Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment
Abstract
1. Introduction
2. Results
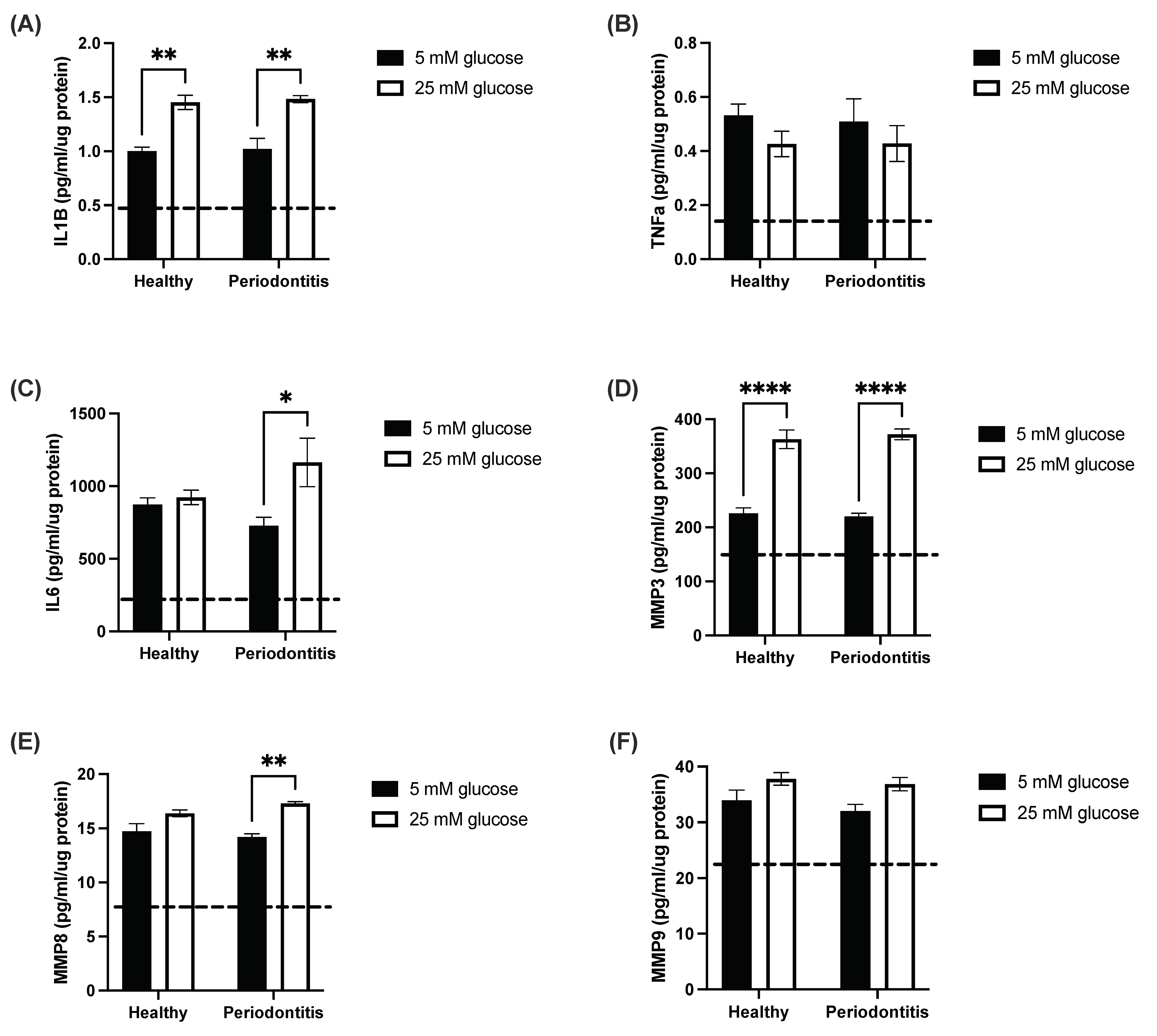
2.1. The Hyperglycemic Microenvironment Impacts Pro-Inflammatory Cytokine and Matrix Metalloproteinase Secretion by a Gingival Coculture Model
2.2. Subgingival Microbiome Highlights Enrichment of Particular ASV
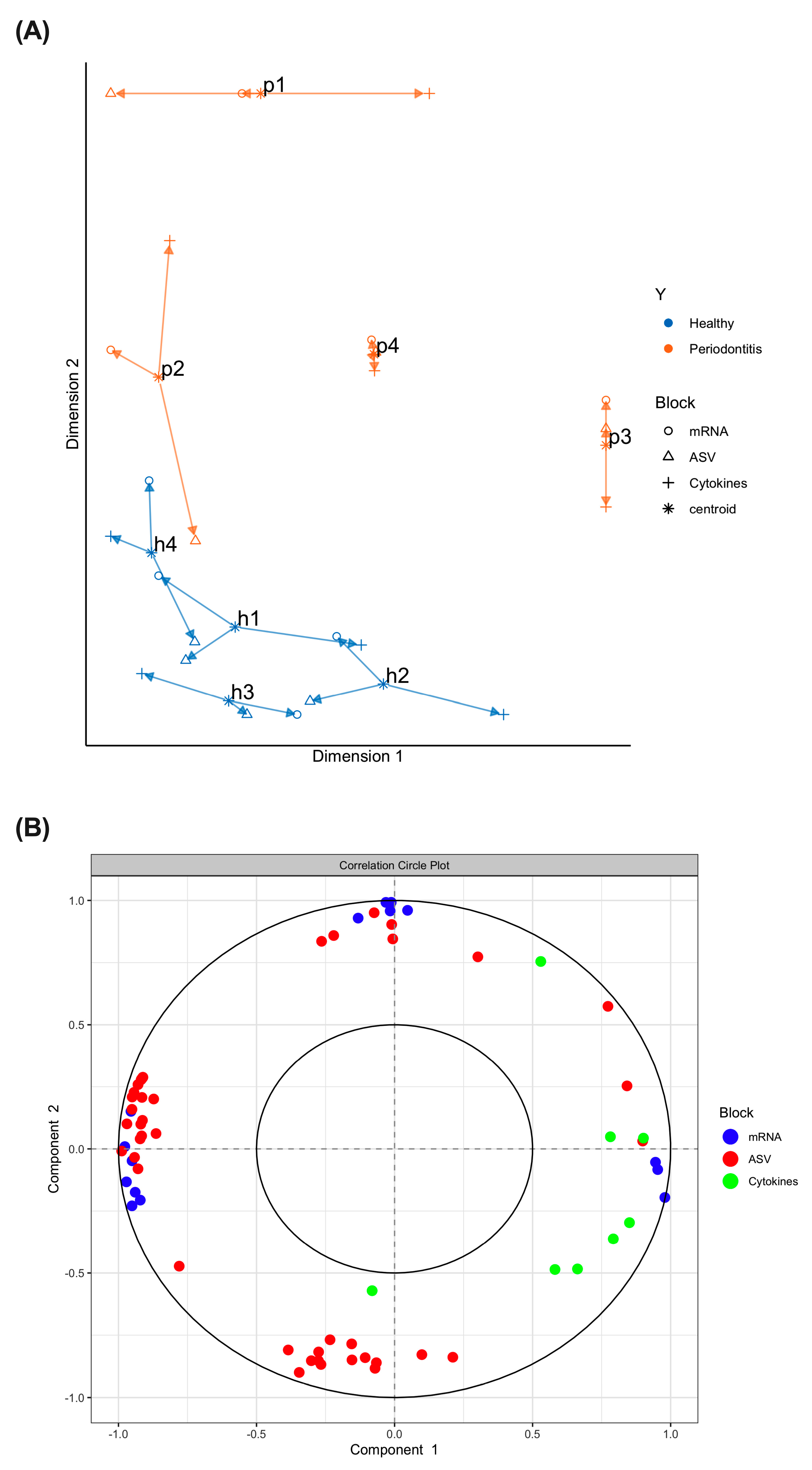
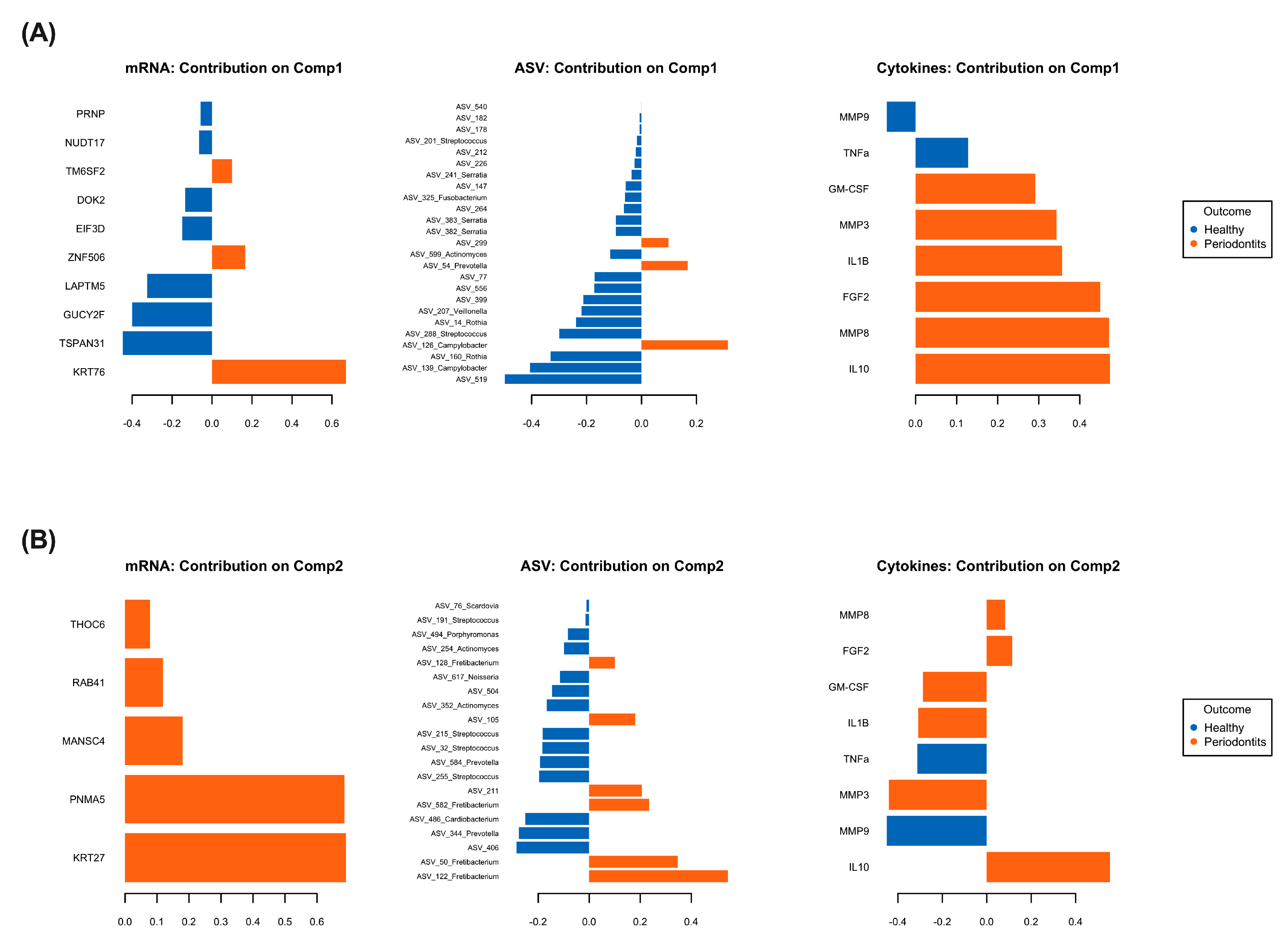
2.3. Multi-Omics Correlations by Computational Biology Analyses Reveal Key Variables Contributing to Periodontitis-Induced Inflammatory Response in a Hyperglycemic Microenvironment
3. Discussion
4. Material and Methods
4.1. Samples Collection
4.2. Coculture Model and Stimulation with the Subgingival Microbiome
4.3. Pro-Inflammatory Cytokines and Matrix Metalloproteinases Measurement
4.4. 16s rRNA Gene Sequencing
4.5. RNA Microarray
4.6. Bioinformatic Analyses and Data Integration
4.6.1. 16s rRNA ASV Analysis Method
4.6.2. Taxonomic Annotation of ASVs
4.6.3. Microarray Analysis
4.6.4. Data Integration (16s, Microarray, Cytokines in Hyperglycemic Microenvironment)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Zukowski, P.; Maciejczyk, M.; Waszkiel, D. Sources of free radicals and oxidative stress in the oral cavity. Arch. Oral. Biol. 2018, 92, 8–17. [Google Scholar] [CrossRef]
- Artese, H.P.; Foz, A.M.; Rabelo Mde, S.; Gomes, G.H.; Orlandi, M.; Suvan, J.; D’Aiuto, F.; Romito, G.A. Periodontal therapy and systemic inflammation in type 2 diabetes mellitus: A meta-analysis. PLoS ONE 2015, 10, e0128344. [Google Scholar] [CrossRef]
- Borgnakke, W.S.; Ylostalo, P.V.; Taylor, G.W.; Genco, R.J. Effect of periodontal disease on diabetes: Systematic review of epidemiologic observational evidence. J. Periodontol. 2013, 84, S135–S152. [Google Scholar] [CrossRef] [PubMed]
- Genco, R.J.; Borgnakke, W.S. Diabetes as a potential risk for periodontitis: Association studies. Periodontology 2020, 83, 40–45. [Google Scholar] [CrossRef]
- Genco, R.J.; Graziani, F.; Hasturk, H. Effects of periodontal disease on glycemic control, complications, and incidence of diabetes mellitus. Periodontology 2020, 83, 59–65. [Google Scholar] [CrossRef]
- Demmer, R.T.; Breskin, A.; Rosenbaum, M.; Zuk, A.; LeDuc, C.; Leibel, R.; Paster, B.; Desvarieux, M.; Jacobs, D.R., Jr.; Papapanou, P.N. The subgingival microbiome, systemic inflammation and insulin resistance: The Oral Infections, Glucose Intolerance and Insulin Resistance Study. J. Clin. Periodontol. 2017, 44, 255–265. [Google Scholar] [CrossRef]
- Demmer, R.T.; Jacobs, D.R., Jr.; Singh, R.; Zuk, A.; Rosenbaum, M.; Papapanou, P.N.; Desvarieux, M. Periodontal Bacteria and Prediabetes Prevalence in ORIGINS: The Oral Infections, Glucose Intolerance, and Insulin Resistance Study. J. Dent. Res. 2015, 94, 201S–211S. [Google Scholar] [CrossRef]
- Demmer, R.T.; Trinh, P.; Rosenbaum, M.; Li, G.; LeDuc, C.; Leibel, R.; Gonzalez, A.; Knight, R.; Paster, B.; Colombo, P.C.; et al. Subgingival Microbiota and Longitudinal Glucose Change: The Oral Infections, Glucose Intolerance and Insulin Resistance Study (ORIGINS). J. Dent. Res. 2019, 98, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Monteiro, M.F.; Dabdoub, S.M.; Miranda, G.L.; Casati, M.Z.; Ribeiro, F.V.; Cirano, F.R.; Pimentel, S.P.; Casarin, R.C.V. Subgingival Host-Microbial Interactions in Hyperglycemic Individuals. J. Dent. Res. 2020, 99, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.C.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M.S. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. J. Periodontol. 2018, 89 (Suppl. 1), S1–S8. [Google Scholar] [CrossRef]
- Graves, D.T.; Ding, Z.; Yang, Y. The impact of diabetes on periodontal diseases. Periodontology 2000 2020, 82, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Lux, R.; Klokkevold, P.; Chang, M.; Barnard, E.; Haake, S.; Li, H. The subgingival microbiome associated with periodontitis in type 2 diabetes mellitus. ISME J. 2020, 14, 519–530. [Google Scholar] [CrossRef]
- Benguigui, C.; Bongard, V.; Ruidavets, J.B.; Chamontin, B.; Sixou, M.; Ferrieres, J.; Amar, J. Metabolic syndrome, insulin resistance, and periodontitis: A cross-sectional study in a middle-aged French population. J. Clin. Periodontol. 2010, 37, 601–608. [Google Scholar] [CrossRef]
- Demmer, R.T.; Squillaro, A.; Papapanou, P.N.; Rosenbaum, M.; Friedewald, W.T.; Jacobs, D.R., Jr.; Desvarieux, M. Periodontal infection, systemic inflammation, and insulin resistance: Results from the continuous National Health and Nutrition Examination Survey (NHANES) 1999–2004. Diabetes Care 2012, 35, 2235–2242. [Google Scholar] [CrossRef]
- Iwamoto, Y.; Nishimura, F.; Nakagawa, M.; Sugimoto, H.; Shikata, K.; Makino, H.; Fukuda, T.; Tsuji, T.; Iwamoto, M.; Murayama, Y. The effect of antimicrobial periodontal treatment on circulating tumor necrosis factor-alpha and glycated hemoglobin level in patients with type 2 diabetes. J. Periodontol. 2001, 72, 774–778. [Google Scholar] [CrossRef]
- Hung, S.L.; Lee, N.G.; Chang, L.Y.; Chen, Y.T.; Lai, Y.L. Stimulatory effects of glucose and Porphyromonas gingivalis lipopolysaccharide on the secretion of inflammatory mediators from human macrophages. J. Periodontol. 2014, 85, 140–149. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, B.; Zhu, R.; Yang, D.; Liu, W.; Wang, Q.; Ji, N.; Chen, Q.; Ding, Y.; Liang, X.; et al. Hyperglycemia accelerates inflammaging in the gingival epithelium through inflammasomes activation. J. Periodontal Res. 2021, 56, 667–678. [Google Scholar] [CrossRef]
- Seneviratne, C.J.; Suriyanarayanan, T.; Widyarman, A.S.; Lee, L.S.; Lau, M.; Ching, J.; Delaney, C.; Ramage, G. Multi-omics tools for studying microbial biofilms: Current perspectives and future directions. Crit. Rev. Microbiol. 2020, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Song, K.E.; Shin, D.S.; Ahn, S.M.; Ha, E.S.; Kim, D.J.; Nam, M.S.; Lee, K.W. Alterations in peripheral blood levels of TIMP-1, MMP-2, and MMP-9 in patients with type-2 diabetes. Diabetes Res. Clin. Pract. 2005, 69, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Uemura, S.; Matsushita, H.; Li, W.; Glassford, A.J.; Asagami, T.; Lee, K.H.; Harrison, D.G.; Tsao, P.S. Diabetes mellitus enhances vascular matrix metalloproteinase activity: Role of oxidative stress. Circ. Res. 2001, 88, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Nowsheen, S.; Aziz, K.; Luo, K.; Deng, M.; Qin, B.; Yuan, J.; Jeganathan, K.B.; Yu, J.; Zhang, H.; Ding, W.; et al. ZNF506-dependent positive feedback loop regulates H2AX signaling after DNA damage. Nat. Commun. 2018, 9, 2736. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, X.; Meng, F.; Zeng, F.; Liu, W.; He, X. PNMA5 accelerated cellular proliferation, invasion and migration in colorectal cancer. Am. J. Transl. Res. 2022, 14, 2231–2243. [Google Scholar]
- Li, C.; Shi, Y.; Zuo, L.; Xin, M.; Guo, X.; Sun, J.; Chen, S.; Zhao, B.; Yang, Z.; Sun, Z.; et al. Identification of Biomarkers Associated with Cancerous Change in Oral Leukoplakia Based on Integrated Transcriptome Analysis. J. Oncol. 2022, 2022, 4599305. [Google Scholar] [CrossRef]
- Sequeira, I.; Neves, J.F.; Carrero, D.; Peng, Q.; Palasz, N.; Liakath-Ali, K.; Lord, G.M.; Morgan, P.R.; Lombardi, G.; Watt, F.M. Immunomodulatory role of Keratin 76 in oral and gastric cancer. Nat. Commun. 2018, 9, 3437. [Google Scholar] [CrossRef]
- Wang, J.; Ouyang, S.; Zhao, S.; Zhang, X.; Cheng, M.; Fan, X.; Cai, Y.; Liao, L. SP1-Mediated Upregulation of circFAM126A Promotes Proliferation and Epithelial-Mesenchymal Transition of Oral Squamous Cell Carcinoma via Regulation of RAB41. Front. Oncol. 2022, 12, 715534. [Google Scholar] [CrossRef]
- Sen, P.; Govaere, O.; Sinioja, T.; McGlinchey, A.; Geng, D.; Ratziu, V.; Bugianesi, E.; Schattenberg, J.M.; Vidal-Puig, A.; Allison, M.; et al. Quantitative modeling of human liver reveals dysregulation of glycosphingolipid pathways in nonalcoholic fatty liver disease. iScience 2022, 25, 104949. [Google Scholar] [CrossRef]
- Zhu, W.; Liang, W.; Lu, H.; Chang, L.; Zhang, J.; Chen, Y.E.; Guo, Y. Myeloid TM6SF2 Deficiency Inhibits Atherosclerosis. Cells 2022, 11, 2877. [Google Scholar] [CrossRef]
- Nakayama, Y.; Matsuda, H.; Itoh, S.; Iwai, Y.; Takai, H.; Mezawa, M.; Yoshino, S.; Ogata, Y. Impact of adjunctive procedures on recombinant human fibroblast growth factor-2-mediated periodontal regeneration therapy: A retrospective study. J. Periodontol. 2021, 92, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Kharkar, V.V.; Kolte, A.P.; Kolte, R.A.; Bawankar, P.V.; Lathiya, V.N.; Bodhare, G.H. Influence of Adjunctive Photodynamic Therapy on Interleukin-6, Interleukin-8, and Interleukin-10 Gingival Crevicular Fluid Levels in Chronic Periodontitis—A Randomized Controlled Trial. Contemp. Clin. Dent. 2021, 12, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontology 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Johnston, W.; Delaney, C.; Rajendran, R.; Butcher, J.; Khan, S.; Bradshaw, D.; Ramage, G.; Culshaw, S. Biofilm-stimulated epithelium modulates the inflammatory responses in co-cultured immune cells. Sci. Rep. 2019, 9, 15779. [Google Scholar] [CrossRef]
- Peyyala, R.; Kirakodu, S.S.; Novak, K.F.; Ebersole, J.L. Oral microbial biofilm stimulation of epithelial cell responses. Cytokine 2012, 58, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Peyyala, R.; Kirakodu, S.S.; Novak, K.F.; Ebersole, J.L. Oral epithelial cell responses to multispecies microbial biofilms. J. Dent. Res. 2013, 92, 235–240. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef]
- Baraniya, D.; Naginyte, M.; Chen, T.; Albandar, J.M.; Chialastri, S.M.; Devine, D.A.; Marsh, P.D.; Al-Hebshi, N.N. Modeling Normal and Dysbiotic Subgingival Microbiomes: Effect of Nutrients. J. Dent. Res. 2020, 99, 695–702. [Google Scholar] [CrossRef]
- Naginyte, M.; Do, T.; Meade, J.; Devine, D.A.; Marsh, P.D. Enrichment of periodontal pathogens from the biofilms of healthy adults. Sci. Rep. 2019, 9, 5491. [Google Scholar] [CrossRef]
- Fine, D.H.; Furgang, D.; Schreiner, H.C.; Goncharoff, P.; Charlesworth, J.; Ghazwan, G.; Fitzgerald-Bocarsly, P.; Figurski, D.H. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: Implications for virulence. Microbiology 1999, 145 Pt 6, 1335–1347. [Google Scholar] [CrossRef]
- Saito, A.; Inagaki, S.; Kimizuka, R.; Okuda, K.; Hosaka, Y.; Nakagawa, T.; Ishihara, K. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 2008, 54, 349–355. [Google Scholar] [CrossRef]
- Sczepanik, F.S.C.; Grossi, M.L.; Casati, M.; Goldberg, M.; Glogauer, M.; Fine, N.; Tenenbaum, H.C. Periodontitis is an inflammatory disease of oxidative stress: We should treat it that way. Periodontology 2000 2020, 84, 45–68. [Google Scholar] [CrossRef]
- Zhao, Z.; Ming, Y.; Li, X.; Tan, H.; He, X.; Yang, L.; Song, J.; Zheng, L. Hyperglycemia Aggravates Periodontitis via Autophagy Impairment and ROS-Inflammasome-Mediated Macrophage Pyroptosis. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Simsek, O.K.; Baser, U.; Ozgunler, O.; Demirci, O.; Aydin, A.F.; Kucukgergin, C.; Yalcin, F. Comparison of oxidative stress markers in the saliva, gingival crevicular fluid, and serum samples of pregnant women with gestational diabetes and healthy pregnant women. J. Periodontal Res. 2023. [Google Scholar] [CrossRef]
- Ausenda, F.; Barbera, E.; Cotti, E.; Romeo, E.; Natto, Z.S.; Valente, N.A. Clinical, microbiological and immunological short, medium and long-term effects of different strains of probiotics as an adjunct to non-surgical periodontal therapy in patients with periodontitis. Systematic review with meta-analysis. Jpn. Dent. Sci. Rev. 2023, 59, 62–103. [Google Scholar] [CrossRef] [PubMed]
- Ben Lagha, A.; Grenier, D. Black tea theaflavins attenuate Porphyromonas gingivalis virulence properties, modulate gingival keratinocyte tight junction integrity and exert anti-inflammatory activity. J. Periodontal Res. 2017, 52, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Ben Lagha, A.; LeBel, G.; Grenier, D. Dual action of highbush blueberry proanthocyanidins on Aggregatibacter actinomycetemcomitans and the host inflammatory response. BMC Complement. Altern. Med. 2018, 18, 10. [Google Scholar] [CrossRef]
- Butera, A.; Gallo, S.; Pascadopoli, M.; Maiorani, C.; Milone, A.; Alovisi, M.; Scribante, A. Paraprobiotics in Non-Surgical Periodontal Therapy: Clinical and Microbiological Aspects in a 6-Month Follow-Up Domiciliary Protocol for Oral Hygiene. Microorganisms 2022, 10. [Google Scholar] [CrossRef]
- Cannizzaro, S.; Maiorani, C.; Scribante, A.; Butera, A. Personalized Treatment of Periodontitis in a Patient with McArdle’s Disease: The Benefits from Probiotics. Case Rep. Dent. 2023, 2023, 5080384. [Google Scholar] [CrossRef] [PubMed]
- Houde, V.; Grenier, D.; Chandad, F. Protective effects of grape seed proanthocyanidins against oxidative stress induced by lipopolysaccharides of periodontopathogens. J. Periodontol. 2006, 77, 1371–1379. [Google Scholar] [CrossRef]
- Kang, M.S.; Park, G.Y.; Lee, A.R. In Vitro Preventive Effect and Mechanism of Action of Weissella cibaria CMU against Streptococcus mutans Biofilm Formation and Periodontal Pathogens. Microorganisms 2023, 11, 962. [Google Scholar] [CrossRef] [PubMed]
- Van Holm, W.; Carvalho, R.; Delanghe, L.; Eilers, T.; Zayed, N.; Mermans, F.; Bernaerts, K.; Boon, N.; Claes, I.; Lebeer, S.; et al. Antimicrobial potential of known and novel probiotics on in vitro periodontitis biofilms. NPJ Biofilms Microbiomes 2023, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Van Holm, W.; Verspecht, T.; Carvalho, R.; Bernaerts, K.; Boon, N.; Zayed, N.; Teughels, W. Glycerol strengthens probiotic effect of Limosilactobacillus reuteri in oral biofilms: A synergistic synbiotic approach. Mol. Oral. Microbiol. 2022, 37, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.A.; Loesche, W.J. Survival of human dental plaque flora in various transport media. Appl. Microbiol. 1972, 24, 638–644. [Google Scholar] [CrossRef]
- Mottillo, E.P.; Desjardins, E.M.; Crane, J.D.; Smith, B.K.; Green, A.E.; Ducommun, S.; Henriksen, T.I.; Rebalka, I.A.; Razi, A.; Sakamoto, K.; et al. Lack of Adipocyte AMPK Exacerbates Insulin Resistance and Hepatic Steatosis through Brown and Beige Adipose Tissue Function. Cell. Metab. 2016, 24, 118–129. [Google Scholar] [CrossRef]
- Sundstrom, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 27 April 2023).
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- MacDonald, J.W. Affycoretools: Functions Useful for Those Doing Repetitive Analyses with Affymetrix GeneChips. R package Version 1.62.0. Available online: https://www.bioconductor.org/packages/release/bioc/html/affycoretools.html (accessed on 27 April 2023).
- MacDonald, J.W. pd.clariom.s.human: Platform Design Info for Affymetrix Clariom_S_Human. R Package Version 3.14.1. 2016. Available online: https://bioconductor.org/packages/release/data/annotation/html/pd.clariom.s.human.html (accessed on 27 April 2023).
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafleur, S.; Bodein, A.; Mbuya Malaïka Mutombo, J.; Mathieu, A.; Joly Beauparlant, C.; Minne, X.; Chandad, F.; Droit, A.; Houde, V.P. Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment. Int. J. Mol. Sci. 2023, 24, 8832. https://doi.org/10.3390/ijms24108832
Lafleur S, Bodein A, Mbuya Malaïka Mutombo J, Mathieu A, Joly Beauparlant C, Minne X, Chandad F, Droit A, Houde VP. Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment. International Journal of Molecular Sciences. 2023; 24(10):8832. https://doi.org/10.3390/ijms24108832
Chicago/Turabian StyleLafleur, Sarah, Antoine Bodein, Joanna Mbuya Malaïka Mutombo, Alban Mathieu, Charles Joly Beauparlant, Xavier Minne, Fatiha Chandad, Arnaud Droit, and Vanessa P. Houde. 2023. "Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment" International Journal of Molecular Sciences 24, no. 10: 8832. https://doi.org/10.3390/ijms24108832
APA StyleLafleur, S., Bodein, A., Mbuya Malaïka Mutombo, J., Mathieu, A., Joly Beauparlant, C., Minne, X., Chandad, F., Droit, A., & Houde, V. P. (2023). Multi-Omics Data Integration Reveals Key Variables Contributing to Subgingival Microbiome Dysbiosis-Induced Inflammatory Response in a Hyperglycemic Microenvironment. International Journal of Molecular Sciences, 24(10), 8832. https://doi.org/10.3390/ijms24108832






