Spatial Gene Expression Analysis Reveals Characteristic Gene Expression Patterns of De Novo Neuroendocrine Prostate Cancer Coexisting with Androgen Receptor Pathway Prostate Cancer
Abstract
1. Introduction
2. Results
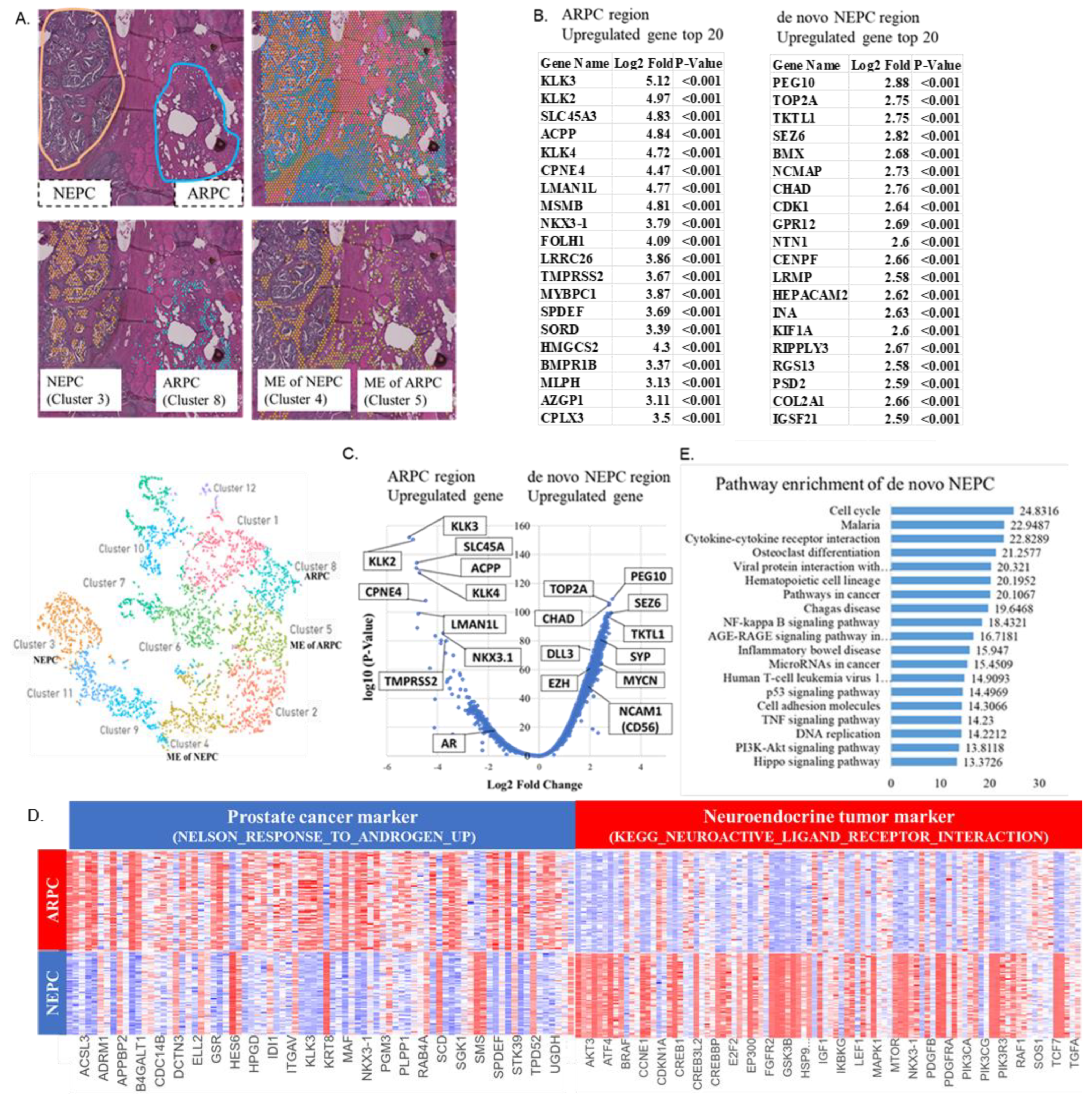
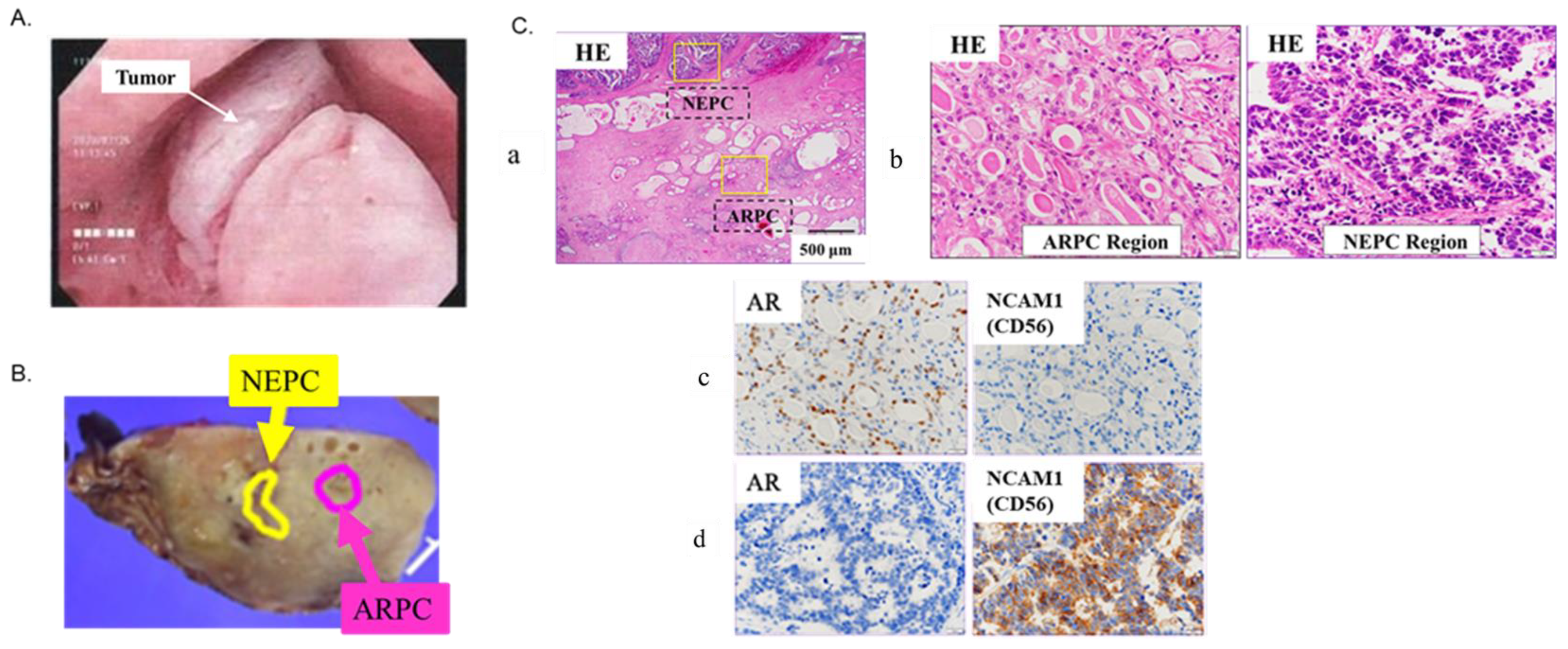
2.1. Localization of De Novo NEPC and ARPC and High-Expression Genes in the Prostate Tissue
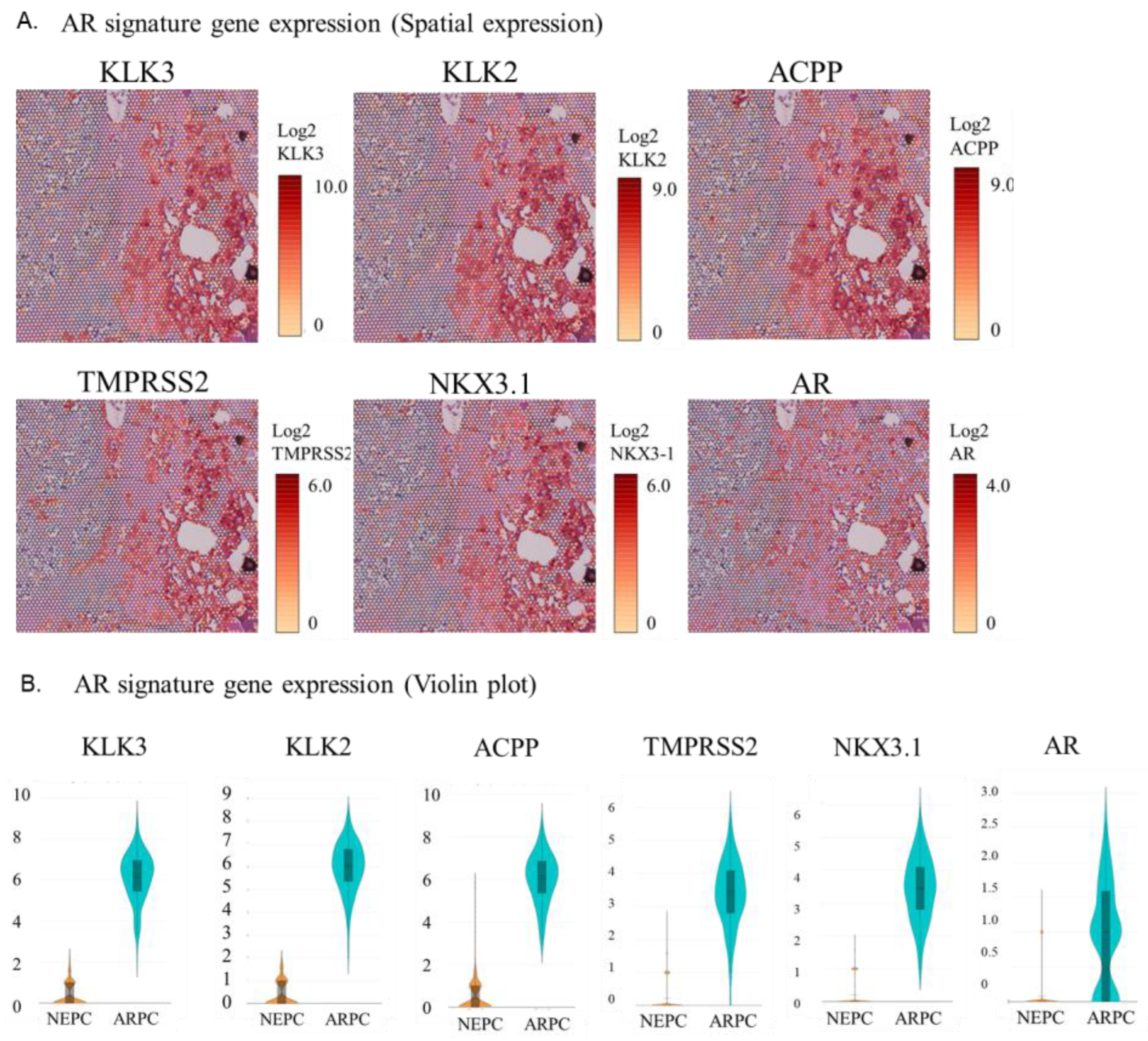
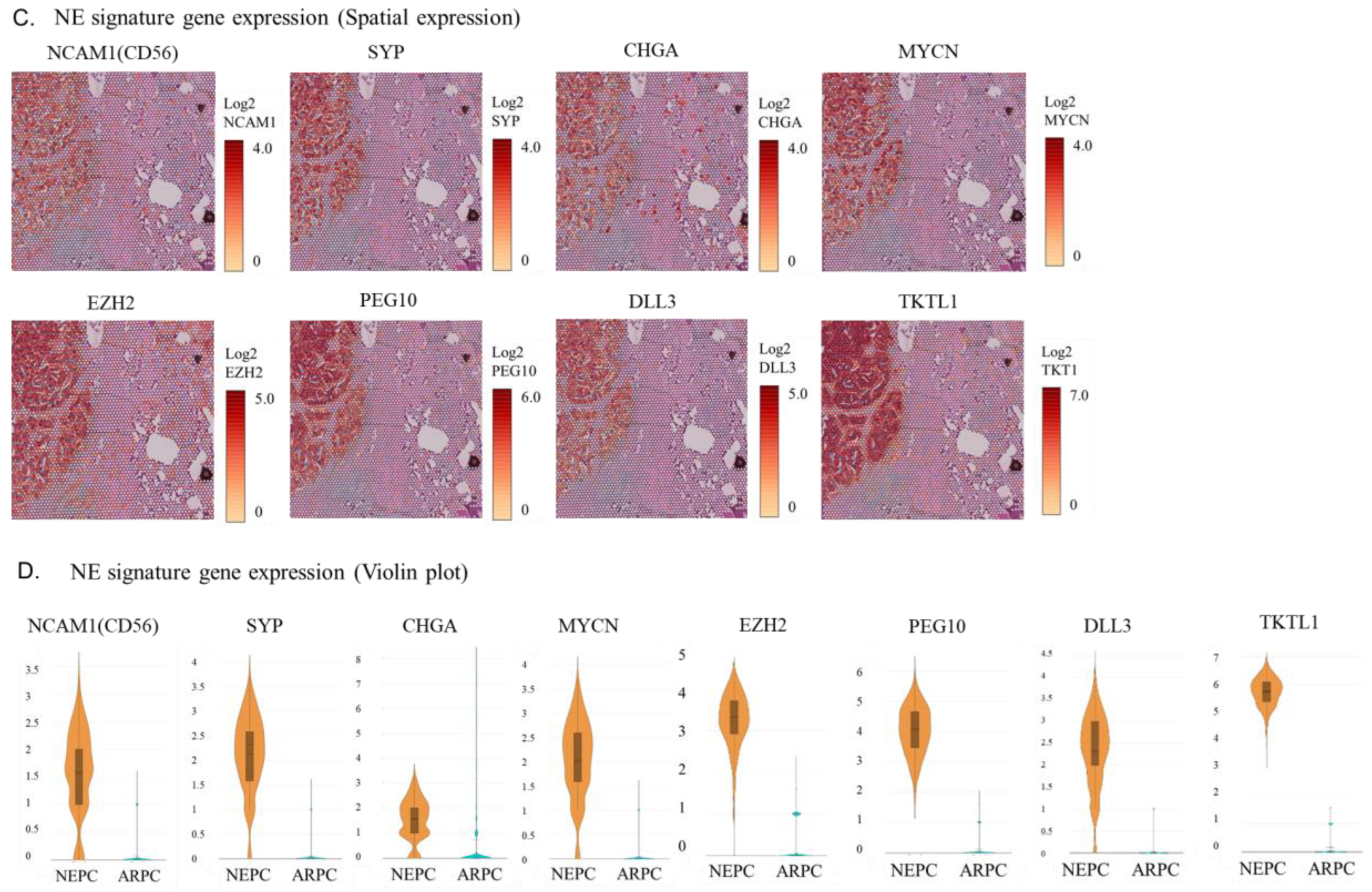
2.2. Visualization of the Expression of AR Signature Genes, NE Signature Genes, and Other Genes in the Tissue
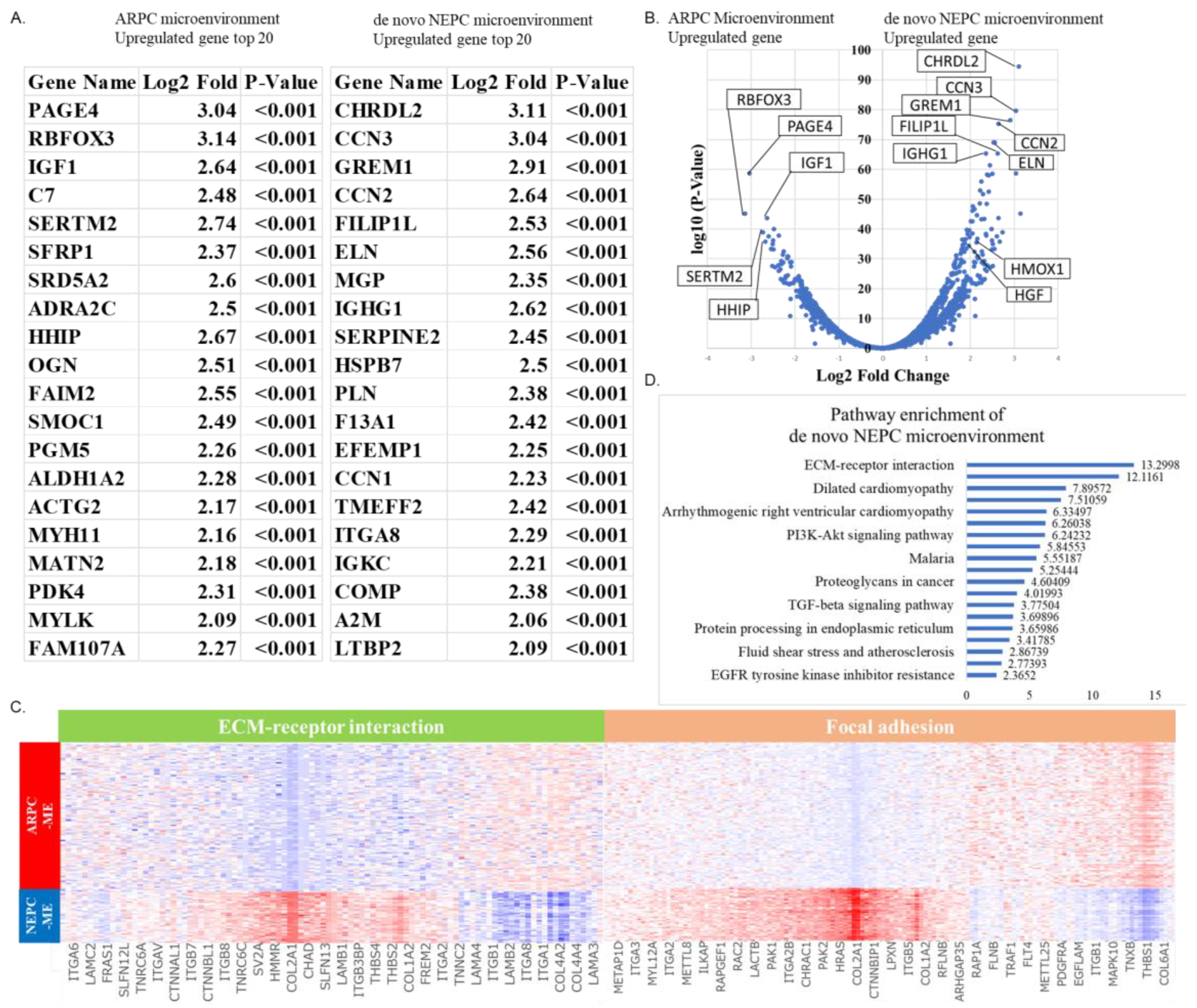
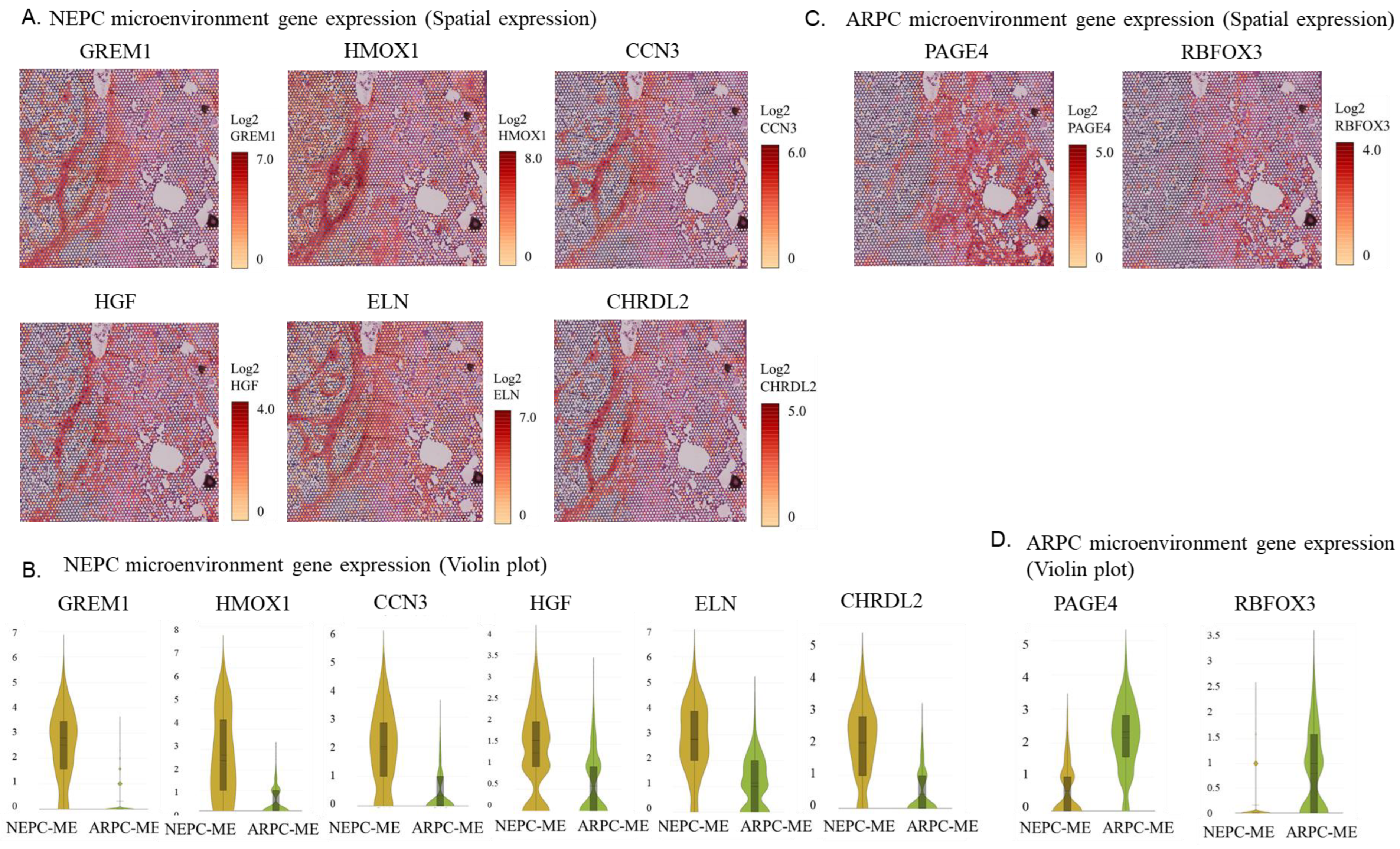
2.3. Spatial Gene Expression Analysis within the Tumor Microenvironment around ARPC and De Novo NEPC Cells
3. Discussion
4. Patient and Methods
4.1. Case Presentation
4.2. Spatial Transcriptomics (CytAssist Visium)
- Number of spots under tissue: 4397; mean reads per spot: 74,832; median genes per spot: 7195; number of reads: 329,038,054; valid barcodes: 94.0%; valid UMIs: 99.9%; sequencing saturation: 48.0%
- UMAP and violin plots were run and plotted using Loupe Browser (10× genomics, Pleasanton, CA, USA). Trajectory analysis and pathway enrichment analysis were performed and plotted using Partek flow software (Partek Incorporated, Chesterfield, MO, USA).
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, R.; Dong, X.; Gleave, M. Molecular Model for Neuroendocrine Prostate Cancer Progression. BJU Int. 2018, 122, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Sargos, P.; Ferretti, L.; Gross-Goupil, M.; Orre, M.; Cornelis, F.; Henriques de Figueiredo, B.; Houédé, N.; Merino, C.; Roubaud, G.; Dallaudiére, B.; et al. Characterization of Prostate Neuroendocrine Cancers and Therapeutic Management: A Literature Review. Prostate Cancer Prostatic Dis. 2014, 17, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.K.; Chugh, N.; Tripathi, M. Neuroendocrine Differentiation of Prostate Cancer-An Intriguing Example of Tumor Evolution at Play. Cancers 2019, 11, 1405. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, M.; Chuen Choi, S.Y.; Wang, Y.; Lin, D.; Zeng, H.; Wang, Y. Genomic alterations in neuroendocrine prostate cancer: A systematic review and meta-analysis. BJUI Compass 2023, 4, 256–265. [Google Scholar] [CrossRef]
- Miyagi, Y.; Sasaki, T.; Fujinami, K.; Sano, J.; Senga, Y.; Miura, T.; Kameda, Y.; Sakuma, Y.; Nakamura, Y.; Harada, M.; et al. ETS Family-associated Gene Fusions in Japanese Prostate Cancer: Analysis of 194 Radical Prostatectomy Samples. Mod. Pathol. 2010, 23, 1492–1498. [Google Scholar] [CrossRef]
- Hong, D.; Dong, B.; Wang, Y.; Huang, H.; Zhang, W.; Lian, B.; Ji, B.; Shi, H.; Qu, M.; Gao, X.; et al. Single-Cell Transcriptional Regulation and Genetic Evolution of Neuroendocrine Prostate Cancer. iScience 2022, 25, 104576. [Google Scholar] [CrossRef]
- Han, M.; Li, F.; Zhang, Y.; Dai, P.; He, J.; Li, Y.; Zhu, Y.; Zheng, J.; Huang, H.; Bai, F.; et al. FOXA2 Drives Lineage Plasticity and KIT Pathway Activation in Neuroendocrine Prostate Cancer. Cancer Cell 2022, 40, 1306–1323.e8. [Google Scholar] [CrossRef]
- Lose, F.; Batra, J.; O’Mara, T.; Fahey, P.; Marquart, L.; Eeles, R.A.; Easton, D.F.; Al Olama, A.A.; Kote-Jarai, Z.; Guy, M.; et al. Common Variation in Kallikrein Genes KLK5, KLK6, KLK12, and KLK13 and Risk of Prostate Cancer and Tumor Aggressiveness. Urol. Oncol. 2013, 31, 635–643. [Google Scholar] [CrossRef]
- Korbakis, D.; Soosaipillai, A.; Diamandis, E.P. Study of Kallikrein-related Peptidase 6 (KLK6) and Its Complex with α1-Antitrypsin in Biological Fluids. Clin. Chem. Lab. Med. 2017, 55, 1385–1396. [Google Scholar] [CrossRef]
- Lotan, T.L.; Gupta, N.S.; Wang, W.; Toubaji, A.; Haffner, M.C.; Chaux, A.; Hicks, J.L.; Meeker, A.K.; Bieberich, C.J.; De Marzo, A.M.; et al. ERG Gene Rearrangements Are Common in Prostatic Small Cell Carcinomas. Mod. Pathol. 2011, 24, 820–828. [Google Scholar] [CrossRef]
- Akamatsu, S.; Wyatt, A.W.; Lin, D.; Lysakowski, S.; Zhang, F.; Kim, S.; Tse, C.; Wang, K.; Mo, F.; Haegert, A.; et al. The Placental Gene PEG10 Promotes Progression of Neuroendocrine Prostate Cancer. Cell Rep. 2015, 12, 922–936. [Google Scholar] [CrossRef] [PubMed]
- Modlin, I.M.; Drozdov, I.; Alaimo, D.; Callahan, S.; Teixiera, N.; Bodei, L.; Kidd, M. A Multianalyte PCR Blood Test Outperforms Single Analyte ELISAs (Chromogranin A, Pancreastatin, Neurokinin A) for Neuroendocrine Tumor Detection. Endocr. Relat. Cancer 2014, 21, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Puca, L.; Gavyert, K.; Sailer, V.; Conteduca, V.; Dardenne, E.; Sigouros, M.; Isse, K.; Kearney, M.; Vosoughi, A.; Fernandez, L.; et al. Delta-like Protein 3 Expression and Therapeutic Targeting in Neuroendocrine Prostate Cancer. Sci. Transl. Med. 2019, 11, eaav0891. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Beltran, H. Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr. Oncol. Rep. 2021, 23, 15. [Google Scholar] [CrossRef]
- Merkens, L.; Sailer, V.; Lessel, D.; Janzen, E.; Greimeier, S.; Kirfel, J.; Perner, S.; Pantel, K.; Werner, S.; von Amsberg, G. Aggressive Variants of Prostate Cancer: Underlying Mechanisms of Neuroendocrine Transdifferentiation. J. Exp. Clin. Cancer Res. 2022, 41, 46. [Google Scholar] [CrossRef]
- Brennen, W.N.; Zhu, Y.; Coleman, I.M.; Dalrymple, S.L.; Antony, L.; Patel, R.A.; Hanratty, B.; Chikarmane, R.; Meeker, A.K.; Zheng, S.L.; et al. Resistance to Androgen Receptor Signaling Inhibition Does Not Necessitate Development of Neuroendocrine Prostate Cancer. JCI Insight 2021, 6, e146827. [Google Scholar] [CrossRef]
- Akamatsu, S.; Inoue, T.; Ogawa, O.; Gleave, M.E. Clinical and Molecular Features of Treatment-related Neuroendocrine Prostate Cancer. Int. J. Urol. 2018, 25, 345–351. [Google Scholar] [CrossRef]
- Aggarwal, R.; Huang, J.; Alumkal, J.J.; Zhang, L.; Feng, F.Y.; Thomas, G.V.; Weinstein, A.S.; Friedl, V.; Zhang, C.; Witte, O.N.; et al. Clinical and Genomic Characterization of Treatment-emergent Small-cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018, 36, 2492–2503. [Google Scholar] [CrossRef]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.S.K.; Varambally, S.; et al. Divergent Clonal Evolution of Castration-resistant Neuroendocrine Prostate Cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef]
- Sen, T.; Gay, C.M.; Byers, L.A. Targeting DNA Damage Repair in Small Cell Lung Cancer and the Biomarker Landscape. Transl. Lung Cancer Res. 2018, 7, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Maekawa, M.; Hieda, M.; Taguchi, T.; Miura, N.; Kikugawa, T.; Saika, T.; Higashiyama, S. SPOP is Essential for DNA-Protein Cross-link Repair in Prostate Cancer Cells: SPOP-dependent Removal of Topoisomerase 2A from the Topoisomerase 2A-DNA Cleavage Complex. Mol. Biol. Cell 2020, 31, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Brady, L.; Nelson, P.S. RISING STARS: Heterogeneity and the Tumor Microenvironment in Neuroendocrine Prostate Cancer. J. Endocrinol. 2022, 256, e220211. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Kim, J.O.; Park, K.S.; Won, M.; Kim, K.E.; Kim, K.K. Transforming Growth Factor-β-induced RBFOX3 Inhibition Promotes Epithelial-mesenchymal Transition of Lung Cancer Cells. Mol. Cells 2016, 39, 625–630. [Google Scholar] [CrossRef]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt Signaling in Colorectal Cancer: Pathogenic Role and Therapeutic Target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef]
- Sun, Y.; Campisi, J.; Higano, C.; Beer, T.M.; Porter, P.; Coleman, I.; True, L.; Nelson, P.S. Treatment-induced Damage to the Tumor Microenvironment Promotes Prostate Cancer Therapy Resistance through WNT16B. Nat. Med. 2012, 18, 1359–1368. [Google Scholar] [CrossRef]
- Ding, X.; Xi, W.; Ji, J.; Cai, Q.; Jiang, J.; Shi, M.; Yu, Y.; Zhu, Z.; Zhang, J. HGF Derived from Cancer-associated Fibroblasts Promotes Vascularization in Gastric Cancer via PI3K/AKT and ERK1/2 Signaling. Oncol. Rep. 2018, 40, 1185–1195. [Google Scholar] [CrossRef]
- Luu Hoang, K.N.; Anstee, J.E.; Arnold, J.N. The Diverse Roles of Heme Oxygenase-1 in Tumor Progression. Front. Immunol. 2021, 12, 658315. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Smid, M.; Iaria, J.; Salvatori, D.C.F.; van Dam, H.; Zhu, H.J.; Martens, J.W.M.; Dijke, P.T. Cancer-associated Fibroblast-derived Gremlin 1 Promotes Breast Cancer Progression. Breast Cancer Res. 2019, 21, 109. [Google Scholar] [CrossRef] [PubMed]
- De Hosson, L.D.; Takkenkamp, T.J.; Kats-Ugurlu, G.; Bouma, G.; Bulthuis, M.; de Vries, E.G.E.; Van Faassen, M.; Kema, I.P.; Walenkamp, A.M.E. Neuroendocrine Tumours and Their Microenvironment. Cancer Immunol. Immunother. 2020, 69, 1449–1459. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, R.; Miura, N.; Kurata, M.; Kitazawa, R.; Kikugawa, T.; Saika, T. Spatial Gene Expression Analysis Reveals Characteristic Gene Expression Patterns of De Novo Neuroendocrine Prostate Cancer Coexisting with Androgen Receptor Pathway Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 8955. https://doi.org/10.3390/ijms24108955
Watanabe R, Miura N, Kurata M, Kitazawa R, Kikugawa T, Saika T. Spatial Gene Expression Analysis Reveals Characteristic Gene Expression Patterns of De Novo Neuroendocrine Prostate Cancer Coexisting with Androgen Receptor Pathway Prostate Cancer. International Journal of Molecular Sciences. 2023; 24(10):8955. https://doi.org/10.3390/ijms24108955
Chicago/Turabian StyleWatanabe, Ryuta, Noriyoshi Miura, Mie Kurata, Riko Kitazawa, Tadahiko Kikugawa, and Takashi Saika. 2023. "Spatial Gene Expression Analysis Reveals Characteristic Gene Expression Patterns of De Novo Neuroendocrine Prostate Cancer Coexisting with Androgen Receptor Pathway Prostate Cancer" International Journal of Molecular Sciences 24, no. 10: 8955. https://doi.org/10.3390/ijms24108955
APA StyleWatanabe, R., Miura, N., Kurata, M., Kitazawa, R., Kikugawa, T., & Saika, T. (2023). Spatial Gene Expression Analysis Reveals Characteristic Gene Expression Patterns of De Novo Neuroendocrine Prostate Cancer Coexisting with Androgen Receptor Pathway Prostate Cancer. International Journal of Molecular Sciences, 24(10), 8955. https://doi.org/10.3390/ijms24108955




