Deregulated Expression of IL-37 in Patients with Bladder Urothelial Cancer: The Diagnostic Potential of the IL-37e Isoform
Abstract
:1. Introduction
2. Results
2.1. IL-37 Levels Are Increased in BLCA versus Non-Cancerous Bladder Tissues
2.2. Increased IL-37 or SIGIRR Expression Are Favorable Prognostic Factors for OS in BLCA Patients
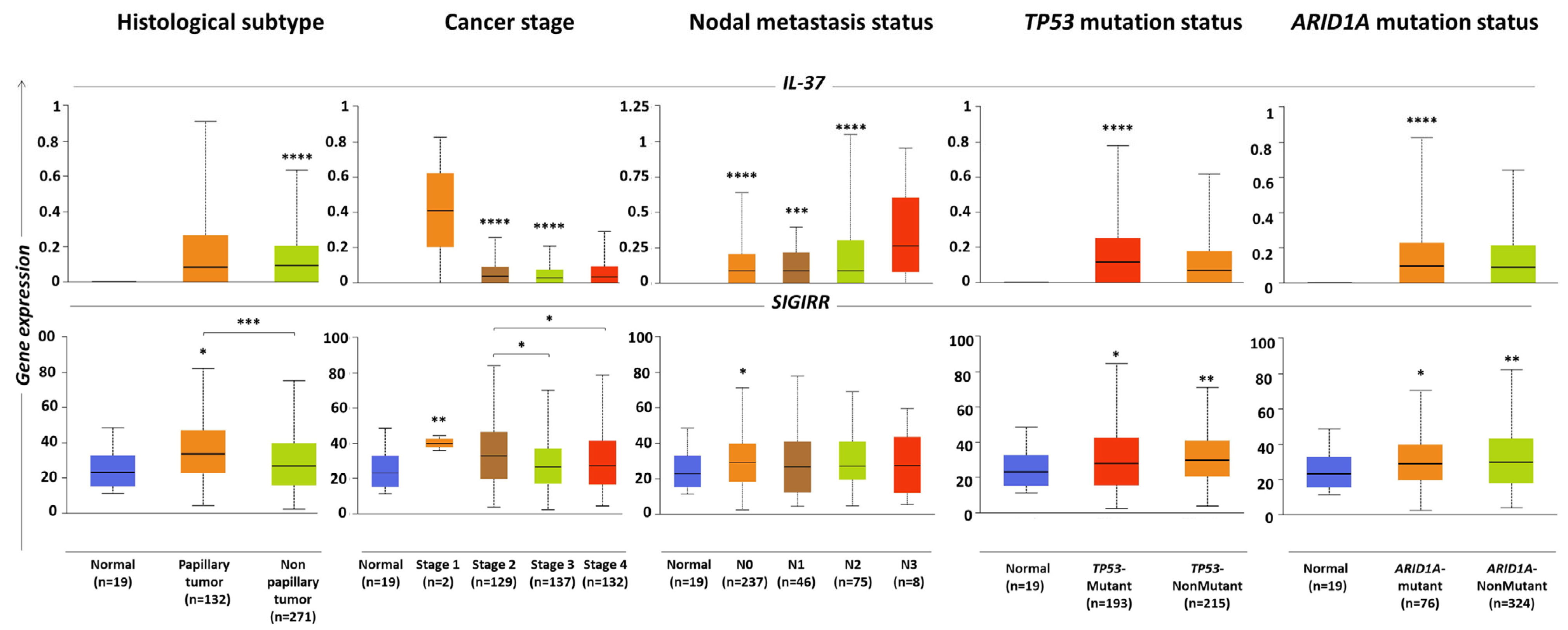
2.3. IL-37 and SIGIRR Levels Associated with Histopathological Parameters of BLCA Tumors
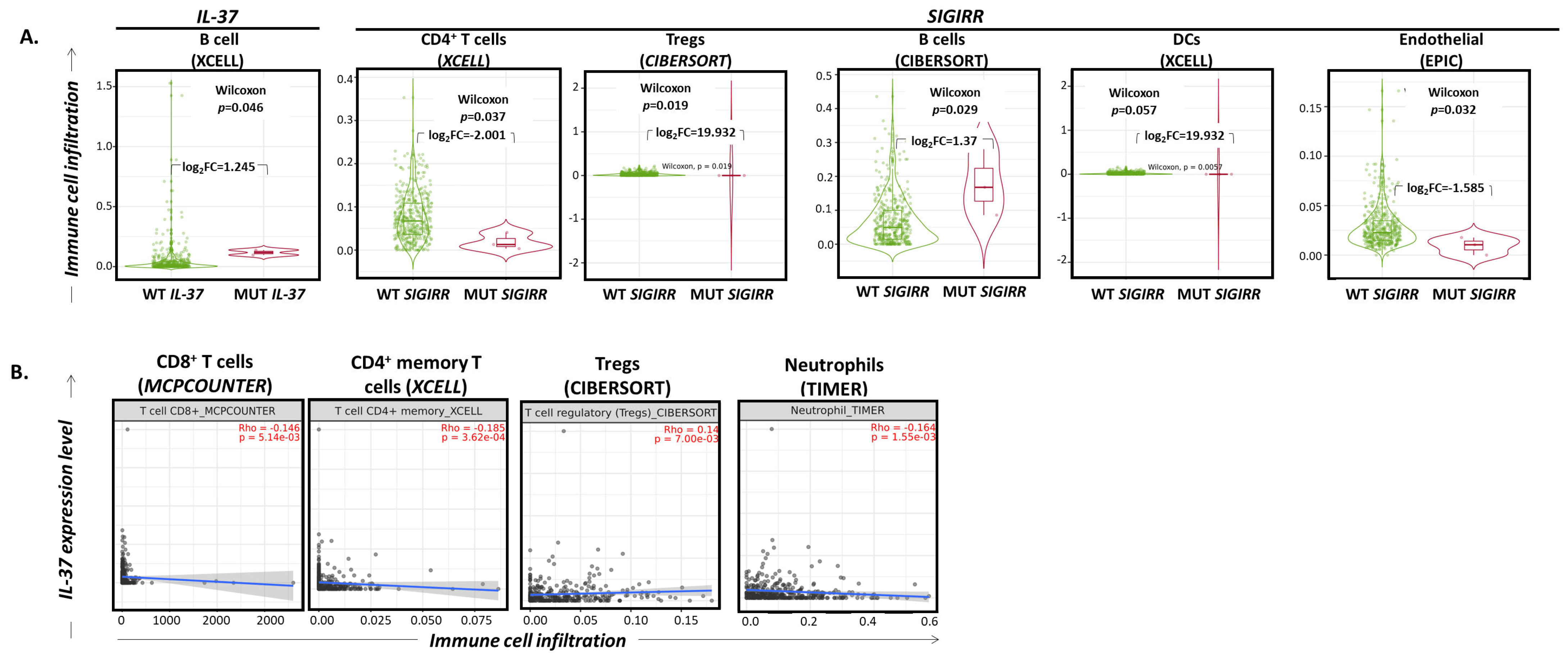
2.4. IL-37 and SIGIRR Gene Alterations Correlate with Altered Infiltration of the BLCA by Certain Immune Cell Subset Levels Associated with Histopathological Parameters of BLCA Tumors
2.5. IL-37 Expression Levels Correlate with Altered Infiltration of the BLCA by Certain Immune Cell Subsets
2.6. Human Bladder Cancer Cells Express IL-37b, c and e Isoforms
2.7. Human BLCA Biopsies Predominantly Express the IL-37e Isoform: Correlation with the Grade of the Tumor
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Assessment of IL-37 and SIGIRR Levels in BLCA versus Non-BLCA Lung Tissues
4.3. Analysis of Associations between IL-37 or SIGIRR Levels and Certain Pathological Characteristics of the BLCA Tumor
4.4. Exploration of Correlations between IL-37 and SIGIRR Gene Alterations or Expression Levels and Immune Cell Infiltration Patterns in BLCA Tumors
4.5. Human Bladder Cancer Biopsies
4.6. Cell Cultures
4.7. Extraction of Total RNA and Reverse Transcription
4.8. Quantitative Real Time PCR (qPCR)
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharaf, N.; Nicklin, M.J.; di Giovine, F.S. Long-Range DNA Interactions at the IL-1/IL-36/IL-37 Gene Cluster (2q13) Are Induced by Activation of Monocytes. Cytokine 2014, 68, 16–22. [Google Scholar] [CrossRef]
- Sims, J.E.; Smith, D.E. The IL-1 Family: Regulators of Immunity. Nat. Rev. Immunol. 2010, 10, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Bufler, P.; Gamboni-Robertson, F.; Azam, T.; Kim, S.-H.; Dinarello, C.A. Interleukin-1 Homologues IL-1F7b and IL-18 Contain Functional MRNA Instability Elements within the Coding Region Responsive to Lipopolysaccharide. Biochem. J. 2004, 381, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Dinarello, C.A. Suppression of Inflammation and Acquired Immunity by IL-37. Immunol. Rev. 2018, 281, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Nold-Petry, C.A.; Lo, C.Y.; Rudloff, I.; Elgass, K.D.; Li, S.; Gantier, M.P.; Lotz-Havla, A.S.; Gersting, S.W.; Cho, S.X.; Lao, J.C.; et al. IL-37 Requires the Receptors IL-18Rα and IL-1R8 (SIGIRR) to Carry out Its Multifaceted Anti-Inflammatory Program upon Innate Signal Transduction. Nat. Immunol. 2015, 16, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Nold, M.F.; Nold-Petry, C.A.; Zepp, J.A.; Palmer, B.E.; Bufler, P.; Dinarello, C.A. IL-37 Is a Fundamental Inhibitor of Innate Immunity. Nat. Immunol. 2010, 11, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Rudloff, I.; Cho, S.X.; Lao, J.C.; Ngo, D.; McKenzie, M.; Nold-Petry, C.A.; Nold, M.F. Monocytes and Dendritic Cells Are the Primary Sources of Interleukin 37 in Human Immune Cells. J. Leukoc. Biol. 2017, 101, 901–911. [Google Scholar] [CrossRef]
- Wald, D.; Qin, J.; Zhao, Z.; Qian, Y.; Naramura, M.; Tian, L.; Towne, J.; Sims, J.E.; Stark, G.R.; Li, X. SIGIRR, a Negative Regulator of Toll-like Receptor-Interleukin 1 Receptor Signaling. Nat. Immunol. 2003, 4, 920–927. [Google Scholar] [CrossRef]
- Moretti, S.; Bozza, S.; Oikonomou, V.; Renga, G.; Casagrande, A.; Iannitti, R.G.; Puccetti, M.; Garlanda, C.; Kim, S.; Li, S.; et al. IL-37 Inhibits Inflammasome Activation and Disease Severity in Murine Aspergillosis. PLoS Pathog. 2014, 10, e1004462. [Google Scholar] [CrossRef]
- Cavalli, G.; Justice, J.N.; Boyle, K.E.; D’Alessandro, A.; Eisenmesser, E.Z.; Herrera, J.J.; Hansen, K.C.; Nemkov, T.; Stienstra, R.; Garlanda, C.; et al. Interleukin 37 Reverses the Metabolic Cost of Inflammation, Increases Oxidative Respiration, and Improves Exercise Tolerance. Proc. Natl. Acad. Sci. USA 2017, 114, 2313–2318. [Google Scholar] [CrossRef]
- Cavalli, G.; Koenders, M.; Kalabokis, V.; Kim, J.; Choon Tan, A.; Garlanda, C.; Mantovani, A.; Dagna, L.; Joosten, L.A.B.; Dinarello, C.A. Treating Experimental Arthritis with the Innate Immune Inhibitor Interleukin-37 Reduces Joint and Systemic Inflammation. Rheumatology (Oxford) 2017, 56, 2256. [Google Scholar] [CrossRef] [PubMed]
- Grimsby, S.; Jaensson, H.; Dubrovska, A.; Lomnytska, M.; Hellman, U.; Souchelnytskyi, S. Proteomics-Based Identification of Proteins Interacting with Smad3: SREBP-2 Forms a Complex with Smad3 and Inhibits Its Transcriptional Activity. FEBS Lett. 2004, 577, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kulk, N.; Nold, M.F.; Gräf, R.; Kim, S.-H.; Reinhardt, D.; Dinarello, C.A.; Bufler, P. The IL-1 Family Member 7b Translocates to the Nucleus and down-Regulates Proinflammatory Cytokines. J. Immunol. Baltim. 2008, 180, 5477–5482. [Google Scholar] [CrossRef] [PubMed]
- Bulau, A.-M.; Nold, M.F.; Li, S.; Nold-Petry, C.A.; Fink, M.; Mansell, A.; Schwerd, T.; Hong, J.; Rubartelli, A.; Dinarello, C.A.; et al. Role of Caspase-1 in Nuclear Translocation of IL-37, Release of the Cytokine, and IL-37 Inhibition of Innate Immune Responses. Proc. Natl. Acad. Sci. USA 2014, 111, 2650–2655. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, Y.; Guo, C.; Wang, L.; Chu, H.; Zhu, F.; Li, Y.; Wang, X.; Wang, Q.; Zhao, W.; et al. IL-37 Isoform D Downregulates pro-Inflammatory Cytokines Expression in a Smad3-Dependent Manner. Cell Death Dis. 2018, 9, 582. [Google Scholar] [CrossRef]
- Su, Z.; Tao, X. Current Understanding of IL-37 in Human Health and Disease. Front. Immunol. 2021, 12, 696605. [Google Scholar] [CrossRef]
- Nascimento, C.; Ferreira, F. Tumor Microenvironment of Human Breast Cancer, and Feline Mammary Carcinoma as a Potential Study Model. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188587. [Google Scholar] [CrossRef]
- Christodoulou, M.-I.; Zaravinos, A. New Clinical Approaches and Emerging Evidence on Immune-Checkpoint Inhibitors as Anti-Cancer Therapeutics: CTLA-4 and PD-1 Pathways and Beyond. Crit. Rev. Immunol. 2019, 39, 379–408. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; He, P.; Han, J.; Sun, C. Interleukin-37 Suppresses Hepatocellular Carcinoma Growth through Inhibiting M2 Polarization of Tumor-Associated Macrophages. Mol. Immunol. 2020, 122, 13–20. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, J.-J.; Zhou, Z.-Q.; Pan, Q.-Z.; Zhu, Q.; Tang, Y.; Xia, J.-C.; Weng, D.-S. IL-37 Induces Anti-Tumor Immunity by Indirectly Promoting Dendritic Cell Recruitment and Activation in Hepatocellular Carcinoma. Cancer Manag. Res. 2019, 11, 6691–6702. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, F.-L.; Hu, Y.-W.; Wang, X.-Y.; Zhao, F.-L.; Zhou, P.; Hu, J.; Xiao, Y.-Y.; Hu, Z.-L.; Guo, M.-F.; et al. Interleukin-37 Promotes Colitis-Associated Carcinogenesis via SIGIRR-Mediated Cytotoxic T Cells Dysfunction. Signal Transduct. Target. Ther. 2022, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Osborne, D.G.; Domenico, J.; Fujita, M. Expression of IL-37 Induces a Regulatory T-Cell-like Phenotype and Function in Jurkat Cells. Cells 2022, 11, 2565. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Zhu, Y.; Teo, H.Y.; Liu, Y.; Song, Y.; Lim, H.Y.; Binte Hanafi, Z.; Angeli, V.; Liu, H. The Indirect Antiangiogenic Effect of IL-37 in the Tumor Microenvironment. J. Leukoc. Biol. 2020, 107, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, M.; Guo, C.; Chu, H.; Li, W.; Chen, X.; Wang, X.; Li, Y.; Jia, Y.; Koussatidjoa, S.; et al. Intracellular Mature IL-37 Suppresses Tumor Metastasis via Inhibiting Rac1 Activation. Oncogene 2018, 37, 1095–1106. [Google Scholar] [CrossRef]
- Christodoulou, P.; Kyriakou, T.-C.; Boutsikos, P.; Andreou, M.; Ji, Y.; Xu, D.; Papageorgis, P.; Christodoulou, M.-I. Aberrant Expression and Prognostic Potential of IL-37 in Human Lung Adenocarcinoma. Biomedicines 2022, 10, 3037. [Google Scholar] [CrossRef]
- Zhu, B.; Luo, J.; Jiang, Y.; Yu, L.; Liu, M.; Fu, J. Prognostic Significance of Nomograms Integrating IL-37 Expression, Neutrophil Level, and MMR Status in Patients with Colorectal Cancer. Cancer Med. 2018, 7, 3682–3694. [Google Scholar] [CrossRef]
- Wei, X.; Li, Y.; Zhang, G.; Wang, N.; Mi, M.; Xin, Y.; Jiang, H.; Sun, C. IL-37 Was Involved in Progress of Acute Myeloid Leukemia Through Regulating IL-6 Expression. Cancer Manag. Res. 2021, 13, 3393–3402. [Google Scholar] [CrossRef]
- Farahani, N.; Mohagheghi, F.; Mosayebi, G.; Ghazavi, A.; Ganji, A. Reduced IL-37 Gene Expression and CD8 T Lymphocytes in Patients with Metastatic Breast Cancer. Breast Dis. 2021, 40, 235–240. [Google Scholar] [CrossRef]
- Ding, L.; Zhao, X.; Zhu, N.; Zhao, M.; Hu, Q.; Ni, Y. The Balance of Serum IL-18/IL-37 Levels Is Disrupted during the Development of Oral Squamous Cell Carcinoma. Surg. Oncol. 2020, 32, 99–107. [Google Scholar] [CrossRef]
- Osborne, D.G.; Domenico, J.; Luo, Y.; Reid, A.L.; Amato, C.; Zhai, Z.; Gao, D.; Ziman, M.; Dinarello, C.A.; Robinson, W.A.; et al. Interleukin-37 Is Highly Expressed in Regulatory T Cells of Melanoma Patients and Enhanced by Melanoma Cell Secretome. Mol. Carcinog. 2019, 58, 1670–1679. [Google Scholar] [CrossRef]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential Scaling of Single-Cell RNA-Seq in the Past Decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Crispen, P.L.; Kusmartsev, S. Mechanisms of Immune Evasion in Bladder Cancer. Cancer Immunol. Immunother. 2020, 69, 3–14. [Google Scholar] [CrossRef]
- Ascione, C.M.; Napolitano, F.; Esposito, D.; Servetto, A.; Belli, S.; Santaniello, A.; Scagliarini, S.; Crocetto, F.; Bianco, R.; Formisano, L. Role of FGFR3 in Bladder Cancer: Treatment Landscape and Future Challenges. Cancer Treat. Rev. 2023, 115, 102530. [Google Scholar] [CrossRef] [PubMed]
- Haghshenas, M.R.; Hosseini, S.R.; Fattahi, M.J.; Malekzadeh, M.; Ariafar, A.; Ghaderi, A. Elevated IL-37 Serum Levels in Patients with Transitional Cell Carcinoma of Bladder. Iran. J. Immunol. 2021, 18, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.Com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder Cancer. Nat. Rev. Dis. Primer 2017, 3, 17022. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.M.; Decastro, G.J.; Steinberg, G.D. Medscape Urothelial Carcinoma of the Bladder: Definition, Treatment and Future Efforts. Nat. Rev. Urol. 2011, 8, 631–642. [Google Scholar] [CrossRef] [PubMed]
- van Rhijn, B.W.G.; Burger, M.; Lotan, Y.; Solsona, E.; Stief, C.G.; Sylvester, R.J.; Witjes, J.A.; Zlotta, A.R. Recurrence and Progression of Disease in Non-Muscle-Invasive Bladder Cancer: From Epidemiology to Treatment Strategy. Eur. Urol. 2009, 56, 430–442. [Google Scholar] [CrossRef]
- Babjuk, M.; Böhle, A.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Hernández, V.; Kaasinen, E.; Palou, J.; Rouprêt, M.; et al. EAU Guidelines on Non-Muscle-Invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur. Urol. 2017, 71, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Franks, L.M.; Rigby, C. Letter: HeLa Cells and RT4 Cells. Science 1975, 188, 168. [Google Scholar] [CrossRef]
- O’Toole, C.M.; Tiptaft, R.C.; Stevens, A. HLA Antigen Expression on Urothelial Cells: Detection by Antibody-Dependent Cell-Mediated Cytotoxicity. Int. J. Cancer 1982, 29, 391–395. [Google Scholar] [CrossRef]
- Liu, H.; Tan, Q.; Geddie, W.R.; Jewett, M.A.S.; Phillips, N.; Ke, D.; Simmons, C.A.; Sun, Y. Biophysical Characterization of Bladder Cancer Cells with Different Metastatic Potential. Cell Biochem. Biophys. 2014, 68, 241–246. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of P53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Lamy, A.; Gobet, F.; Laurent, M.; Blanchard, F.; Varin, C.; Moulin, C.; Andreou, A.; Frebourg, T.; Pfister, C. Molecular Profiling of Bladder Tumors Based on the Detection of FGFR3 and TP53 Mutations. J. Urol. 2006, 176, 2686–2689. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Rentsch, C.A.; Müller, D.C.; Ruiz, C.; Bubendorf, L. Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma: A Step Closer to Clinical Translation? Eur. Urol. 2017, 72, 960–961. [Google Scholar] [CrossRef]
- Lang, G.A.; Iwakuma, T.; Suh, Y.-A.; Liu, G.; Rao, V.A.; Parant, J.M.; Valentin-Vega, Y.A.; Terzian, T.; Caldwell, L.C.; Strong, L.C.; et al. Gain of Function of a P53 Hot Spot Mutation in a Mouse Model of Li-Fraumeni Syndrome. Cell 2004, 119, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A Resource for Therapeutic Biomarker Discovery in Cancer Cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Cazier, J.-B.; Rao, S.R.; McLean, C.M.; Walker, A.K.; Wright, B.J.; Jaeger, E.E.M.; Kartsonaki, C.; Marsden, L.; Yau, C.; Camps, C.; et al. Whole-Genome Sequencing of Bladder Cancers Reveals Somatic CDKN1A Mutations and Clinicopathological Associations with Mutation Burden. Nat. Commun. 2014, 5, 3756. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Bourn, J.; Cekanova, M. Mutations of P53 Decrease Sensitivity to the Anthracycline Treatments in Bladder Cancer Cells. Oncotarget 2018, 9, 28514–28531. [Google Scholar] [CrossRef]
- Rehman, H.; Chandrashekar, D.S.; Balabhadrapatruni, C.; Nepal, S.; Balasubramanya, S.A.H.; Shelton, A.K.; Skinner, K.R.; Ma, A.-H.; Rao, T.; Agarwal, S.; et al. ARID1A-Deficient Bladder Cancer Is Dependent on PI3K Signaling and Sensitive to EZH2 and PI3K Inhibitors. JCI Insight 2022, 7, e155899. [Google Scholar] [CrossRef]
- Hodges, C.; Kirkland, J.G.; Crabtree, G.R. The Many Roles of BAF (MSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026930. [Google Scholar] [CrossRef]
- Mathur, R. ARID1A Loss in Cancer: Towards a Mechanistic Understanding. Pharmacol. Ther. 2018, 190, 15–23. [Google Scholar] [CrossRef]
- Loskog, A.; Ninalga, C.; Paul-Wetterberg, G.; de la Torre, M.; Malmström, P.-U.; Tötterman, T.H. Human Bladder Carcinoma Is Dominated by T-Regulatory Cells and Th1 Inhibitory Cytokines. J. Urol. 2007, 177, 353–358. [Google Scholar] [CrossRef]
- Tsai, Y.-S.; Jou, Y.-C.; Tung, C.-L.; Lin, C.-T.; Shen, C.-H.; Chen, S.-Y.; Tsai, H.-T.; Lai, C.-L.; Wu, C.-L.; Tzai, T.-S. Loss of Nuclear Prothymosin-α Expression Is Associated with Disease Progression in Human Superficial Bladder Cancer. Virchows Arch. Int. J. Pathol. 2014, 464, 717–724. [Google Scholar] [CrossRef]
- Miyake, M.; Tatsumi, Y.; Gotoh, D.; Ohnishi, S.; Owari, T.; Iida, K.; Ohnishi, K.; Hori, S.; Morizawa, Y.; Itami, Y.; et al. Regulatory T Cells and Tumor-Associated Macrophages in the Tumor Microenvironment in Non-Muscle Invasive Bladder Cancer Treated with Intravesical Bacille Calmette-Guérin: A Long-Term Follow-Up Study of a Japanese Cohort. Int. J. Mol. Sci. 2017, 18, 2186. [Google Scholar] [CrossRef] [PubMed]
- Baras, A.S.; Drake, C.; Liu, J.-J.; Gandhi, N.; Kates, M.; Hoque, M.O.; Meeker, A.; Hahn, N.; Taube, J.M.; Schoenberg, M.P.; et al. The Ratio of CD8 to Treg Tumor-Infiltrating Lymphocytes Is Associated with Response to Cisplatin-Based Neoadjuvant Chemotherapy in Patients with Muscle Invasive Urothelial Carcinoma of the Bladder. Oncoimmunology 2016, 5, e1134412. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, M.F.; Schneider, A.K.; Cesson, V.; Dartiguenave, F.; Lucca, I.; Jichlinski, P.; Nardelli-Haefliger, D.; Derré, L. Conventional and PD-L1-Expressing Regulatory T Cells Are Enriched During BCG Therapy and May Limit Its Efficacy. Eur. Urol. 2018, 74, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.K.; Chevalier, M.F.; Derré, L. The Multifaceted Immune Regulation of Bladder Cancer. Nat. Rev. Urol. 2019, 16, 613–630. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Powles, T.; Shi, L.; Zhang, L.; Ingersoll, M.A.; Lu, Y.-J. Bladder Cancer, a Unique Model to Understand Cancer Immunity and Develop Immunotherapy Approaches. J. Pathol. 2019, 249, 151–165. [Google Scholar] [CrossRef]
- Ferro, M.; Babă, D.-F.; de Cobelli, O.; Musi, G.; Lucarelli, G.; Terracciano, D.; Porreca, A.; Busetto, G.M.; Del Giudice, F.; Soria, F.; et al. Neutrophil Percentage-to-Albumin Ratio Predicts Mortality in Bladder Cancer Patients Treated with Neoadjuvant Chemotherapy Followed by Radical Cystectomy. Future Sci. OA 2021, 7, FSO709. [Google Scholar] [CrossRef]
- Christodoulou, M.I.; Kapsogeorgou, E.K.; Moutsopoulos, H.M. Characteristics of the Minor Salivary Gland Infiltrates in Sjögren’s Syndrome. J. Autoimmun. 2010, 34, 400–407. [Google Scholar] [CrossRef]
- Christodoulou, M.I.; Kapsogeorgou, E.K.; Moutsopoulos, N.M.; Moutsopoulos, H.M. Foxp3+ T-Regulatory Cells in Sjogren’s Syndrome: Correlation with the Grade of the Autoimmune Lesion and Certain Adverse Prognostic Factors. Am. J. Pathol. 2008, 173, 1389–1396. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
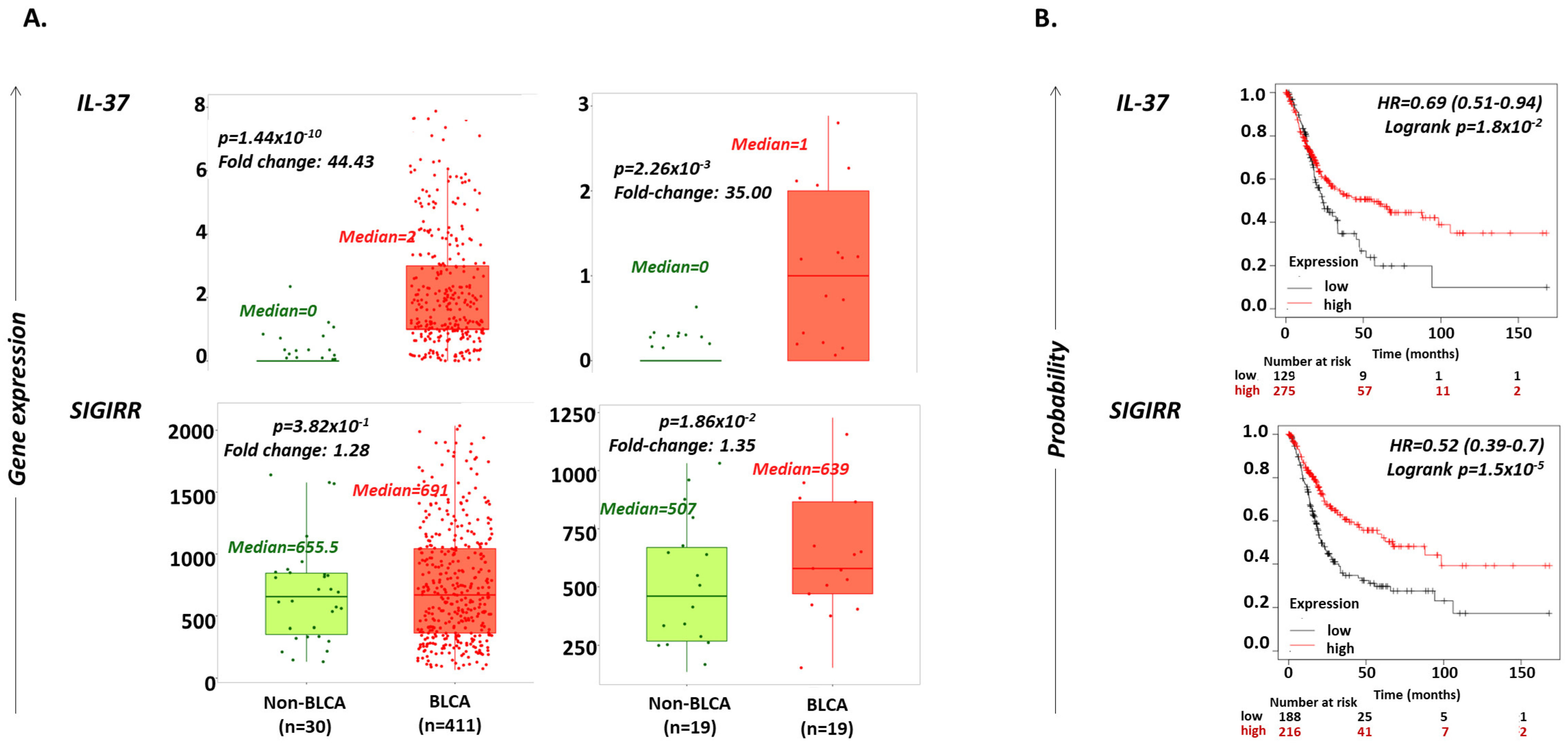

| IL-37 | SIGIRR | |||||||
|---|---|---|---|---|---|---|---|---|
| Non-BLCA vs. BLCA | ||||||||
| n | Levels (Median; Range) | Fold Change | p-Value | n | Levels (Median; Range) | Fold Change | p-Value | |
| Non-BLCA (normal) | 30 | 0 | 30 | 655.5 | ||||
| BLCA | 411 | 2 | 2 | 1.44 × 10−10 | 411 | 691 | 1.05 | 3.82 × 10−1 |
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| Primary tumor | 408 | 0.09 (0–0.73) | 0.09 | NS | 408 | 29.31 (2.30–80.10) | 1.27 | 9.46 × 10−3 |
| Tumors vs. adjacent normal | ||||||||
| Adjacent normal | 19 | 0 | 19 | 507 | ||||
| Tumor | 19 | 1 | 1 | 2.26 × 10−3 | 19 | 639 | 1.26 | 1.86 × 10−2 |
| BLCA histological subtype | ||||||||
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| Papillary | 132 | 0.08 (0–0.91) | 0.08 | NS | 132 | 33.48 (4.25–82.13) | 1.45 | 2.88 × 10−2 |
| Non-papillary | 271 | 0.09 (0–0.63) | 0.09 | 2.06 × 10−10 | 271 | 26.84 (2.30–75.22) | 1.16 | NS |
| Cancer stage | ||||||||
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| Stage 1 | 2 | 1.03 (0–2.06) | 1.03 | NS | 2 | 40.08 (35.86–44.30) | 1.73 | NS |
| Stage 2 | 129 | 0.1 (0–0.64) | 0.1 | 6.38 × 10−5 | 129 | 33.13 (3.81–84.25) | 1.43 | 9.77 × 10−4 |
| Stage 3 | 137 | 0.08 (0–0.52) | 0.08 | 4.02 × 10−6 | 137 | 26.84 (2.30–70.21) | 1.16 | NS |
| Stage 4 | 132 | 0.09 (0–0.73) | 0.09 | NS | 132 | 27.23 (4.33–78.70) | 1.18 | NS |
| Nodal metastasis status | ||||||||
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| N0 | 237 | 0.09 (0–0.64) | 0.09 | 1.83 × 10−8 | 237 | 29.31 (2.30–71.13) | 1.27 | 1.45 × 10−2 |
| N1 | 46 | 0.09 (0–0.4) | 0.09 | 1.82 × 10−4 | 46 | 26.85 (4.33–77.7) | 1.16 | NS |
| N2 | 75 | 0.09 (0–1.05) | 0.09 | 1.62 × 10−7 | 75 | 27.16 (4.53–69.18) | 1.18 | NS |
| N3 | 8 | 0.26 (0–0.95) | 0.26 | NS | 8 | 27.39 (5.29–59.47) | 1.19 | NS |
| TP53 mutation status | ||||||||
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| Mutant | 193 | 0.116 (0–0.779) | 0.116 | 5.64 × 10−14 | 193 | 27.84 (2.3–84.45) | 1.20 | 2.45 × 10−2 |
| Non-mutant | 215 | 0.069 (0–0616) | 0.069 | NS | 215 | 29.68 (3.67–71.13) | 1.28 | 6.38 × 10−3 |
| ARID1A mutation status | ||||||||
| Normal | 19 | 0 (0–0) | 19 | 23.11 (11.29–48.4) | ||||
| Mutant | 76 | 0.096 (0–0.824) | 0.096 | 3.63 × 10−5 | 76 | 28.93 (2.3–70.52) | 1.25 | 1.80 × 10−2 |
| Non-mutant | 324 | 0.091 (0–0.641) | 0.091 | NS | 324 | 29.69 (3.81–82.13) | 1.28 | 8.96 × 10−3 |
| N | Median (Range) | Fold Change | p-Value | ||
|---|---|---|---|---|---|
| Total BLCA biopsies | |||||
| Low-grade | 18 | 0.14 (0.007–25.18) | High- over low-grade: | 2.79 | 0.0269 |
| High-grade | 49 | 0.39 (0.024–50.83) | |||
| Grade 1 | 6 | 0.03 (0.007–1.88) | Grade 2 over Grade 1 | 11.67 | 0.049 |
| Grade 2 | 16 | 0.35 (0.029–25.18) | Grade 3 over Grade 2 | 0.94 | NS |
| Grade 3 | 45 | 0.33 (0.024–50.85) | Grade 3 over Grade 1 | 11.00 | 0.017 |
| Non-muscle-invasive BLCA biopsies | |||||
| Low-grade | 18 | 0.14 (0.007–25.18) | High- over low-grade: | 2.79 | 0.0269 |
| High-grade | 16 | 0.39 (0.024–50.83) | |||
| Grade 1 | 6 | 0.03 (0.007–1.88) | Grade 2 over Grade 1 | 11.67 | 0.049 |
| Grade 2 | 16 | 0.34 (0.029–25.18) | Grade 3 over Grade 2 | 0.94 | NS |
| Grade 3 | 12 | 0.35 (0.026–11.34) | Grade 3 over Grade 1 | 11.00 | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papasavva, M.; Amvrosiou, S.; Pilala, K.-M.; Soureas, K.; Christodoulou, P.; Ji, Y.; Stravodimos, K.; Xu, D.; Scorilas, A.; Avgeris, M.; et al. Deregulated Expression of IL-37 in Patients with Bladder Urothelial Cancer: The Diagnostic Potential of the IL-37e Isoform. Int. J. Mol. Sci. 2023, 24, 9258. https://doi.org/10.3390/ijms24119258
Papasavva M, Amvrosiou S, Pilala K-M, Soureas K, Christodoulou P, Ji Y, Stravodimos K, Xu D, Scorilas A, Avgeris M, et al. Deregulated Expression of IL-37 in Patients with Bladder Urothelial Cancer: The Diagnostic Potential of the IL-37e Isoform. International Journal of Molecular Sciences. 2023; 24(11):9258. https://doi.org/10.3390/ijms24119258
Chicago/Turabian StylePapasavva, Maria, Styliana Amvrosiou, Katerina-Marina Pilala, Konstantinos Soureas, Panayiota Christodoulou, Yuan Ji, Konstantinos Stravodimos, Damo Xu, Andreas Scorilas, Margaritis Avgeris, and et al. 2023. "Deregulated Expression of IL-37 in Patients with Bladder Urothelial Cancer: The Diagnostic Potential of the IL-37e Isoform" International Journal of Molecular Sciences 24, no. 11: 9258. https://doi.org/10.3390/ijms24119258






