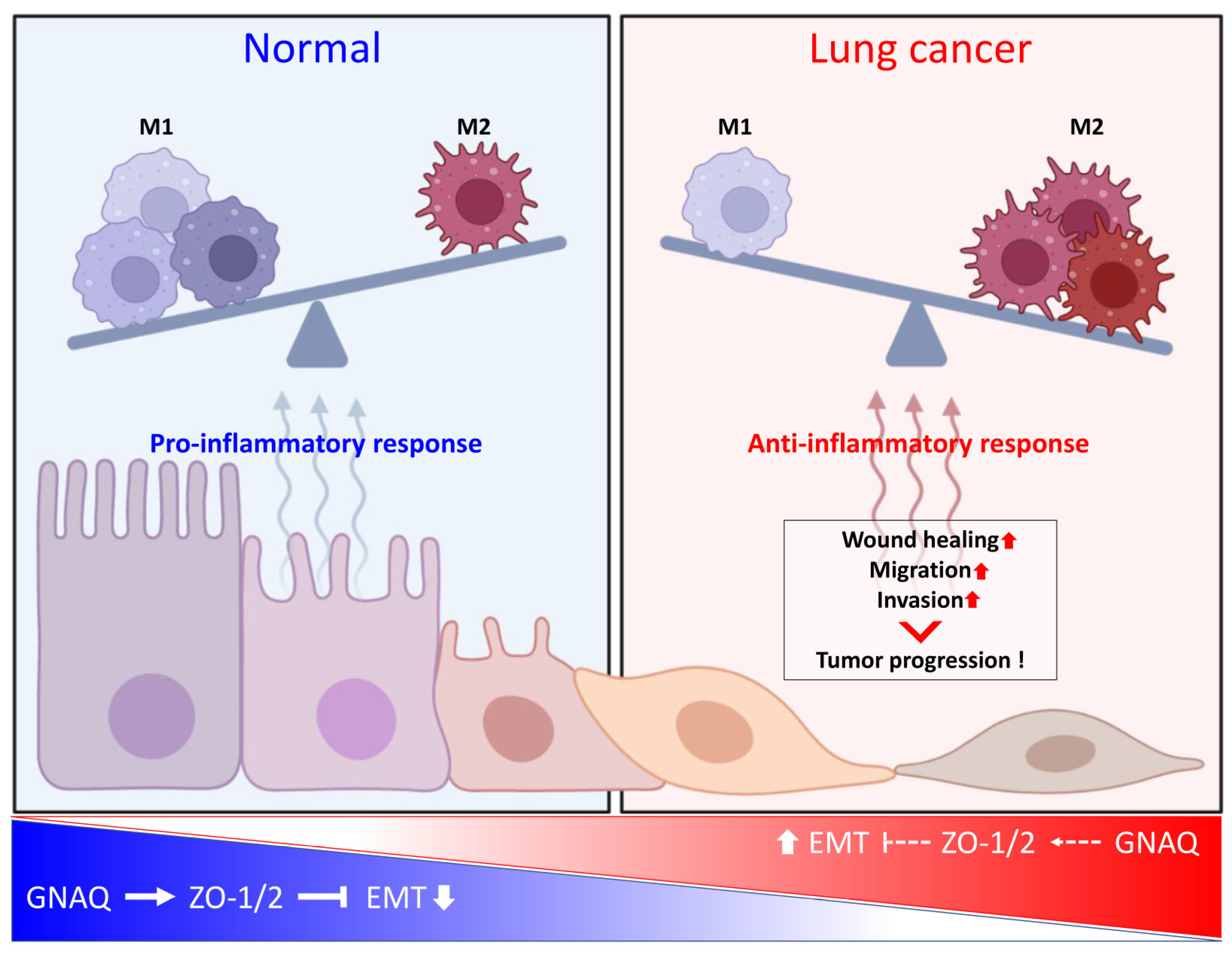
3. Discussion
4. Materials and Methods
4.1. Gene Expression Profiling Using Public Databases
4.2. Cell Culture
4.3. Transfection and Treatment
4.4. Quantitative Real-Time PCR
4.5. Western Blot Analysis
4.6. Proliferation Assay with Cell Counting Kit-8
4.7. Clonogenic Assay
4.8. Wound Healing Assay
4.9. Migration and Invasion Assay
4.10. Co-Culture Procedures for M2 Polarization
4.11. Macrophage Phenotypic Analysis by Flow Cytometry
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Roointan, A.; Ahmad Mir, T.; Ibrahim Wani, S.; Mati Ur, R.; Hussain, K.K.; Ahmed, B.; Abrahim, S.; Savardashtaki, A.; Gandomani, G.; Gandomani, M.; et al. Early detection of lung cancer biomarkers through biosensor technology: A review. J. Pharm. Biomed. Anal. 2019, 164, 93–103. [Google Scholar] [CrossRef]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Watkins, G.; Mansel, R.E.; Jiang, W.G. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur. J. Cancer 2004, 40, 2717–2725. [Google Scholar] [CrossRef]
- Ram, A.K.; Pottakat, B.; Vairappan, B. Increased systemic zonula occludens 1 associated with inflammation and independent biomarker in patients with hepatocellular carcinoma. BMC Cancer 2018, 18, 572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Zhang, H.; Tu, F.; Qiang, Y.; Nie, C. Decreased expression of ZO-1 is associated with tumor metastases in liver cancer. Oncol. Lett. 2019, 17, 1859–1864. [Google Scholar] [CrossRef] [PubMed]
- Masahiko, I.; Mikio, F. Direct Binding of Three Tight Junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH Termini of Claudins. J. Cell Biol. 1999, 147, 1351–1363. [Google Scholar]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Wettschureck, N.; Moers, A.; Wallenwein, B.; Parlow, A.F.; Maser-Gluth, C.; Offermanns, S. Loss of Gq/11 family G proteins in the nervous system causes pituitary somatotroph hypoplasia and dwarfism in mice. Mol. Cell. Biol. 2005, 25, 1942–1948. [Google Scholar] [CrossRef]
- Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013, 70, 631–659. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Bouchalova, P.; Bouchal, P. Current methods for studying metastatic potential of tumor cells. Cancer Cell Int. 2022, 22, 394. [Google Scholar] [CrossRef]
- Choi, J.Y.; Lee, Y.S.; Shim, D.M.; Lee, Y.K.; Seo, S.W. GNAQ knockdown promotes bone metastasis through epithelial-mesenchymal transition in lung cancer cells. Bone Jt. Res. 2021, 10, 310–320. [Google Scholar] [CrossRef]
- Bauer, H.; Zweimueller-Mayer, J.; Steinbacher, P.; Lametschwandtner, A.; Bauer, H.C. The dual role of zonula occludens (ZO) proteins. J. Biomed. Biotechnol. 2010, 2010, 402593. [Google Scholar] [CrossRef]
- Ebnet, K.; Schulz, C.U.; Meyer Zu Brickwedde, M.K.; Pendl, G.G.; Vestweber, D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 2000, 275, 27979–27988. [Google Scholar] [CrossRef]
- Masahiko, I.; Morita, K. Characterization of ZO-2 as a MAGUK Family Member Associated with Tight as well as Adherens Junctions with a Binding Affinity to Occludin and a Catenin. J. Biol. Chem. 1999, 274, 5981–5986. [Google Scholar]
- Lynne, A.; Daniel, A. Molecular Characterization and Tissue Distribution of ZO-2, A Tight Junction Protein Homologous to ZO-1 and the Drosophila Discs-Large Tumor Suppressor Protein. J. Cell Biol. 1994, 124, 949–961. [Google Scholar]
- You, X.; Li, M.; Cai, H.; Zhang, W.; Hong, Y.; Gao, W.; Liu, Y.; Liang, X.; Wu, T.; Chen, F.; et al. Calcium Binding Protein S100A16 Expedites Proliferation, Invasion and Epithelial-Mesenchymal Transition Process in Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 736929. [Google Scholar] [CrossRef]
- Karacz, C.M.; Yan, J.; Zhu, H.; Gerber, D.E. Timing, Sites, and Correlates of Lung Cancer Recurrence. Clin. Lung Cancer 2020, 21, 127–135.e123. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Bersteom, G.; Blank, J.; Jhon, D. Phospholipase C-beta 1 is a GTPase-activating protein for Gq/11, its physiologic regulator. Cell 1992, 70, 411–418. [Google Scholar] [CrossRef]
- Silva-Rodriguez, P.; Fernandez-Diaz, D.; Bande, M.; Pardo, M.; Loidi, L.; Blanco-Teijeiro, M.J. GNAQ and GNA11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma. Cancers 2022, 14, 3066. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Niyazi, M.; Niyazi, I.; Belka, C. Counting colonies of clonogenic assays by using densitometric software. Radiat. Oncol. 2007, 2, 4. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.S.; Lee, S.I.; Choi, Y.R.; Kim, J.; Eun, J.W.; Song, K.S.; Jeong, J.-Y. GNAQ-Regulated ZO-1 and ZO-2 Act as Tumor Suppressors by Modulating EMT Potential and Tumor-Repressive Microenvironment in Lung Cancer. Int. J. Mol. Sci. 2023, 24, 8801. https://doi.org/10.3390/ijms24108801
Kim HS, Lee SI, Choi YR, Kim J, Eun JW, Song KS, Jeong J-Y. GNAQ-Regulated ZO-1 and ZO-2 Act as Tumor Suppressors by Modulating EMT Potential and Tumor-Repressive Microenvironment in Lung Cancer. International Journal of Molecular Sciences. 2023; 24(10):8801. https://doi.org/10.3390/ijms24108801
Chicago/Turabian StyleKim, Hyung Seok, Su In Lee, Yu Rim Choi, Jiyun Kim, Jung Woo Eun, Kyoung Seob Song, and Jee-Yeong Jeong. 2023. "GNAQ-Regulated ZO-1 and ZO-2 Act as Tumor Suppressors by Modulating EMT Potential and Tumor-Repressive Microenvironment in Lung Cancer" International Journal of Molecular Sciences 24, no. 10: 8801. https://doi.org/10.3390/ijms24108801
APA StyleKim, H. S., Lee, S. I., Choi, Y. R., Kim, J., Eun, J. W., Song, K. S., & Jeong, J.-Y. (2023). GNAQ-Regulated ZO-1 and ZO-2 Act as Tumor Suppressors by Modulating EMT Potential and Tumor-Repressive Microenvironment in Lung Cancer. International Journal of Molecular Sciences, 24(10), 8801. https://doi.org/10.3390/ijms24108801







