Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition
Abstract
:1. Introduction
2. CDKs and Cell Cycle
3. LncRNAs and Cell Cycle
3.1. LncRNA Profile
3.2. Cell Cycle Regulated by LncRNAs
4. MiRNAs and Cell Cycle
4.1. MiRNA Profile
4.2. Cell Cycle Regulated by MiRNAs
5. NcRNAs and CDKI Treatment
5.1. CDKIs and Cancer Treatment
5.2. LncRNA and CDKI Clinical Efficiency
5.3. MiRNA and CDKI Clinical Efficiency
6. Perspective and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, M.; Schade, A.E.; Branigan, T.B.; Müller, G.A.; DeCaprio, J.A. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 2022, 47, 1009–1022. [Google Scholar] [CrossRef]
- Wang, L.; Chen, H.; Wang, C.; Hu, Z.; Yan, S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc. Natl. Acad. Sci. USA 2018, 115, E3837–E3845. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Layseca, P.; Streuli, C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014, 34, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Beach, D.; Shapiro, G.I. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016, 6, 353–367. [Google Scholar] [CrossRef]
- Gong, X.; Litchfield, L.M.; Webster, Y.; Chio, L.C.; Wong, S.S.; Stewart, T.R.; Dowless, M.; Dempsey, J.; Zeng, Y.; Torres, R.; et al. Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell 2017, 32, 761–776.e6. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016, 13, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Ro, J.; André, F.; Loi, S.; Verma, S.; Iwata, H.; Harbeck, N.; Loibl, S.; Huang Bartlett, C.; Zhang, K.; et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2015, 373, 209–219. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Hart, L.; Campone, M.; Petrakova, K.; Winer, E.P.; Janni, W.; et al. Overall Survival with Ribociclib plus Letrozole in Advanced Breast Cancer. N. Engl. J. Med. 2022, 386, 942–950. [Google Scholar] [CrossRef]
- Johnston, S.R.D.; Harbeck, N.; Hegg, R.; Toi, M.; Martin, M.; Shao, Z.M.; Zhang, Q.Y.; Martinez Rodriguez, J.L.; Campone, M.; Hamilton, E.; et al. Abemaciclib Combined With Endocrine Therapy for the Adjuvant Treatment of HR+, HER2-, Node-Positive, High-Risk, Early Breast Cancer (monarchE). J. Clin. Oncol. 2020, 38, 3987–3998. [Google Scholar] [CrossRef]
- Wang, M.Q.; Zhu, W.J.; Gao, P. New insights into long non-coding RNAs in breast cancer: Biological functions and therapeutic prospects. Exp. Mol. Pathol. 2021, 120, 104640. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Saw, P.E.; Xu, X.; Chen, J.; Song, E.W. Non-coding RNAs: The new central dogma of cancer biology. Sci. China Life Sci. 2021, 64, 22–50. [Google Scholar] [CrossRef]
- Barr, A.R.; Heldt, F.S.; Zhang, T.; Bakal, C.; Novák, B. A Dynamical Framework for the All-or-None G1/S Transition. Cell Syst. 2016, 2, 27–37. [Google Scholar] [CrossRef]
- Hume, S.; Dianov, G.L.; Ramadan, K. A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 2020, 48, 12483–12501. [Google Scholar] [CrossRef]
- Coffman, J.A. Cell cycle development. Dev. Cell 2004, 6, 321–327. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Pruitt, S.C.; Hershberger, P.A.; Witkiewicz, A.K.; Goodrich, D.W. Cell Cycle and Beyond: Exploiting New RB1 Controlled Mechanisms for Cancer Therapy. Trends Cancer 2019, 5, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Dimova, D.K.; Stevaux, O.; Frolov, M.V.; Dyson, N.J. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes. Dev. 2003, 17, 2308–2320. [Google Scholar] [CrossRef] [PubMed]
- Swaffer, M.P.; Jones, A.W.; Flynn, H.R.; Snijders, A.P.; Nurse, P. CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell 2016, 167, 1750–1761.e16. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Orlando, D.A.; Lin, C.Y.; Bernard, A.; Wang, J.Y.; Socolar, J.E.; Iversen, E.S.; Hartemink, A.J.; Haase, S.B. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 2008, 453, 944–947. [Google Scholar] [CrossRef]
- Lavoie, J.N.; L’Allemain, G.; Brunet, A.; Müller, R.; Pouysségur, J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 1996, 271, 20608–20616. [Google Scholar] [CrossRef]
- Hwang, H.C.; Clurman, B.E. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005, 24, 2776–2786. [Google Scholar] [CrossRef]
- Clijsters, L.; Ogink, J.; Wolthuis, R. The spindle checkpoint, APC/C(Cdc20), and APC/C(Cdh1) play distinct roles in connecting mitosis to S phase. J. Cell Biol. 2013, 201, 1013–1026. [Google Scholar] [CrossRef]
- Carnero, A.; Hannon, G.J. The INK4 family of CDK inhibitors. Curr. Top. Microbiol. Immunol. 1998, 227, 43–55. [Google Scholar] [CrossRef]
- Vidal, A.; Koff, A. Cell-cycle inhibitors: Three families united by a common cause. Gene 2000, 247, 1–15. [Google Scholar] [CrossRef]
- Bury, M.; Le Calvé, B.; Ferbeyre, G.; Blank, V.; Lessard, F. New Insights into CDK Regulators: Novel Opportunities for Cancer Therapy. Trends Cell Biol. 2021, 31, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K.; Zhang, J.; Cheng, L.; Shapiro, D.N.; Winoto, A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol. Cell Biol. 1995, 15, 2682–2688. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J.; Beach, D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 1994, 371, 257–261. [Google Scholar] [CrossRef]
- Guan, K.L.; Jenkins, C.W.; Li, Y.; Nichols, M.A.; Wu, X.; O’Keefe, C.L.; Matera, A.G.; Xiong, Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes. Dev. 1994, 8, 2939–2952. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Roussel, M.F.; Kato, J.Y.; Ashmun, R.A.; Sherr, C.J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol. Cell Biol. 1995, 15, 2672–2681. [Google Scholar] [CrossRef]
- Jeffrey, P.D.; Tong, L.; Pavletich, N.P. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes. Dev. 2000, 14, 3115–3125. [Google Scholar] [CrossRef]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef]
- Kamb, A.; Gruis, N.A.; Weaver-Feldhaus, J.; Liu, Q.; Harshman, K.; Tavtigian, S.V.; Stockert, E.; Day, R.S., 3rd; Johnson, B.E.; Skolnick, M.H. A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264, 436–440. [Google Scholar] [CrossRef]
- Roussel, M.F. The INK4 family of cell cycle inhibitors in cancer. Oncogene 1999, 18, 5311–5317. [Google Scholar] [CrossRef]
- Serrano, M.; Lee, H.; Chin, L.; Cordon-Cardo, C.; Beach, D.; DePinho, R.A. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996, 85, 27–37. [Google Scholar] [CrossRef]
- Kim, W.Y.; Sharpless, N.E. The regulation of INK4/ARF in cancer and aging. Cell 2006, 127, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Roberts, J.M. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes. Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hengst, L.; Göpfert, U.; Lashuel, H.A.; Reed, S.I. Complete inhibition of Cdk/cyclin by one molecule of p21(Cip1). Genes. Dev. 1998, 12, 3882–3888. [Google Scholar] [CrossRef]
- Lacy, E.R.; Filippov, I.; Lewis, W.S.; Otieno, S.; Xiao, L.; Weiss, S.; Hengst, L.; Kriwacki, R.W. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004, 11, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Blain, S.W. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle 2008, 7, 892–898. [Google Scholar] [CrossRef]
- Bencivenga, D.; Stampone, E.; Roberti, D.; Della Ragione, F.; Borriello, A. p27(Kip1), an Intrinsically Unstructured Protein with Scaffold Properties. Cells 2021, 10, 2254. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhang, P.; Harper, J.W.; Elledge, S.J.; Leder, P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995, 82, 675–684. [Google Scholar] [CrossRef]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Matheson, C.J.; Backos, D.S.; Reigan, P. Targeting WEE1 Kinase in Cancer. Trends Pharmacol. Sci. 2016, 37, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Geenen, J.J.J.; Schellens, J.H.M. Molecular Pathways: Targeting the Protein Kinase Wee1 in Cancer. Clin. Cancer Res. 2017, 23, 4540–4544. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Proudfoot, N.J. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 2022, 23, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 1008, pp. 1–46. [Google Scholar] [CrossRef]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Susumu, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef]
- Ali, S.A.; Peffers, M.J.; Ormseth, M.J.; Jurisica, I.; Kapoor, M. The non-coding RNA interactome in joint health and disease. Nat. Rev. Rheumatol. 2021, 17, 692–705. [Google Scholar] [CrossRef]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2017, 18, 558–576. [Google Scholar] [CrossRef]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Ghosh, S. Disease-Associated SNPs in Inflammation-Related lncRNAs. Front. Immunol. 2019, 10, 420. [Google Scholar] [CrossRef]
- Gao, P.; Wei, G.H. Genomic Insight into the Role of lncRNA in Cancer Susceptibility. Int. J. Mol. Sci. 2017, 18, 1239. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. X-chromosome inactivation: Closing in on proteins that bind Xist RNA. Trends Genet. 2002, 18, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Liu, Y.; Liu, R.; Zhang, K.; Zhang, Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015, 11, 137–148. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef]
- Lefevre, P.; Witham, J.; Lacroix, C.E.; Cockerill, P.N.; Bonifer, C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell 2008, 32, 129–139. [Google Scholar] [CrossRef]
- Khyzha, N.; Khor, M.; DiStefano, P.V.; Wang, L.; Matic, L.; Hedin, U.; Wilson, M.D.; Maegdefessel, L.; Fish, J.E. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 16410–16419. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, B.L.; Rong, L.J.; Ye, L.; Xu, H.J.; Zhou, Y.; Yan, X.J.; Liu, W.D.; Zhu, B.; Wang, L.; et al. Roles of PTBP1 in alternative splicing, glycolysis, and oncogensis. J. Zhejiang Univ. Sci. B 2020, 21, 122–136. [Google Scholar] [CrossRef]
- Yang, M.; Lu, H.; Liu, J.; Wu, S.; Kim, P.; Zhou, X. lncRNAfunc: A knowledgebase of lncRNA function in human cancer. Nucleic Acids Res. 2022, 50, D1295–D1306. [Google Scholar] [CrossRef]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef]
- Lee, E.S.; Wolf, E.J.; Ihn, S.S.J.; Smith, H.W.; Emili, A.; Palazzo, A.F. TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res. 2020, 48, 11645–11663. [Google Scholar] [CrossRef] [PubMed]
- Zuckerman, B.; Ulitsky, I. Predictive models of subcellular localization of long RNAs. RNA 2019, 25, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Vlachos, I.S.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016, 44, D231–D238. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, H.; Li, F.; Heindl, L.M.; He, X.; Yu, J.; Yang, J.; Ge, S.; Ruan, J.; Jia, R.; et al. Long Non-coding RNA LINC-PINT Suppresses Cell Proliferation and Migration of Melanoma via Recruiting EZH2. Front. Cell Dev. Biol. 2019, 7, 350. [Google Scholar] [CrossRef]
- Bukhari, I.; Khan, M.R.; Hussain, M.A.; Thorne, R.F.; Yu, Y.; Zhang, B.; Zheng, P.; Mi, Y. PINTology: A short history of the lncRNA LINC-PINT in different diseases. Wiley Interdiscip. Rev. RNA 2022, 13, e1705. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, L.; Huo, X.S.; Yuan, J.H.; Xu, D.; Yuan, S.X.; Zhu, N.; Zhou, W.P.; Yang, G.S.; Wang, Y.Z.; et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54, 1679–1689. [Google Scholar] [CrossRef]
- Pasmant, E.; Laurendeau, I.; Héron, D.; Vidaud, M.; Vidaud, D.; Bièche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Zangouei, A.S.; Zangoue, M.; Taghehchian, N.; Zangooie, A.; Rahimi, H.R.; Saburi, E.; Alavi, M.S.; Moghbeli, M. Cell cycle related long non-coding RNAs as the critical regulators of breast cancer progression and metastasis. Biol. Res. 2023, 56, 1. [Google Scholar] [CrossRef]
- Li, Q.; Bian, Y.; Li, Q. Down-Regulation of TMPO-AS1 Induces Apoptosis in Lung Carcinoma Cells by Regulating miR-143-3p/CDK1 Axis. Technol. Cancer Res. Treat. 2021, 20, 1533033820948880. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Song, Y.; Xu, W.; Zhu, Y. The CDK1-Related lncRNA and CXCL8 Mediated Immune Resistance in Lung Adenocarcinoma. Cells 2022, 11, 2688. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Huang, X.; Dong, W.; Zhu, X.; Li, M.; Cui, N. Long non-coding RNA LINC00630 facilitates hepatocellular carcinoma progression through recruiting transcription factor E2F1 to up-regulate cyclin-dependent kinase 2 expression. Hum. Exp. Toxicol. 2021, 40, S257–S268. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhang, Y.; Li, J.; Ni, Z.; Tao, Z.; You, Q.; He, Z.; Huang, D.; Zheng, S. Low expression of long non-coding RNA ARAP1-AS1 can inhibit lung cancer proliferation by inducing G0/G1 cell cycle organization. J. Thorac. Dis. 2020, 12, 7326–7336. [Google Scholar] [CrossRef] [PubMed]
- Luan, T.; Zhang, T.-Y.; Lv, Z.-H.; Guan, B.-X.; Xu, J.-Y.; Li, J.; Li, M.-X.; Hu, S.-L. The lncRNA ALMS1-IT1 may promote malignant progression of lung adenocarcinoma via AVL9-mediated activation of the cyclin-dependent kinase pathway. Febs Open Bio 2021, 11, 1504–1515. [Google Scholar] [CrossRef]
- Sun, Y.; Niu, X.; Wang, G.; Qiao, X.; Chen, L.; Zhong, M. A Novel lncRNA ENST00000512916 Facilitates Cell Proliferation, Migration and Cell Cycle Progression in Ameloblastoma. Oncotargets Ther. 2020, 13, 1519–1531. [Google Scholar] [CrossRef]
- Su, L.; Han, D.; Wu, J.; Huo, X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumor Biol. 2016, 37, 3925–3931. [Google Scholar] [CrossRef]
- Tang, J.; Huang, F.; Wang, H.; Cheng, F.; Pi, Y.; Zhao, J.; Li, Z. Knockdown of TPT1-AS1 inhibits cell proliferation, cell cycle G1/S transition, and epithelial-mesenchymal transition in gastric cancer. Bosn. J. Basic Med. Sci. 2021, 21, 39–46. [Google Scholar] [CrossRef]
- Lin, X.; Yang, M.; Xia, T.; Guo, J. Increased expression of long noncoding RNA ABHD11-AS1 in gastric cancer and its clinical significance. Med. Oncol. 2014, 31, 42. [Google Scholar] [CrossRef]
- Wu, D.D.; Chen, X.; Sun, K.X.; Wang, L.L.; Chen, S.; Zhao, Y. Role of the lncRNA ABHD11-AS(1) in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol. Cancer 2017, 16, 138. [Google Scholar] [CrossRef]
- Chen, M.; Li, J.; Zhuang, C.; Cai, Z. Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of bladder cancer. Oncotarget 2017, 8, 28176–28186. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, L.-L.; Chen, S.; Zong, Z.-H.; Guan, X.; Zhao, Y. LncRNA ABHD11-AS1 promotes the development of endometrial carcinoma by targeting cyclin D1. J. Cell. Mol. Med. 2018, 22, 3955–3964. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Barwal, T.S.; Khandelwal, A.; Malhotra, A.; Rana, M.K.; Rana, A.P.S.; Imyanitov, E.N.; Vasquez, K.M.; Jain, A. LncRNA ZFAS1 inhibits triple-negative breast cancer by targeting STAT3. Biochimie 2021, 182, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qi, X.; Li, Z.; Jin, S.; Xie, Y.; Zhong, H. lncRNA CADM1-AS1 inhibits cell-cycle progression and invasion via PTEN/AKT/GSK-3β axis in hepatocellular carcinoma. Cancer Manag. Res. 2019, 11, 3813–3828. [Google Scholar] [CrossRef]
- Liu, X.; Li, D.; Zhang, W.; Guo, M.; Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012, 31, 4415–4427. [Google Scholar] [CrossRef]
- Hollander, M.C.; Alamo, I.; Fornace, A.J., Jr. A novel DNA damage-inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res. 1996, 24, 1589–1593. [Google Scholar] [CrossRef]
- Liang, S.; Gong, X.; Zhang, G.; Huang, G.; Lu, Y.; Li, Y. The lncRNA XIST interacts with miR-140/miR-124/iASPP axis to promote pancreatic carcinoma growth. Oncotarget 2017, 8, 113701–113718. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Wu, S. LncRNA-CCDC144NL-AS1 Promotes the Development of Hepatocellular Carcinoma by Inducing WDR5 Expression via Sponging miR-940. J. Hepatocell. Carcinoma 2021, 8, 333–348. [Google Scholar] [CrossRef]
- Jin, X.; Ge, L.P.; Li, D.Q.; Shao, Z.M.; Di, G.H.; Xu, X.E.; Jiang, Y.Z. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Mol. Cancer 2020, 19, 87. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Zheng, H.; Li, C.; Xiong, J.; Wang, W.; Bao, H.; Jin, H.; Liang, P. Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2019, 38, 380. [Google Scholar] [CrossRef]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs--the micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes. Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar] [CrossRef]
- Raisch, J.; Darfeuille-Michaud, A.; Nguyen, H.T. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol. 2013, 19, 2985–2996. [Google Scholar] [CrossRef]
- Chen, K.; Rajewsky, N. Deep conservation of microRNA-target relationships and 3’UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Lin, H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Xu, H.; He, J.H.; Xiao, Z.D.; Zhang, Q.Q.; Chen, Y.Q.; Zhou, H.; Qu, L.H. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010, 52, 1431–1442. [Google Scholar] [CrossRef]
- Young, D.D.; Connelly, C.M.; Grohmann, C.; Deiters, A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 2010, 132, 7976–7981. [Google Scholar] [CrossRef]
- Scott, G.K.; Goga, A.; Bhaumik, D.; Berger, C.E.; Sullivan, C.S.; Benz, C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007, 282, 1479–1486. [Google Scholar] [CrossRef]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Xu, W.; Zhou, P.; Gao, P.; Jiang, S.; Lobie, P.E.; Zhu, T. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 2013, 288, 18121–18133. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.; Park, S.M.; Radjabi, A.R.; Schickel, R.; Kistner, E.O.; Jewell, D.A.; Feig, C.; Lengyel, E.; Peter, M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 11400–11405. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ye, Y.; Jiao, D.; Qiao, J.; Cui, S.; Liu, Z. miR-155 and miR-31 are differentially expressed in breast cancer patients and are correlated with the estrogen receptor and progesterone receptor status. Oncol. Lett. 2012, 4, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Guo, Y.; Song, H.; Xiao, B.; Sun, W.; Liu, Z.; Yu, X.; Xia, T.; Cui, L.; Guo, J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 2013, 518, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhu, Y.; Xiong, Y.; Ge, Y.Y.; Yun, J.P.; Zhuang, S.M. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009, 50, 113–121. [Google Scholar] [CrossRef]
- Lobert, S.; Jefferson, B.; Morris, K. Regulation of β-tubulin isotypes by micro-RNA 100 in MCF7 breast cancer cells. Cytoskeleton 2011, 68, 355–362. [Google Scholar] [CrossRef]
- Gong, Y.; He, T.; Yang, L.; Yang, G.; Chen, Y.; Zhang, X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci. Rep. 2015, 5, 11650. [Google Scholar] [CrossRef]
- Li, H.; Jiang, M.; Cui, M.; Feng, G.; Dong, J.; Li, Y.; Xiao, H.; Fan, S. MiR-365 enhances the radiosensitivity of non-small cell lung cancer cells through targeting CDC25A. Biochem. Biophys. Res. Commun. 2019, 512, 392–398. [Google Scholar] [CrossRef]
- Kim, M.H.; Ham, O.; Lee, S.Y.; Choi, E.; Lee, C.Y.; Park, J.H.; Lee, J.; Seo, H.H.; Seung, M.; Choi, E.; et al. MicroRNA-365 inhibits the proliferation of vascular smooth muscle cells by targeting cyclin D1. J. Cell. Biochem. 2014, 115, 1752–1761. [Google Scholar] [CrossRef]
- Luo, Q.; Li, X.; Li, J.; Kong, X.; Zhang, J.; Chen, L.; Huang, Y.; Fang, L. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int. J. Oncol. 2013, 43, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008, 14, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.K.; Zhao, G.Y.; Tian, L.Y.; Liu, L.; Yan, K.; Ma, Y.L.; Ji, Z.W.; Li, X.X.; Han, K.; Gao, J.; et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol. Rep. 2012, 28, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Bandi, N.; Vassella, E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol. Cancer 2011, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef]
- Han, Z.; Li, Q.; Wang, Y.; Wang, L.; Li, X.; Ge, N.; Wang, Y.; Guo, C. Niclosamide Induces Cell Cycle Arrest in G1 Phase in Head and Neck Squamous Cell Carcinoma Through Let-7d/CDC34 Axis. Front. Pharmacol. 2018, 9, 1544. [Google Scholar] [CrossRef]
- Yin, J.; Hu, W.; Pan, L.; Fu, W.; Dai, L.; Jiang, Z.; Zhang, F.; Zhao, J. let-7 and miR-17 promote self-renewal and drive gefitinib resistance in non-small cell lung cancer. Oncol. Rep. 2019, 42, 495–508. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, S.; Zhang, X.; Wang, L.; Zhang, X.; Yan, B.; Zhao, J.; Yang, A.; Zhang, R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2014, 443, 1078–1084. [Google Scholar] [CrossRef]
- Sanchez, N.; Gallagher, M.; Lao, N.; Gallagher, C.; Clarke, C.; Doolan, P.; Aherne, S.; Blanco, A.; Meleady, P.; Clynes, M.; et al. MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including Skp2 and Psme3. PLoS ONE 2013, 8, e65671. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Cao, W.; Li, X.; Xie, C.; Geng, S.; Zhu, M.; Liang, Z.; Zhu, J.; Zhu, W.; et al. miR-19 targeting of PTEN mediates butyl benzyl phthalate-induced proliferation in both ER(+) and ER(-) breast cancer cells. Toxicol. Lett. 2018, 295, 124–133. [Google Scholar] [CrossRef]
- Hackl, M.; Brunner, S.; Fortschegger, K.; Schreiner, C.; Micutkova, L.; Mück, C.; Laschober, G.T.; Lepperdinger, G.; Sampson, N.; Berger, P.; et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 2010, 9, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, C.; Wang, M.; Li, Z.; Casimiro, M.C.; Liu, M.; Wu, K.; Whittle, J.; Ju, X.; Hyslop, T.; et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008, 182, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Fiorino, A.; Zoni, E.; Crippa, E.; Reid, J.F.; Gariboldi, M.; Pierotti, M.A. The Effects of miR-20a on p21: Two Mechanisms Blocking Growth Arrest in TGF-β-Responsive Colon Carcinoma. J. Cell. Physiol. 2015, 230, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007, 282, 2135–2143. [Google Scholar] [CrossRef]
- Qin, Y.E.; Duan, L.; He, Y.; Yuan, C.; Wang, T.; Yuan, D.; Zhang, C.; Liu, C. Saturated Fatty Acids Promote Hepatocytic Senecence through Regulation of miR-34a/Cyclin-Dependent Kinase 6. Mol. Nutr. Food Res. 2020, 64, e2000383. [Google Scholar] [CrossRef]
- Zhao, J.; Lammers, P.; Torrance, C.J.; Bader, A.G. TP53-independent function of miR-34a via HDAC1 and p21(CIP1/WAF1.). Mol. Ther. 2013, 21, 1678–1686. [Google Scholar] [CrossRef]
- Agirre, X.; Vilas-Zornoza, A.; Jiménez-Velasco, A.; Martin-Subero, J.I.; Cordeu, L.; Gárate, L.; San José-Eneriz, E.; Abizanda, G.; Rodríguez-Otero, P.; Fortes, P.; et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009, 69, 4443–4453. [Google Scholar] [CrossRef]
- Nakamachi, Y.; Kawano, S.; Takenokuchi, M.; Nishimura, K.; Sakai, Y.; Chin, T.; Saura, R.; Kurosaka, M.; Kumagai, S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009, 60, 1294–1304. [Google Scholar] [CrossRef]
- Kawano, S.; Nakamachi, Y. miR-124a as a key regulator of proliferation and MCP-1 secretion in synoviocytes from patients with rheumatoid arthritis. Ann. Rheum. Dis. 2011, 70 (Suppl. 1), i88–i91. [Google Scholar] [CrossRef]
- Kim, J.K.; Noh, J.H.; Jung, K.H.; Eun, J.W.; Bae, H.J.; Kim, M.G.; Chang, Y.G.; Shen, Q.; Park, W.S.; Lee, J.Y.; et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology 2013, 57, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, J.; Pan, T.; Zhou, J.; Gong, W.; Liu, N.; Fu, Z.; You, Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010, 1312, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Luo, J.; Cai, Q.; Pan, Q.; Zeng, H.; Guo, Z.; Dong, W.; Huang, J.; Lin, T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int. J. Cancer 2011, 128, 1758–1769. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-24 regulates CDKN1B/p27 expression in prostate cancer. Prostate 2016, 76, 637–648. [Google Scholar] [CrossRef]
- Lu, J.; He, M.L.; Wang, L.; Chen, Y.; Liu, X.; Dong, Q.; Chen, Y.C.; Peng, Y.; Yao, K.T.; Kung, H.F.; et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011, 71, 225–233. [Google Scholar] [CrossRef]
- Trohatou, O.; Zagoura, D.; Orfanos, N.K.; Pappa, K.I.; Marinos, E.; Anagnou, N.P.; Roubelakis, M.G. miR-26a Mediates Adipogenesis of Amniotic Fluid Mesenchymal Stem/Stromal Cells via PTEN, Cyclin E1, and CDK6. Stem Cells Dev. 2017, 26, 482–494. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, J.; Zhang, Y.; Yang, L.; Wang, J.; Ni, J.; Cui, D.; Yu, C.; Cai, Z. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol. Ther. 2011, 19, 1521–1528. [Google Scholar] [CrossRef]
- Zhou, X.; Xia, Y.; Li, L.; Zhang, G. MiR-101 inhibits cell growth and tumorigenesis of Helicobacter pylori related gastric cancer by repression of SOCS2. Cancer Biol. Ther. 2015, 16, 160–169. [Google Scholar] [CrossRef]
- Li, M.; Tian, L.; Ren, H.; Chen, X.; Wang, Y.; Ge, J.; Wu, S.; Sun, Y.; Liu, M.; Xiao, H. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J. Transl. Med. 2015, 13, 271. [Google Scholar] [CrossRef]
- Shang, A.; Lu, W.-Y.; Yang, M.; Zhou, C.; Zhang, H.; Cai, Z.-X.; Wang, W.-W.; Wang, W.-X.; Wu, G.-Q. miR-9 induces cell arrest and apoptosis of oral squamous cell carcinoma via CDK 4/6 pathway. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1754–1762. [Google Scholar] [CrossRef]
- O’Loghlen, A.; Brookes, S.; Martin, N.; Rapisarda, V.; Peters, G.; Gil, J. CBX7 and miR-9 are part of an autoregulatory loop controlling p16(INK) (4a). Aging Cell 2015, 14, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, J.; Tian, S.; Luo, J. miR-9 depletion suppresses the proliferation of osteosarcoma cells by targeting p16. Int. J. Oncol. 2019, 54, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Jun, G.J.; Zhong, G.G.; Ming, Z.S. miR-218 inhibits the proliferation of glioma U87 cells through the inactivation of the CDK6/cyclin D1/p21(Cip1/Waf1) pathway. Oncol. Lett. 2015, 9, 2743–2749. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Zeng, C.; Lu, X.; He, X.; Zhang, R.; Qiu, Q.; Zheng, G.; Jia, X.; Liu, H.; He, Z. miR-218 suppresses gastric cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis in a feedback loop. Cancer Lett. 2017, 403, 175–185. [Google Scholar] [CrossRef]
- Giannakakis, A.; Sandaltzopoulos, R.; Greshock, J.; Liang, S.; Huang, J.; Hasegawa, K.; Li, C.; O’Brien-Jenkins, A.; Katsaros, D.; Weber, B.L.; et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 2008, 7, 255–264. [Google Scholar] [CrossRef]
- Kang, H.; Yu, H.; Zeng, L.; Ma, H.; Cao, G. LncRNA Rian reduces cardiomyocyte pyroptosis and alleviates myocardial ischemia-reperfusion injury by regulating by the miR-17-5p/CCND1 axis. Hypertens. Res. 2022, 45, 976–989. [Google Scholar] [CrossRef]
- Wang, Q.; Han, J.; Xu, P.; Jian, X.; Huang, X.; Liu, D. Silencing of LncRNA SNHG16 Downregulates Cyclin D1 (CCND1) to Abrogate Malignant Phenotypes in Oral Squamous Cell Carcinoma (OSCC) Through Upregulating miR-17-5p. Cancer Manag. Res. 2021, 13, 1831–1841. [Google Scholar] [CrossRef]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, A.R.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef]
- Pickering, M.T.; Stadler, B.M.; Kowalik, T.F. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene 2009, 28, 140–145. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Trompeter, H.-I.; Abbad, H.; Iwaniuk, K.M.; Hafner, M.; Renwick, N.; Tuschl, T.; Schira, J.; Mueller, H.W.; Wernet, P. MicroRNAs MiR-17, MiR-20a, and MiR-106b Act in Concert to Modulate E2F Activity on Cell Cycle Arrest during Neuronal Lineage Differentiation of USSC. PLoS ONE 2011, 6, e16138. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhu, X.; Meehan, B.; Venneti, S.; Martinez, D.; Morin, G.; Maïga, R.I.; Chen, H.; Papadakis, A.I.; Johnson, R.M.; et al. SMARCB1 loss induces druggable cyclin D1 deficiency via upregulation of MIR17HG in atypical teratoid rhabdoid tumors. J. Pathol. 2020, 252, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.; Iwasaki, M.; Somervaille, T.C.; Ficara, F.; Carico, C.; Arnold, C.; Chen, C.Z.; Cleary, M.L. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010, 70, 3833–3842. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Li, Z.; Chen, P.; He, C.; Cao, D.; Elkahloun, A.; Lu, J.; Pelloso, L.A.; Wunderlich, M.; Huang, H.; et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 3710–3715. [Google Scholar] [CrossRef]
- Brockway, S.; Zeleznik-Le, N.J. WEE1 is a validated target of the microRNA miR-17-92 cluster in leukemia. Cancer Genet. 2015, 208, 279–287. [Google Scholar] [CrossRef]
- Tai, L.; Huang, C.J.; Choo, K.B.; Cheong, S.K.; Kamarul, T. Oxidative Stress Down-Regulates MiR-20b-5p, MiR-106a-5p and E2F1 Expression to Suppress the G1/S Transition of the Cell Cycle in Multipotent Stromal Cells. Int. J. Med. Sci. 2020, 17, 457–470. [Google Scholar] [CrossRef]
- Zhang, A.; Hao, J.; Wang, K.; Huang, Q.; Yu, K.; Kang, C.; Wang, G.; Jia, Z.; Han, L.; Pu, P. Down-regulation of miR-106b suppresses the growth of human glioma cells. J. Neuro-Oncol. 2013, 112, 179–189. [Google Scholar] [CrossRef]
- Kan, T.; Sato, F.; Ito, T.; Matsumura, N.; David, S.; Cheng, Y.; Agarwal, R.; Paun, B.C.; Jin, Z.; Olaru, A.V.; et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 2009, 136, 1689–1700. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Kroesen, B.J.; Koster, R.; Halsema, N.; de Jong, D.; de Jong, S.; Poppema, S.; Kluiver, J.; Diepstra, A.; van den Berg, A. MiR-17/106b seed family regulates p21 in Hodgkin’s lymphoma. J. Pathol. 2011, 225, 609–617. [Google Scholar] [CrossRef]
- Lee, K.H.; Chen, Y.L.; Yeh, S.D.; Hsiao, M.; Lin, J.T.; Goan, Y.G.; Lu, P.J. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009, 28, 3360–3370. [Google Scholar] [CrossRef]
- Guo, X.; Guo, L.; Ji, J.; Zhang, J.; Zhang, J.; Chen, X.; Cai, Q.; Li, J.; Gu, Q.; Liu, B.; et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 2010, 398, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lizé, M.; Pilarski, S.; Dobbelstein, M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010, 17, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Gu, X.; Li, Z.; Xiang, J.; Chen, Z. miR-449b inhibits the proliferation of SW1116 colon cancer stem cells through downregulation of CCND1 and E2F3 expression. Oncol. Rep. 2013, 30, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, B.; Wang, C.; Luo, Y.; Zhao, M.; Chen, P. Long noncoding RNA FOXD2-AS1 promotes glioma cell cycle progression and proliferation through the FOXD2-AS1/miR-31/CDK1 pathway. J. Cell. Biochem. 2019, 120, 19784–19795. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Quan, J.; Chen, F.; Pan, X.; Zhuang, C.; Xiong, T.; Zhuang, C.; Li, J.; Huang, X.; Ye, J.; et al. MiR-31-5p acts as a tumor suppressor in renal cell carcinoma by targeting cyclin-dependent kinase 1 (CDK1). Biomed. Pharmacother. 2019, 111, 517–526. [Google Scholar] [CrossRef]
- Malhas, A.; Saunders, N.J.; Vaux, D.J. The nuclear envelope can control gene expression and cell cycle progression via miRNA regulation. Cell Cycle 2010, 9, 531–539. [Google Scholar] [CrossRef]
- Zhu, W.; Yu, Y.; Fang, K.; Xiao, S.; Ni, L.; Yin, C.; Huang, X.; Wang, X.; Zhang, Y.; Le, H.B.; et al. miR-31/QKI-5 axis facilitates cell cycle progression of non-small-cell lung cancer cells by interacting and regulating p21 and CDK4/6 expressions. Cancer Med. 2023, 12, 4590–4604. [Google Scholar] [CrossRef]
- Chu, K.; Gao, G.; Yang, X.; Ren, S.; Li, Y.; Wu, H.; Huang, Y.; Zhou, C. miR-512-5p induces apoptosis and inhibits glycolysis by targeting p21 in non-small cell lung cancer cells. Int. J. Oncol. 2016, 48, 577–586. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Zeng, W.; He, J.; Mao, X. Upregulation of MircoRNA-370 Induces Proliferation in Human Prostate Cancer Cells by Downregulating the Transcription Factor FOXO1. PLoS ONE 2012, 7, e45825. [Google Scholar] [CrossRef]
- Jiping, Z.; Ming, F.; Lixiang, W.; Xiuming, L.; Yuqun, S.; Han, Y.; Zhifang, L.; Yundong, S.; Shili, L.; Chunyan, C.; et al. MicroRNA-212 inhibits proliferation of gastric cancer by directly repressing retinoblastoma binding protein 2. J. Cell Biochem. 2013, 114, 2666–2672. [Google Scholar] [CrossRef]
- Zhao, J.L.; Zhang, L.; Guo, X.; Wang, J.H.; Zhou, W.; Liu, M.; Li, X.; Tang, H. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life 2015, 67, 380–394. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Jin, F.Y.; Xia, R.; Kong, R.; Li, J.H.; Xu, T.P.; Liu, Y.W.; Zhang, E.B.; Liu, X.H.; De, W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 2014, 14, 319. [Google Scholar] [CrossRef]
- Delgir, S.; Ilkhani, K.; Safi, A.; Rahmati, Y.; Montazari, V.; Zaynali-Khasraghi, Z.; Seif, F.; Bastami, M.; Alivand, M.R. The expression of miR-513c and miR-3163 was downregulated in tumor tissues compared with normal adjacent tissue of patients with breast cancer. BMC Med. Genom. 2021, 14, 180. [Google Scholar] [CrossRef]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar] [CrossRef]
- Yamashita, R.; Sato, M.; Kakumu, T.; Hase, T.; Yogo, N.; Maruyama, E.; Sekido, Y.; Kondo, M.; Hasegawa, Y. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015, 4, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, W.; Wang, Y. MiR-222 regulates the progression of oral squamous cell carcinoma by targeting CDKN1B. Am. J. Transl. Res. 2022, 14, 5215–5227. [Google Scholar] [PubMed]
- Liao, L.; Chen, J.; Zhang, C.; Guo, Y.; Liu, W.; Liu, W.; Duan, L.; Liu, Z.; Hu, J.; Lu, J. LncRNA NEAT1 Promotes High Glucose-Induced Mesangial Cell Hypertrophy by Targeting miR-222-3p/CDKN1B Axis. Front. Mol. Biosci. 2020, 7, 627827. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Y.; Yang, Y.; Liu, J.; Song, Y.; Cao, Y.; Chen, X.; Yang, W.; Wang, F.; Gao, J.; et al. MicroRNA-222 Controls Human Pancreatic Cancer Cell Line Capan-2 Proliferation by P57 Targeting. J. Cancer 2015, 6, 1230–1235. [Google Scholar] [CrossRef]
- Im, W.R.; Lee, H.-S.; Lee, Y.-S.; Lee, J.-S.; Jang, H.-J.; Kim, S.-Y.; Park, J.-L.; Lee, Y.; Kim, M.S.; Lee, J.M.; et al. A Regulatory Noncoding RNA, nc886, Suppresses Esophageal Cancer by Inhibiting the AKT Pathway and Cell Cycle Progression. Cells 2020, 9, 801. [Google Scholar] [CrossRef]
- Fort, R.S.; Mathó, C.; Geraldo, M.V.; Ottati, M.C.; Yamashita, A.S.; Saito, K.C.; Leite, K.R.M.; Méndez, M.; Maedo, N.; Méndez, L.; et al. Nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth. BMC Cancer 2018, 18, 127. [Google Scholar] [CrossRef]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Zhang, W.; Wang, X.; Shi, Y.; Yang, F.; Xie, H.; Zhou, W.; Wang, S.; Guan, X. c-myc regulates the sensitivity of breast cancer cells to palbociclib via c-myc/miR-29b-3p/CDK6 axis. Cell Death Dis. 2020, 11, 760. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.Y.; Wei, M.; Liu, Y.Y.; Di, Z.L.; Li, S.Z. miR-497/MIR497HG inhibits glioma cell proliferation by targeting CCNE1 and the miR-588/TUSC1 axis. Oncol. Rep. 2021, 46, 255. [Google Scholar] [CrossRef] [PubMed]
- Hoareau-Aveilla, C.; Quelen, C.; Congras, A.; Caillet, N.; Labourdette, D.; Dozier, C.; Brousset, P.; Lamant, L.; Meggetto, F. miR-497 suppresses cycle progression through an axis involving CDK6 in ALK-positive cells. Haematologica 2019, 104, 347–359. [Google Scholar] [CrossRef]
- Chen, J.; Feilotter, H.E.; Paré, G.C.; Zhang, X.; Pemberton, J.G.; Garady, C.; Lai, D.; Yang, X.; Tron, V.A. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am. J. Pathol. 2010, 176, 2520–2529. [Google Scholar] [CrossRef]
- Andrikopoulou, A.; Shalit, A.; Zografos, E.; Koutsoukos, K.; Korakiti, A.-M.; Liontos, M.; Dimopoulos, M.-A.; Zagouri, F. MicroRNAs as Potential Predictors of Response to CDK4/6 Inhibitor Treatment. Cancers 2021, 13, 4114. [Google Scholar] [CrossRef]
- Gao, J.; Ma, S.; Yang, F.; Chen, X.; Wang, W.; Zhang, J.; Li, Y.; Wang, T.; Shan, L. miR-193b exhibits mutual interaction with MYC, and suppresses growth and metastasis of osteosarcoma. Oncol. Rep. 2020, 44, 139–155. [Google Scholar] [CrossRef]
- Kaukoniemi, K.M.; Rauhala, H.E.; Scaravilli, M.; Latonen, L.; Annala, M.; Vessella, R.L.; Nykter, M.; Tammela, T.L.; Visakorpi, T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015, 4, 1417–1425. [Google Scholar] [CrossRef]
- Bustos, M.A.; Ono, S.; Marzese, D.M.; Oyama, T.; Iida, Y.; Cheung, G.; Nelson, N.; Hsu, S.C.; Yu, Q.; Hoon, D.S.B. MiR-200a Regulates CDK4/6 Inhibitor Effect by Targeting CDK6 in Metastatic Melanoma. J. Invest. Dermatol. 2017, 137, 1955–1964. [Google Scholar] [CrossRef]
- Georgantas, R.W., 3rd; Streicher, K.; Luo, X.; Greenlees, L.; Zhu, W.; Liu, Z.; Brohawn, P.; Morehouse, C.; Higgs, B.W.; Richman, L.; et al. MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment. Cell Melanoma Res. 2014, 27, 275–286. [Google Scholar] [CrossRef]
- Dahiya, N.; Sherman-Baust, C.A.; Wang, T.L.; Davidson, B.; Shih Ie, M.; Zhang, Y.; Wood, W., 3rd; Becker, K.G.; Morin, P.J. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE 2008, 3, e2436. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yao, Q.; Hou, Y.; Xu, M.; Liu, S.; Yang, L.; Zhang, L.; Xu, H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed. Pharmacother. 2013, 67, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Fabris, L.; Lovat, F.; Berton, S.; Segatto, I.; D’Andrea, S.; Ivan, C.; Cascione, L.; Calin, G.A.; Croce, C.M.; et al. Contact inhibition modulates intracellular levels of miR-223 in a p27kip1-dependent manner. Oncotarget 2014, 5, 1185–1197. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Q.; Yang, P.; Long, G. Restoration of microRNA-212 causes a G0/G1 cell cycle arrest and apoptosis in adult T-cell leukemia/lymphoma cells by repressing CCND3 expression. J. Investig. Med. 2017, 65, 82–87. [Google Scholar] [CrossRef]
- Guo, S.L.; Ye, H.; Teng, Y.; Wang, Y.L.; Yang, G.; Li, X.B.; Zhang, C.; Yang, X.; Yang, Z.Z.; Yang, X. Akt-p53-miR-365-cyclin D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN deficiency. Nat. Commun. 2013, 4, 2544. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fu, H.; Sun, F.; Zhang, H.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008, 36, 5391–5404. [Google Scholar] [CrossRef] [PubMed]
- Linsley, P.S.; Schelter, J.; Burchard, J.; Kibukawa, M.; Martin, M.M.; Bartz, S.R.; Johnson, J.M.; Cummins, J.M.; Raymond, C.K.; Dai, H.; et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 2007, 27, 2240–2252. [Google Scholar] [CrossRef]
- Qin, X.; Wang, X.; Wang, Y.; Tang, Z.; Cui, Q.; Xi, J.; Li, Y.S.; Chien, S.; Wang, N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. USA 2010, 107, 3240–3244. [Google Scholar] [CrossRef]
- Sun, F.; Fu, H.; Liu, Q.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008, 582, 1564–1568. [Google Scholar] [CrossRef]
- Zheng, S.Z.; Sun, P.; Wang, J.P.; Liu, Y.; Gong, W.; Liu, J. MiR-34a overexpression enhances the inhibitory effect of doxorubicin on HepG2 cells. World J. Gastroenterol. 2019, 25, 2752–2762. [Google Scholar] [CrossRef]
- Yang, J.; Song, Q.; Cai, Y.; Wang, P.; Wang, M.; Zhang, D. RLIP76-dependent suppression of PI3K/AKT/Bcl-2 pathway by miR-101 induces apoptosis in prostate cancer. Biochem. Biophys. Res. Commun. 2015, 463, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Dai, Z.; Ma, Y.; Wang, Z.; Liu, X.; Wang, X. MicroRNA-101 inhibits cell proliferation and induces apoptosis by targeting EYA1 in breast cancer. Int. J. Mol. Med. 2016, 37, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xie, D.; Zhang, H. MicroRNA-101-3p advances cisplatin sensitivity in bladder urothelial carcinoma through targeted silencing EZH2. J. Cancer 2019, 10, 2628–2634. [Google Scholar] [CrossRef] [PubMed]
- Aguda, B.D.; Kim, Y.; Piper-Hunter, M.G.; Friedman, A.; Marsh, C.B. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. USA 2008, 105, 19678–19683. [Google Scholar] [CrossRef]
- Bueno, M.J.; Gómez de Cedrón, M.; Laresgoiti, U.; Fernández-Piqueras, J.; Zubiaga, A.M.; Malumbres, M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol. Cell Biol. 2010, 30, 2983–2995. [Google Scholar] [CrossRef]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors. Int. J. Mol. Sci. 2021, 22, 543. [Google Scholar] [CrossRef]
- Yang, X.; Feng, M.; Jiang, X.; Wu, Z.; Li, Z.; Aau, M.; Yu, Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes. Dev. 2009, 23, 2388–2393. [Google Scholar] [CrossRef]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar] [CrossRef]
- Hydbring, P.; Wang, Y.; Fassl, A.; Li, X.; Matia, V.; Otto, T.; Choi, Y.J.; Sweeney, K.E.; Suski, J.M.; Yin, H.; et al. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer Cell 2017, 31, 576–590.e578. [Google Scholar] [CrossRef]
- Glover, D.M.; Hagan, I.M.; Tavares, A.A. Polo-like kinases: A team that plays throughout mitosis. Genes. Dev. 1998, 12, 3777–3787. [Google Scholar] [CrossRef]
- Shi, W.; Alajez, N.M.; Bastianutto, C.; Hui, A.B.; Mocanu, J.D.; Ito, E.; Busson, P.; Lo, K.W.; Ng, R.; Waldron, J.; et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int. J. Cancer 2010, 126, 2036–2048. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yu, J.; Han, T.S.; Park, S.Y.; Namkoong, B.; Kim, D.H.; Hur, K.; Yoo, M.W.; Lee, H.J.; Yang, H.K.; et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009, 37, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Ivanovska, I.; Ball, A.S.; Diaz, R.L.; Magnus, J.F.; Kibukawa, M.; Schelter, J.M.; Kobayashi, S.V.; Lim, L.; Burchard, J.; Jackson, A.L.; et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell Biol. 2008, 28, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Kim, H.H.; Abdelmohsen, K.; Kuwano, Y.; Pullmann, R., Jr.; Srikantan, S.; Subrahmanyam, R.; Martindale, J.L.; Yang, X.; Ahmed, F.; et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE 2008, 3, e1864. [Google Scholar] [CrossRef] [PubMed]
- Giglio, S.; Cirombella, R.; Amodeo, R.; Portaro, L.; Lavra, L.; Vecchione, A. MicroRNA miR-24 promotes cell proliferation by targeting the CDKs inhibitors p27Kip1 and p16INK4a. J. Cell Physiol. 2013, 228, 2015–2023. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Z.; Ge, Q.; Hu, J.; Li, F.; Hu, J.; Xu, H.; Ye, Z.; Li, L.C. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett. 2014, 588, 4654–4664. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zhang, Y.; Li, J. Upregulation of MiR-196a promotes cell proliferation by downregulating p27(kip1) in laryngeal cancer. Biol. Res. 2016, 49, 40. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Russo, L.; Pallante, P.; De Martino, I.; Ferraro, A.; Leone, V.; Borbone, E.; Petrocca, F.; Alder, H.; Croce, C.M.; et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer 2007, 14, 791–798. [Google Scholar] [CrossRef]
- Yu, Z.; Willmarth, N.E.; Zhou, J.; Katiyar, S.; Wang, M.; Liu, Y.; McCue, P.A.; Quong, A.A.; Lisanti, M.P.; Pestell, R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8231–8236. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, L.; Wang, C.; Ju, X.; Wang, M.; Chen, K.; Loro, E.; Li, Z.; Zhang, Y.; Wu, K.; et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat. Commun. 2013, 4, 2812. [Google Scholar] [CrossRef]
- Mughal, M.J.; Bhadresha, K.; Kwok, H.F. CDK inhibitors from past to present: A new wave of cancer therapy. Semin. Cancer Biol. 2023, 88, 106–122. [Google Scholar] [CrossRef]
- Finn, R.S.; Martin, M.; Rugo, H.S.; Jones, S.; Im, S.A.; Gelmon, K.; Harbeck, N.; Lipatov, O.N.; Walshe, J.M.; Moulder, S.; et al. Palbociclib and Letrozole in Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Turner, N.C.; Bondarenko, I.; Ro, J.; Im, S.A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S.; et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016, 17, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Blackwell, K.L.; André, F.; Winer, E.P.; et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Duso, B.A.; Trapani, D.; Viale, G.; Criscitiello, C.; D’Amico, P.; Belli, C.; Mazzarella, L.; Locatelli, M.; Minchella, I.; Curigliano, G. Clinical efficacy of ribociclib as a first-line therapy for HR-positive, advanced breast cancer. Expert. Opin. Pharmacother. 2018, 19, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Sledge, G.W., Jr.; Toi, M.; Neven, P.; Sohn, J.; Inoue, K.; Pivot, X.; Burdaeva, O.; Okera, M.; Masuda, N.; Kaufman, P.A.; et al. MONARCH 2: Abemaciclib in Combination With Fulvestrant in Women With HR+/HER2- Advanced Breast Cancer Who Had Progressed While Receiving Endocrine Therapy. J. Clin. Oncol. 2017, 35, 2875–2884. [Google Scholar] [CrossRef]
- Rader, J.; Russell, M.R.; Hart, L.S.; Nakazawa, M.S.; Belcastro, L.T.; Martinez, D.; Li, Y.; Carpenter, E.L.; Attiyeh, E.F.; Diskin, S.J.; et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin. Cancer Res. 2013, 19, 6173–6182. [Google Scholar] [CrossRef]
- Puyol, M.; Martín, A.; Dubus, P.; Mulero, F.; Pizcueta, P.; Khan, G.; Guerra, C.; Santamaría, D.; Barbacid, M. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell 2010, 18, 63–73. [Google Scholar] [CrossRef]
- Finn, R.S.; Dering, J.; Conklin, D.; Kalous, O.; Cohen, D.J.; Desai, A.J.; Ginther, C.; Atefi, M.; Chen, I.; Fowst, C.; et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009, 11, R77. [Google Scholar] [CrossRef]
- Lu, J. Palbociclib: A first-in-class CDK4/CDK6 inhibitor for the treatment of hormone-receptor positive advanced breast cancer. J. Hematol. Oncol. 2015, 8, 98. [Google Scholar] [CrossRef]
- Fry, D.W.; Harvey, P.J.; Keller, P.R.; Elliott, W.L.; Meade, M.; Trachet, E.; Albassam, M.; Zheng, X.; Leopold, W.R.; Pryer, N.K.; et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004, 3, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, S.L.; Harris, P.S.; Venkataraman, S.; Alimova, I.; Birks, D.K.; Donson, A.M.; Foreman, N.K.; Vibhakar, R. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J. Neurooncol. 2013, 111, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Rascon, K.; Flajc, G.; De Angelis, C.; Liu, X.; Trivedi, M.V.; Ekinci, E. Ribociclib in HR+/HER2− Advanced or Metastatic Breast Cancer Patients. Ann. Pharmacother. 2019, 53, 501–509. [Google Scholar] [CrossRef]
- Jansen, V.M.; Bhola, N.E.; Bauer, J.A.; Formisano, L.; Lee, K.M.; Hutchinson, K.E.; Witkiewicz, A.K.; Moore, P.D.; Estrada, M.V.; Sánchez, V.; et al. Kinome-Wide RNA Interference Screen Reveals a Role for PDK1 in Acquired Resistance to CDK4/6 Inhibition in ER-Positive Breast Cancer. Cancer Res. 2017, 77, 2488–2499. [Google Scholar] [CrossRef]
- Wu, T.; Chen, Z.; To, K.K.W.; Fang, X.; Wang, F.; Cheng, B.; Fu, L. Effect of abemaciclib (LY2835219) on enhancement of chemotherapeutic agents in ABCB1 and ABCG2 overexpressing cells in vitro and in vivo. Biochem. Pharmacol. 2017, 124, 29–42. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Petrakova, K.; Blackwell, K.L.; Winer, E.P.; et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann. Oncol. 2018, 29, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Toi, M.; Campone, M.; Sohn, J.; Paluch-Shimon, S.; Huober, J.; Park, I.H.; Trédan, O.; Chen, S.C.; Manso, L.; et al. MONARCH 3: Abemaciclib As Initial Therapy for Advanced Breast Cancer. J. Clin. Oncol. 2017, 35, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.; Martin, M.; Di Leo, A.; Im, S.A.; Awada, A.; Forrester, T.; Frenzel, M.; Hardebeck, M.C.; Cox, J.; Barriga, S.; et al. MONARCH 3 final PFS: A randomized study of abemaciclib as initial therapy for advanced breast cancer. NPJ Breast Cancer 2019, 5, 5. [Google Scholar] [CrossRef]
- Gao, J.J.; Cheng, J.; Bloomquist, E.; Sanchez, J.; Wedam, S.B.; Singh, H.; Amiri-Kordestani, L.; Ibrahim, A.; Sridhara, R.; Goldberg, K.B.; et al. CDK4/6 inhibitor treatment for patients with hormone receptor-positive, HER2-negative, advanced or metastatic breast cancer: A US Food and Drug Administration pooled analysis. Lancet Oncol. 2020, 21, 250–260. [Google Scholar] [CrossRef]
- Gao, J.J.; Cheng, J.; Prowell, T.M.; Bloomquist, E.; Tang, S.; Wedam, S.B.; Royce, M.; Krol, D.; Osgood, C.; Ison, G.; et al. Overall survival in patients with hormone receptor-positive, HER2-negative, advanced or metastatic breast cancer treated with a cyclin-dependent kinase 4/6 inhibitor plus fulvestrant: A US Food and Drug Administration pooled analysis. Lancet Oncol. 2021, 22, 1573–1581. [Google Scholar] [CrossRef]
- Bolzacchini, E.; Pomero, F.; Fazio, M.; Civitelli, C.; Fabro, G.; Pellegrino, D.; Giordano, M.; Squizzato, A. Risk of venous and arterial thromboembolic events in women with advanced breast cancer treated with CDK 4/6 inhibitors: A systematic review and meta-analysis. Thromb. Res. 2021, 208, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; An, H.J.; Kim, S.K.; Lee, S.A.; Kim, S.; Lim, S.M.; Kim, G.M.; Sohn, J.; Moon, Y.W. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int. J. Cancer 2019, 145, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.; Migliaccio, I.; Bonechi, M.; Biagioni, C.; Romagnoli, D.; De Luca, F.; Galardi, F.; Risi, E.; De Santo, I.; Benelli, M.; et al. Mechanisms of Resistance to CDK4/6 Inhibitors: Potential Implications and Biomarkers for Clinical Practice. Front. Oncol. 2019, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.Y.; Shu, S.; Kwon, M.; Jovanović, B.; Murphy, K.; Gulvady, A.; Fassl, A.; Trinh, A.; Kuang, Y.; Heavey, G.A.; et al. Acquired resistance to combined BET and CDK4/6 inhibition in triple-negative breast cancer. Nat. Commun. 2020, 11, 2350. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, B.; Guo, J.; Shao, H.; Del Priore, I.S.; Chang, Q.; Kudo, R.; Li, Z.; Razavi, P.; Liu, B.; et al. INK4 Tumor Suppressor Proteins Mediate Resistance to CDK4/6 Kinase Inhibitors. Cancer Discov. 2022, 12, 356–371. [Google Scholar] [CrossRef]
- Ismail, A.; Bandla, S.; Reveiller, M.; Toia, L.; Zhou, Z.; Gooding, W.E.; Kalatskaya, I.; Stein, L.; D’Souza, M.; Litle, V.R.; et al. Early G₁ cyclin-dependent kinases as prognostic markers and potential therapeutic targets in esophageal adenocarcinoma. Clin. Cancer Res. 2011, 17, 4513–4522. [Google Scholar] [CrossRef]
- Dean, J.L.; McClendon, A.K.; Hickey, T.E.; Butler, L.M.; Tilley, W.D.; Witkiewicz, A.K.; Knudsen, E.S. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle 2012, 11, 2756–2761. [Google Scholar] [CrossRef]
- Dean, J.L.; Thangavel, C.; McClendon, A.K.; Reed, C.A.; Knudsen, E.S. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene 2010, 29, 4018–4032. [Google Scholar] [CrossRef]
- Heilmann, A.M.; Perera, R.M.; Ecker, V.; Nicolay, B.N.; Bardeesy, N.; Benes, C.H.; Dyson, N.J. CDK4/6 and IGF1 receptor inhibitors synergize to suppress the growth of p16INK4A-deficient pancreatic cancers. Cancer Res. 2014, 74, 3947–3958. [Google Scholar] [CrossRef]
- Condorelli, R.; Spring, L.; O’Shaughnessy, J.; Lacroix, L.; Bailleux, C.; Scott, V.; Dubois, J.; Nagy, R.J.; Lanman, R.B.; Iafrate, A.J.; et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann. Oncol. 2018, 29, 640–645. [Google Scholar] [CrossRef]
- Chandarlapaty, S.; Razavi, P. Cyclin E mRNA: Assessing Cyclin-Dependent Kinase (CDK) Activation State to Elucidate Breast Cancer Resistance to CDK4/6 Inhibitors. J. Clin. Oncol. 2019, 37, 1148–1150. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Bhatt, T.; Dickler, M.; Giri, D.; Scaltriti, M.; Baselga, J.; Rosen, N.; Chandarlapaty, S. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017, 36, 2255–2264. [Google Scholar] [CrossRef] [PubMed]
- Min, A.; Kim, J.E.; Kim, Y.J.; Lim, J.M.; Kim, S.; Kim, J.W.; Lee, K.H.; Kim, T.Y.; Oh, D.Y.; Bang, Y.J.; et al. Cyclin E overexpression confers resistance to the CDK4/6 specific inhibitor palbociclib in gastric cancer cells. Cancer Lett. 2018, 430, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Al-Qasem, A.J.; Alves, C.L.; Ehmsen, S.; Tuttolomondo, M.; Terp, M.G.; Johansen, L.E.; Vever, H.; Hoeg, L.V.A.; Elias, D.; Bak, M.; et al. Co-targeting CDK2 and CDK4/6 overcomes resistance to aromatase and CDK4/6 inhibitors in ER+ breast cancer. NPJ Precis. Oncol. 2022, 6, 68. [Google Scholar] [CrossRef]
- Patnaik, A.; Rosen, L.S.; Tolaney, S.M.; Tolcher, A.W.; Goldman, J.W.; Gandhi, L.; Papadopoulos, K.P.; Beeram, M.; Rasco, D.W.; Hilton, J.F.; et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov. 2016, 6, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Lorusso, P.M.; Demichele, A.; Abramson, V.G.; Courtney, R.; Randolph, S.S.; Shaik, M.N.; Wilner, K.D.; O’Dwyer, P.J.; Schwartz, G.K. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin. Cancer Res. 2012, 18, 568–576. [Google Scholar] [CrossRef]
- Nourbakhsh, M.; Hauser, H. Constitutive silencing of IFN-beta promoter is mediated by NRF (NF-kappaB-repressing factor), a nuclear inhibitor of NF-kappaB. EMBO J. 1999, 18, 6415–6425. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Herrera-Abreu, M.T.; Palafox, M.; Asghar, U.; Rivas, M.A.; Cutts, R.J.; Garcia-Murillas, I.; Pearson, A.; Guzman, M.; Rodriguez, O.; Grueso, J.; et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016, 76, 2301–2313. [Google Scholar] [CrossRef]
- Kruer, T.L.; Dougherty, S.M.; Reynolds, L.; Long, E.; de Silva, T.; Lockwood, W.W.; Clem, B.F. Expression of the lncRNA Maternally Expressed Gene 3 (MEG3) Contributes to the Control of Lung Cancer Cell Proliferation by the Rb Pathway. PLoS ONE 2016, 11, e0166363. [Google Scholar] [CrossRef]
- Yu, Y.; Liao, H.; Xie, R.; Zhang, Y.; Zheng, R.; Chen, J.; Zhang, B. Overexpression of miRNA-3613-3p Enhances the Sensitivity of Triple Negative Breast Cancer to CDK4/6 Inhibitor Palbociclib. Front. Oncol. 2020, 10, 590813. [Google Scholar] [CrossRef] [PubMed]
- Citron, F.; Segatto, I.; Vinciguerra, G.L.R.; Musco, L.; Russo, F.; Mungo, G.; D’Andrea, S.; Mattevi, M.C.; Perin, T.; Schiappacassi, M.; et al. Downregulation of miR-223 Expression Is an Early Event during Mammary Transformation and Confers Resistance to CDK4/6 Inhibitors in Luminal Breast Cancer. Cancer Res. 2020, 80, 1064–1077. [Google Scholar] [CrossRef] [PubMed]
- Baldassari, F.; Zerbinati, C.; Galasso, M.; Corrà, F.; Minotti, L.; Agnoletto, C.; Previati, M.; Croce, C.M.; Volinia, S. Screen for MicroRNA and Drug Interactions in Breast Cancer Cell Lines Points to miR-126 as a Modulator of CDK4/6 and PIK3CA Inhibitors. Front. Genet. 2018, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Cornell, L.; Wander, S.A.; Visal, T.; Wagle, N.; Shapiro, G.I. MicroRNA-Mediated Suppression of the TGF-β Pathway Confers Transmissible and Reversible CDK4/6 Inhibitor Resistance. Cell Rep. 2019, 26, 2667–2680.e2667. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Qiu, M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem. Biophys. Res. Commun. 2018, 495, 1594–1600. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.X.; Zhang, M.Y.; Liu, K.; Liu, J.; Zhang, Z.L.; Fu, L. LncRNA SNHG15 promotes proliferation and migration of lung cancer via targeting microRNA-211-3p. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6838–6844. [Google Scholar] [CrossRef]
- Zhang, J.H.; Wei, H.W.; Yang, H.G. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1720–1724. [Google Scholar]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef]
- Lee, Y.; Liu, J.; Patel, S.; Cloughesy, T.; Lai, A.; Farooqi, H.; Seligson, D.; Dong, J.; Liau, L.; Becker, D.; et al. Genomic landscape of meningiomas. Brain Pathol. 2010, 20, 751–762. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef]
- Lu, K.H.; Li, W.; Liu, X.H.; Sun, M.; Zhang, M.L.; Wu, W.Q.; Xie, W.P.; Hou, Y.Y. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013, 13, 461. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ren, Z.; Sun, P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 2012, 113, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010, 70, 2350–2358. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef]
- Liu, J.; Wan, L.; Lu, K.; Sun, M.; Pan, X.; Zhang, P.; Lu, B.; Liu, G.; Wang, Z. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PLoS ONE 2015, 10, e0114586. [Google Scholar] [CrossRef]
- Huang, Q.; Shen, Y.J.; Hsueh, C.Y.; Guo, Y.; Zhang, Y.F.; Li, J.Y.; Zhou, L. miR-17-5p drives G2/M-phase accumulation by directly targeting CCNG2 and is related to recurrence of head and neck squamous cell carcinoma. BMC Cancer 2021, 21, 1074. [Google Scholar] [CrossRef]
- Lee, S.; Schmitt, C.A. The dynamic nature of senescence in cancer. Nat. Cell. Biol. 2019, 21, 94–101. [Google Scholar] [CrossRef]
- Dang, F.; Nie, L.; Wei, W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell. Death Differ. 2021, 28, 427–438. [Google Scholar] [CrossRef]
- Diaz-Moralli, S.; Tarrado-Castellarnau, M.; Miranda, A.; Cascante, M. Targeting cell cycle regulation in cancer therapy. Pharmacol. Ther. 2013, 138, 255–271. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, K. MiR-369-3p inhibits tumorigenesis of hepatocellular carcinoma by binding to PAX6. J. Biol. Regul. Homeost. Agents 2020, 34, 917–926. [Google Scholar] [CrossRef]
- Geng, Z.; Chen, H.; Zou, G.; Yuan, L.; Liu, P.; Li, B.; Zhang, K.; Jing, F.; Nie, X.; Liu, T.; et al. Human Amniotic Fluid Mesenchymal Stem Cell-Derived Exosomes Inhibit Apoptosis in Ovarian Granulosa Cell via miR-369-3p/YAF2/PDCD5/p53 Pathway. Oxid. Med. Cell Longev. 2022, 2022, 3695848. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Reiter, L.; Picotti, P.; Lange, V.; Bogan, E.; Hurschler, B.A.; Blenkiron, C.; Lehrbach, N.J.; Ding, X.C.; Weiss, M.; et al. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat. Methods 2010, 7, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Chang, E.J.; Drapkin, B.J.; Cross, F.R.; Chait, B.T.; Rout, M.P. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 2004, 14, 699–711. [Google Scholar] [CrossRef]
- Yu, R.; Hu, Y.; Zhang, S.; Li, X.; Tang, M.; Yang, M.; Wu, X.; Li, Z.; Liao, X.; Xu, Y.; et al. LncRNA CTBP1-DT-encoded microprotein DDUP sustains DNA damage response signalling to trigger dual DNA repair mechanisms. Nucleic Acids Res. 2022, 50, 8060–8079. [Google Scholar] [CrossRef]
- Taheri, M.; Shoorei, H.; Tondro Anamag, F.; Ghafouri-Fard, S.; Dinger, M.E. LncRNAs and miRNAs participate in determination of sensitivity of cancer cells to cisplatin. Exp. Mol. Pathol. 2021, 123, 104602. [Google Scholar] [CrossRef]
- Wen, D.; Peng, Y.; Lin, F.; Singh, R.K.; Mahato, R.I. Micellar Delivery of miR-34a Modulator Rubone and Paclitaxel in Resistant Prostate Cancer. Cancer Res. 2017, 77, 3244–3254. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 47. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Bencivenga, D.; Stampone, E.; Vastante, A.; Barahmeh, M.; Della Ragione, F.; Borriello, A. An Unanticipated Modulation of Cyclin-Dependent Kinase Inhibitors: The Role of Long Non-Coding RNAs. Cells 2022, 11, 1346. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G. miR-34—A microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, 120. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Rui, M.; Qu, Y.; Gao, T.; Ge, Y.; Feng, C.; Xu, X. Simultaneous delivery of anti-miR21 with doxorubicin prodrug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int. J. Nanomed. 2017, 12, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
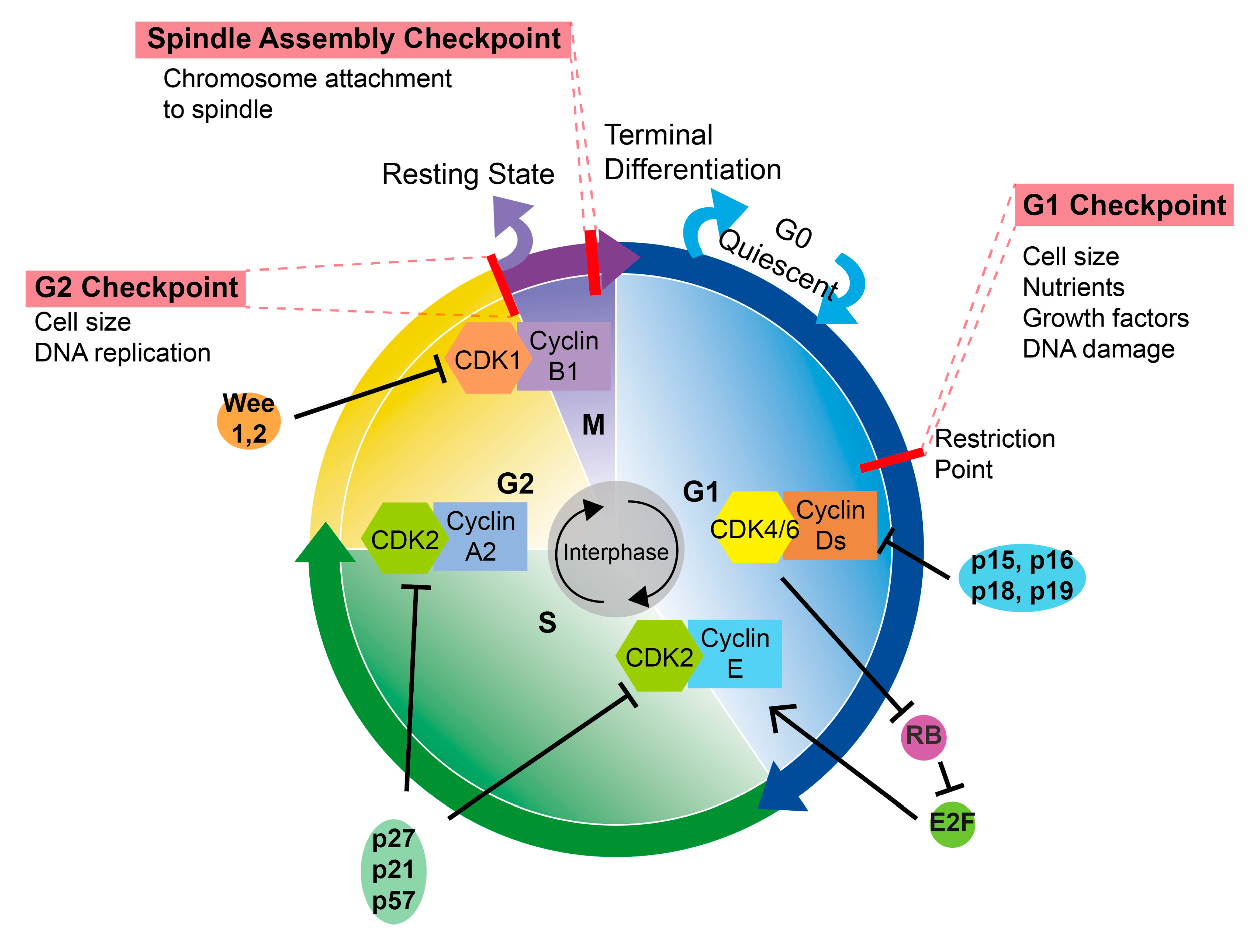
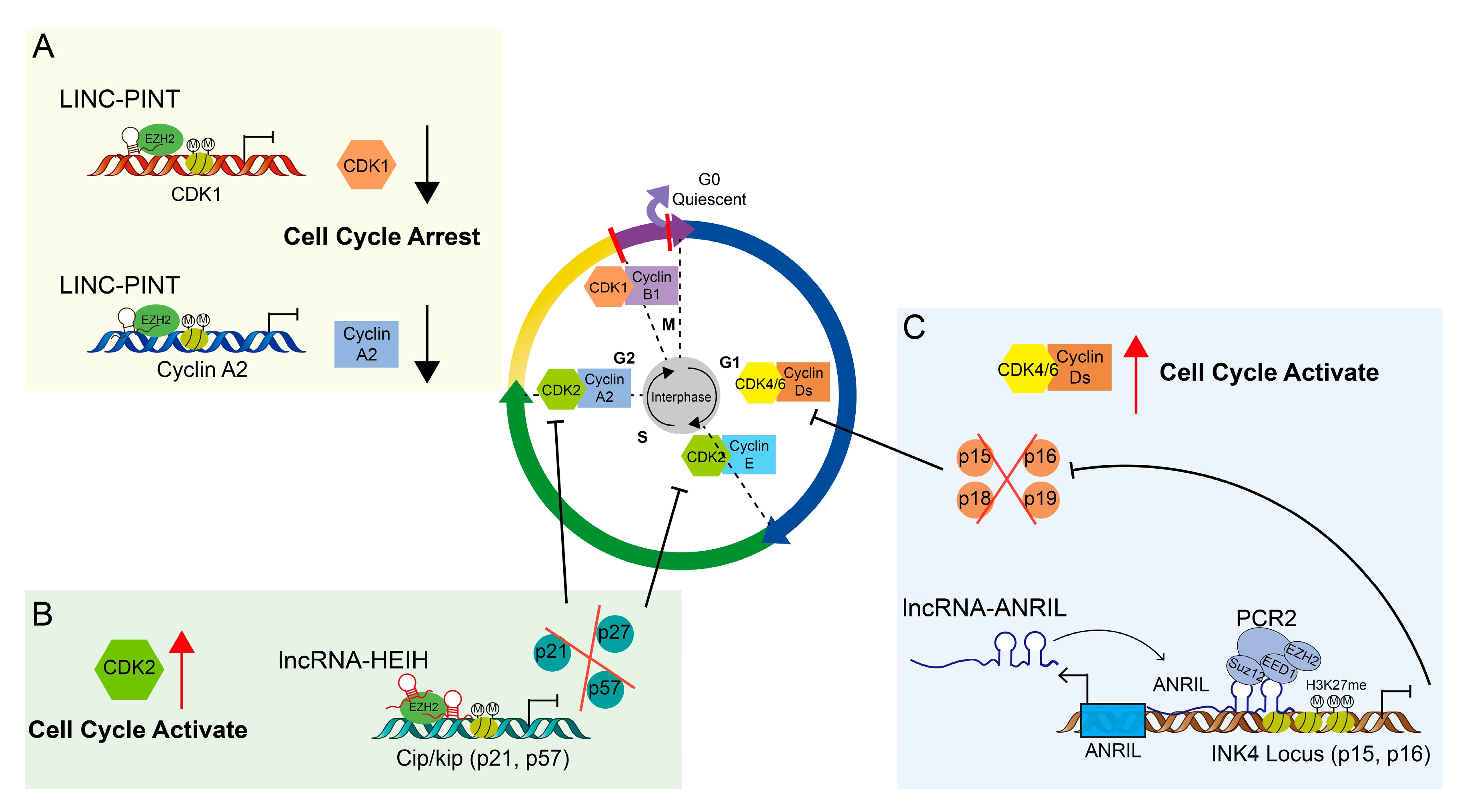
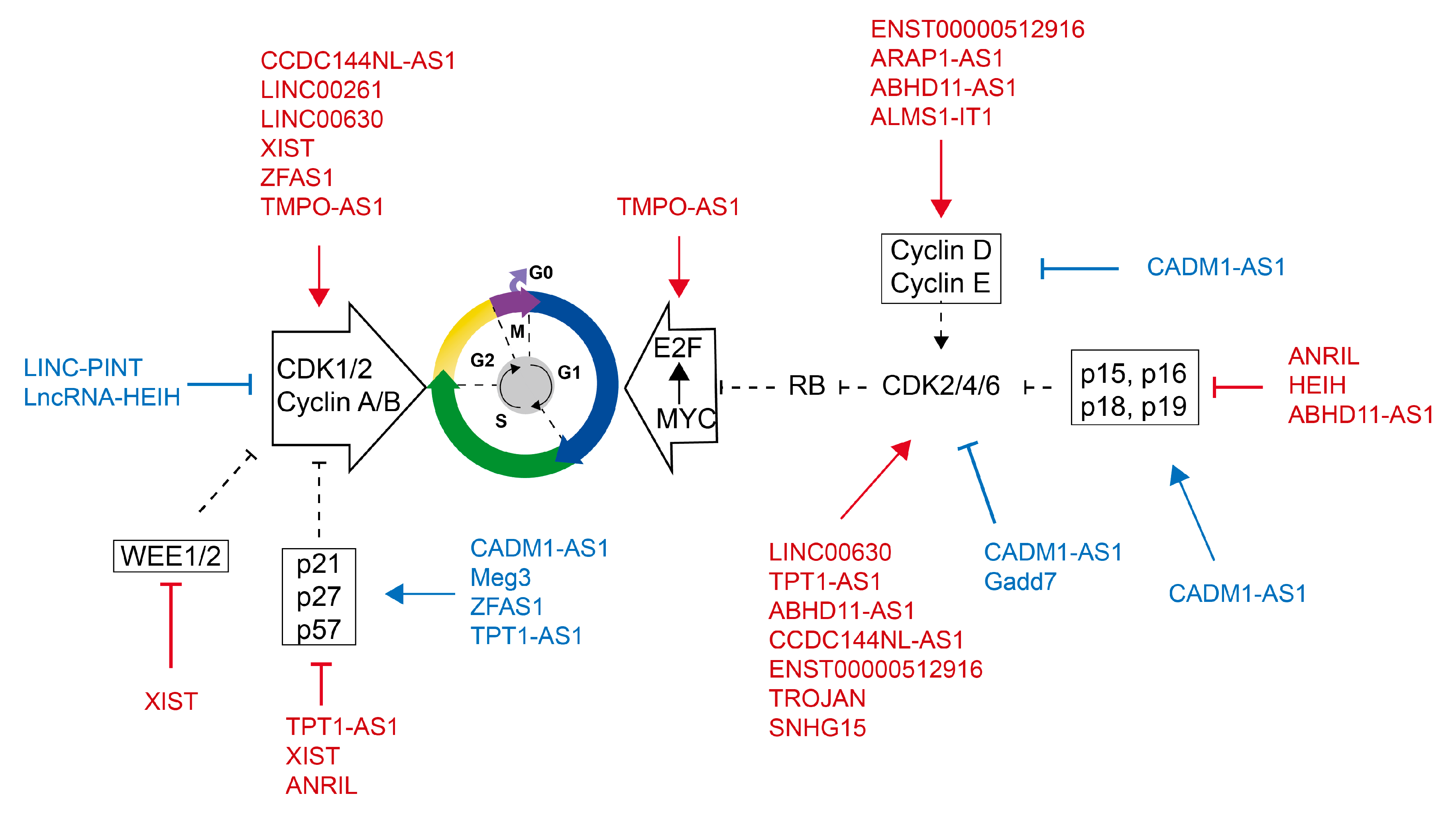
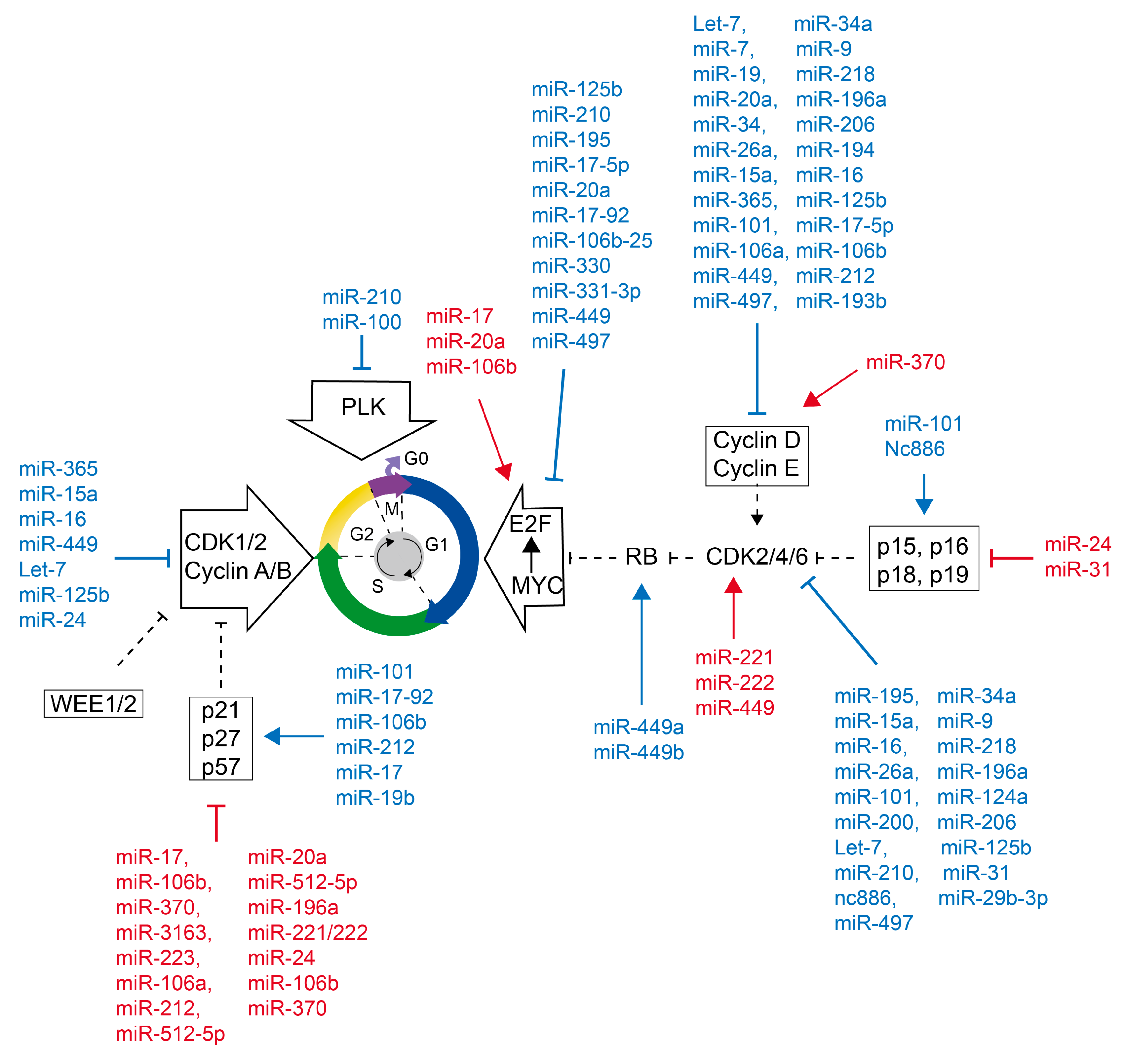

| LncRNA | Target in Cell Cycle (Phase) | Deregulation in Cancer | References |
|---|---|---|---|
| MEG3 | Promotion of p27 by miR-3163/Skp2 mRNA translation. | Downregulated in BLCA, BRCA, CCA, COCA, OVCA. Upregulated in small cell lung cancer. | [88] |
| LINC-PINT | Suppression of CDK1 and cyclin A2 with PRC2 and EZH2 (G2/M). | Downregulated in HCC, BLCA, BRCA, MM, T and B-ALL. Upregulated in COCA. | [75,76] |
| HEIH | Suppression of p15, p16, p21 and p57 with EZH2 (G0/G1). | Upregulated in HCC. Downregulated in BRCA, CRCA, OVCA, PRCA. | [77] |
| ANRIL | Suppression of p21, p14, p15, p16 upregulated cyclin G1 by targeting miR-181a-5p (G0/G1, G2/M). | Upregulated in glioma, LSCC, GC, EOC, HNSC, NSCLC, ESCC, CRCA. | [78,79,80] |
| LINC00630 | Promotion of CDK2 transcription (S). | Upregulated in HCC, CRCA. | [84] |
| ZFAS1 | Promotion of p21 and p27 expression; destabilization of p53; promotion of CDK1/cyclin B1 complex by interaction (G1/S). | Upregulated in CRCA, BLCA, GC, HCC, PRCA. Downregulated in OVCA, basal-like BRCA. | [94] |
| TMPO-AS1 | Promotion of CDK1. Promotion proliferation by sponging miR-326 and targeted by E2F1 (G0/G1, G2/M). | Upregulated in BLCA, basal-like BRCA, CCA, CRCA, LUCA, PRCA, HCC, OVCA. | [82] |
| ARAP1-AS1 | Promotion of cyclin D1 (G0/G1). | Upregulated in LUCA. | [85] |
| CADM1-AS1 | Promotion of p15, p21, p27. Suppression of cyclin Ds, cyclin Es, CDK2, CDK4, CDK6 (G0/G1). | Downregulated in HCC. | [95] |
| TPT1-AS1 | Promotion of CDK4, cyclin D1, suppression of p21 (G1/S). | Upregulated in GC, HCC. Downregulated in BLCA, BRCA, LUCA, OVCA, PRCA. | [89] |
| ABHD11-AS1 | Promotion of Cyclin D1, cyclin E1, CDK1, CDK2, CDK4. Suppression of p16 (G1/S). | Upregulated in EC, PCA, GC, LUCA. Downregulated in CRCA. | [90,91,92,93] |
| CCDC144NL-AS1 | Promotion of CDK1, CKD2 and CDK4 (G0-1/S) through miR-940/WDR5. | Upregulated in CRCA, HCC. Downregulated in LUCA. | [99] |
| LINC00261 | Co-regulation with CDK1 (G0/G1; G2/M). | Downregulated in choriocarcinoma, HCC, BRCA, PRCA, CRCA, GC, LUCA. | [83] |
| Gadd7 | Suppression of CDK6 mRNA degradation (G1/S). | Induced by DNA damage and oxidative stress. | [96,97] |
| ALMS1- IT1 | ALMS1-IT1/AVL9 promote CDK pathway. | Upregulated in small cell lung cancer, CCA, CRCA, OVCA. Downregulated in LUCA. | [86] |
| XIST | Promotion of cyclin D1, CDK1 and p53, suppression of p21 (G1). Suppression of Wee1 (G2). | Upregulated in PCA, CRCA, ESCC. Downregulated in OVCA, NSCLC, HCC, BRCA, BLCA. | [63,98] |
| ENST00000512916 | Promotion of CDK2/4/6 and cyclin D1 (G1). | Upregulated in AB. | [87] |
| TROJAN | Promotion of CDK2. | Upregulated in ER-positive BRCA. | [100] |
| SNHG15 | Promotion of CDK6 by miR-627 sponging. | Upregulated in BLCA, TNBC, CRCA, GC, LUCA, PRCA. | [101] |
| miRNA | Target in Cell Cycle (Phase) | Deregulation in Cancer | References |
|---|---|---|---|
| miR-195 | Suppression of CDK6, cyclin E1, CDK4, cyclin D1, cyclin D3 (G1/S). | Downregulated in NSCLC, GC, HCC, BLCA. Upregulated in BRCA, CLL, ACA. | [126,127] |
| miR-100 | Targeting and suppressing PLK1; suppression of reduced β-tubulin I, IIA, IIB and V mRNA; suppression of p27 (G2/M). | Upregulated in BRCA, PCA. Downregulated in PRCA, OVCA, BLCA. | [128,129] |
| miR-365 | Suppression cyclin D1 and CDC25A, promotion of p27 (G1/S). | Downregulated in NSCLC, BRCA, OVCA. | [130,131] |
| miR-15a | Suppression of CDK1, CDK2, CDK6, cyclin D1, cyclin D2 and cyclin E1 (G1/S). | Downregulated in HCC, BRCA, OS, NSCLC, PCA, PRCA. Upregulated in CLL. | [132,133,134,135] |
| miR-16 | Suppression of CDK1, CDK2, CDK6, cyclin D1, cyclin D2 and cyclin E1 (G1/S). | Downregulated in PRCA, GC, LUCA. Upregulated in PCA, HCC. | [133,134,135] |
| Let-7 | Suppression of CDK4, Cyclin D1, cyclin D3, Cyclin A and Cyclin B translation (G1). | Downregulation in OVCA, LUCA, NSCLC, BRCA, CRCA, PRCA, HNSC. | [136,137,138,139] |
| miR-7 | Suppression of p53, CDC42, Cyclin D, cyclin E1 (G0-1/S). | Downregulated in BRCA, CCA, MG, HCC. Upregulated in HNSC, LUCA. | [136,140,141] |
| miR-19 | Suppression of Cyclin Ds, promotion of p21 (G1/S). | downregulated in MG. | [142,143] |
| miR-20a | Suppression of E2F2 and E2F3 translation; Suppression cyclin D1 mRNA transcription; Suppression of p21 (G1/S). | Upregulated in HCC, CRCA, PRCA, PCA, LUCA. Downregulated in BRCA, HCC. | [144,145,146] |
| miR-34 | Suppression of Cyclin D1, CDK4, CDK6 translation; promotion of p21 (G1/S). | Downregulated in PCA, OVCA, NSCLC, HCC. Upregulated in CRCA. | [147,148] |
| miR-124a | Suppression CDK2, CDK6, cyclin D1, cyclin D2 translation (G1). | Downregulated in all cancer. | [149,150,151] |
| miR-125b | Suppression cyclin D1, CDK6, promotion of p21 (G1/S). | Downregulated in PRCA, LUCA, HCC, BRCA, CCA, CRCA. Upregulated in PCA, BLCA. | [152,153,154] |
| miR-24 | Suppression of p27, p16, p21(G2/M). | Downregulated in PRCA, PTC. Upregulated in LUCA, PCA, CRCA. | [154,155] |
| miR-26a | Suppression of cyclin D2, cyclin D3, cyclin E2, CDK4/6; promotion of p14. | Downregulated in PTC, LUCA, HCC, BRCA, BLCA. | [156,157,158] |
| miR-101 | Suppression of CDK2, CDK4, CDK6, cyclin D2, cyclin D3 and cyclin E2; Promotion of p14, p16, p21 and p27 (G1). | Downregulated in LUCA, PRCA, HCC, LSCC, OVCA, BLCA. | [159,160] |
| miR-9 | Suppression of CDK6, cyclin D1; promotion p16 (G0/G1). | Downregulated in HSCC. | [161,162,163] |
| miR-218 | Suppression CDK6, cyclin D1; promotion of p14, p16. | Downregulated in MG, LUCA, CCA. Upregulated in PRCA. | [164,165] |
| miR-210 | Suppression of CDK4, CDK6; promotion of p27 (G1/S). | Upregulated in PRCA, LUCA, PCA, HNSC, BRCA. | [166] |
| miR-17-5p | Suppression of cyclin D1; promotion of p21 (G2/M). | Upregulated in bladder cancer, HCC, BRCA, CRCA, LUCA. | [144,167,168,169] |
| miR-17-92 cluster (MIR17HG) | Suppression of E2F1, cyclin D1. Promotion of p21 (G1/S). | Upregulated in BL, CRCA, LUCA, BLCA, BRCA, PRCA, PCA, HCC. | [170,171,172,173,174,175,176] |
| miR-106a | Suppression of p21, cyclin D1 (G1/S). | Upregulated in LUCA, HCC, PCA, PRCA, CRCA. | [143,177] |
| miR-106b | Suppression of cyclin D1, E2F1, p21 (G1/S). | upregulated in HCC, GC, CRCA. Downregulated in OVCA. | [172,178,179,180] |
| miR-330 | Suppression of E2F1. | Downregulated in OSCC, PRCA. | [181] |
| miR-331-3p | Suppression of E2F1. | Downregulated in HNSC, OSCC. Upregulated in AML. | [182] |
| miR-449 | Promotion of CDK6 and CDC25A; Suppression E2F1, E2F3, cyclin D1, cyclin A2. | Downregulated in PRCA. | [183,184] |
| miR-31 | Suppression of CDK1, CDK4, CDK6; promotion of p21. | Upregulated in HNSC, CRCA, OSCC, LUCA. Downregulated in PRCA, BRCA, BLCA. | [185,186,187,188] |
| miR-512-5p | Suppression of p21. | Downregulated in GC. | [189] |
| miR-370 | Promotion of p27 and p21 (G1/S). | Upregulated in PRCA. Downregulated in OSCC. | [190] |
| miR-212 | Promotion of p21 and p27; suppression cyclin D3 (G1/S). | Upregulated in LUCA, PCA, HCC. | [191,192] |
| miR-196a | Suppression of p27. | Upregulated in BRCA, PDAC, ESCC, CRCA. | [193] |
| miR-3163 | Promotion of p27 (G0/G1). | Downregulated in BRCA. | [88,194] |
| miR-221 | Suppression of p27 and p57, promotion of CDK2 (G2/M). | Downregulated in PRCA, NSCLC. Upregulated in HCC, PCA, BLCA, CRCA, OVCA, BRCA, OSCC. | [195,196] |
| miR-222 | Suppression of p27 (G1/S). | Downregulated in PRCA. Upregulated in PCA, NSCLC, HCC, GC. | [197,198,199] |
| Nc886 | Promotion of CDKN2A and CDKN2C, suppression of CDK4 (G1/S). | Downregulated in ESCC, GC, PRCA. | [200,201] |
| miR-29b-3p | Suppression of CDK6 (G1). | Downregulated in PRCA, LUCA, HNSC, CLL, AML. Upregulated in BRCA, CRCA, PCA. | [202,203] |
| miR-497 | Suppression of CDK6, E2F3 and cyclin E1 (G1). | Downregulated in PRCA, LUCA, CRCA, BRCA. | [204,205] |
| miR-193b | Suppression of cyclin D1 by targeting 3′UTR of mRNA (G1/S). | Downregulated in BRCA, PRCA, OS, MM. | [206,207,208,209] |
| miR-200 | Suppression of CDK6 (G1). | Downregulated in CRCA, HCC, OV, BRCA, MM (all cancer) | [210] |
| miR-206 | Suppression of CDK4, cyclin D1, cyclin C translation (G1). | Downregulated in BRCA, OVCA. | [211,212] |
| miR-223 | Suppression of p21, p27 (G1). E2F1 binds to the miR-223 promoter and inhibits transcription. | Downregulated in AML, HCC, CLL. Upregulated in BLCA, PCA, CRCA, PRCA. | [213,214] |
| ncRNA | Tumor Type | CDKIs | Sample | Function and Mechanism | References |
|---|---|---|---|---|---|
| MEG3 | LC | Palbociclib | cell lines | Palbociclib increase MEG3 expression. Silencing MEG3 expression rescue palbociclib-mediated decrease in cell proliferation. | [281] |
| TROJAN | ER-positive BC, TNBC | Palbociclib | cell lines | TROJAN inhibit NKRF/RELA and upregulate CDK2 expression, which promote cell proliferation and resistance to a CDK4/6 inhibition in ER+ breast cancer. | [100] |
| SNHG15 | GBM, BC, LC, HCC | Palbociclib | cell lines | SNHG15 upregulated CDK6 by sponging inhibit miR-627-5p. palbociclib treatment increased miR-627-5p expression and reduced SNHG15 and CDK6 expression. | [101] |
| miR-3613-3p | BC | Palbociclib | clinical samples; PDTX; cell lines | MiR-3613-3p induce G1 cell cycle arrest and enhance TNBC sensitivity to palbociclib. | [282] |
| miR-29b-3p | HER-negative BC | Palbociclib | clinical samples; PDTX; cell lines | MiR-29b downregulate CDK6 and induce cell cycle arrest at G1 phase. Activated c-Myc downregulate miR-29b, further activate CDK6 and resistance to palbociclib. | [203] |
| miR-497 | ALCL | Palbociclib | cell lines | Hypermethylation repressed miR-497 expression. miR-497 inhibited CDK6, E2F3 and cyclin E1 (G1) caused cell cycle arrest. higher miR-497 expression cell more sensitive to palbociclib | [205] |
| miR-193b | PCA | Palbociclib | cell lines | MiR-193b downregulate cyclin D1 by targeting 3′UTR of mRNA. palbociclib has no effect on cells with high miR-193b expression. | [209] |
| miR-200a | MM | Palbociclib | cell lines | MiR-200 reduces CDK6 expression and reduces melanoma response to palbociclib. | [210] |
| miR-223 | lum BC; HER2-positive BC | Palbociclib | transgenic mouse model; clinical samples; cell lines | Palbociclib restrain E2F1 activity and restore miR-223 expression. miR-223 deficiency induces luminal breast cancer resistance to palbociclib. | [283] |
| miR-126 | lum BC; HER2-positive BC | Ribociclib | cell lines. | MiR-126 targets PI3K/AKT/MTOR pathway, affects cell cycle and mitosis. miR-125 preserving leukemia stem cell quiescence and promoting chemotherapy resistance. | [284] |
| MIR17HG | GBM, AT/RT | Palbociclib; Ribociclib | cell lines | MIR17HG reduce cyclin D1 expression and sensitizes ATRT cells to palbociclib treatment. | [173] |
| miR-432-5p | ER-positive BC | palbociclib; Ribociclib | cell lines | Palbociclib resistant cells increased miR-432-5p and CDK6 expression. In palbociclib resistance breast cancers, miR-432-5p is higher expressed. | [285] |
| miR-9 | HER2-positive BC; TNBC | Ribociclib; | cell lines | Suppression of CDK6 and enhance the activity of ribociclib. | [284] |
| miR-326 | HER2-positive BC | Ribociclib | cell lines | Suppression of CDK6 and enhance the activity of ribociclib. | [82] |
| miR-124a | ALL | Palbociclib; PD-0332991 | cell lines | MiR-124a was down-regulated in ALL by hypermethylation of the promoter and histone modifications. MiR-124 negatively regulates CDK6 and pRB. ALL patients with overexpressed miR-124a benefit from CDK6 inhibition. | [149] |
| miR-9 | ALL | Palbociclib | cell lines | Epigenetic downregulation of MIR9 induced upregulation of CDK6. | [174] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Q.; Huang, T. Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition. Int. J. Mol. Sci. 2023, 24, 8939. https://doi.org/10.3390/ijms24108939
Hu Q, Huang T. Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition. International Journal of Molecular Sciences. 2023; 24(10):8939. https://doi.org/10.3390/ijms24108939
Chicago/Turabian StyleHu, Qingyi, and Tao Huang. 2023. "Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition" International Journal of Molecular Sciences 24, no. 10: 8939. https://doi.org/10.3390/ijms24108939
APA StyleHu, Q., & Huang, T. (2023). Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition. International Journal of Molecular Sciences, 24(10), 8939. https://doi.org/10.3390/ijms24108939




