Insights into the Alcyoneusvirus Adsorption Complex
Abstract
1. Introduction
2. Results
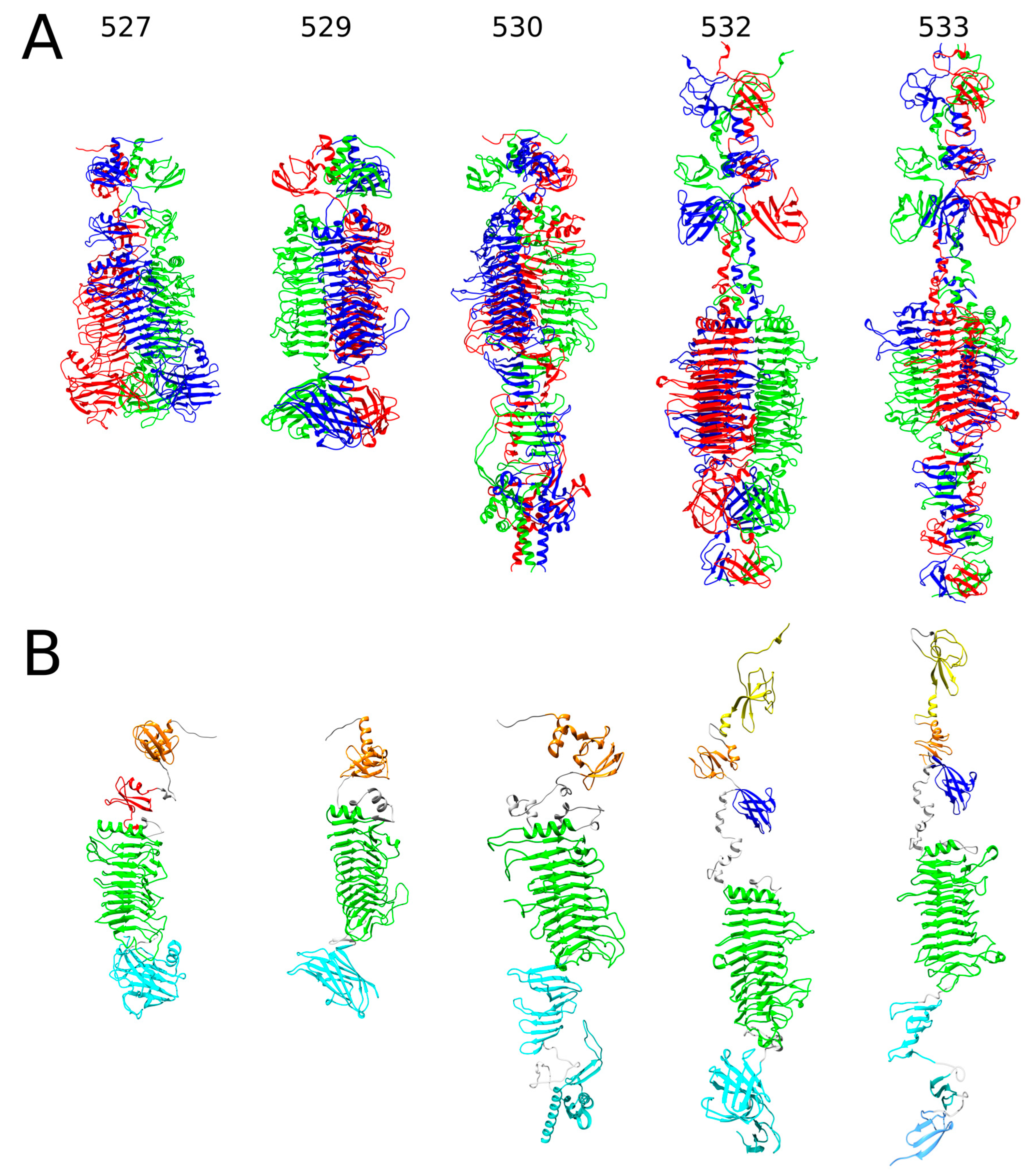
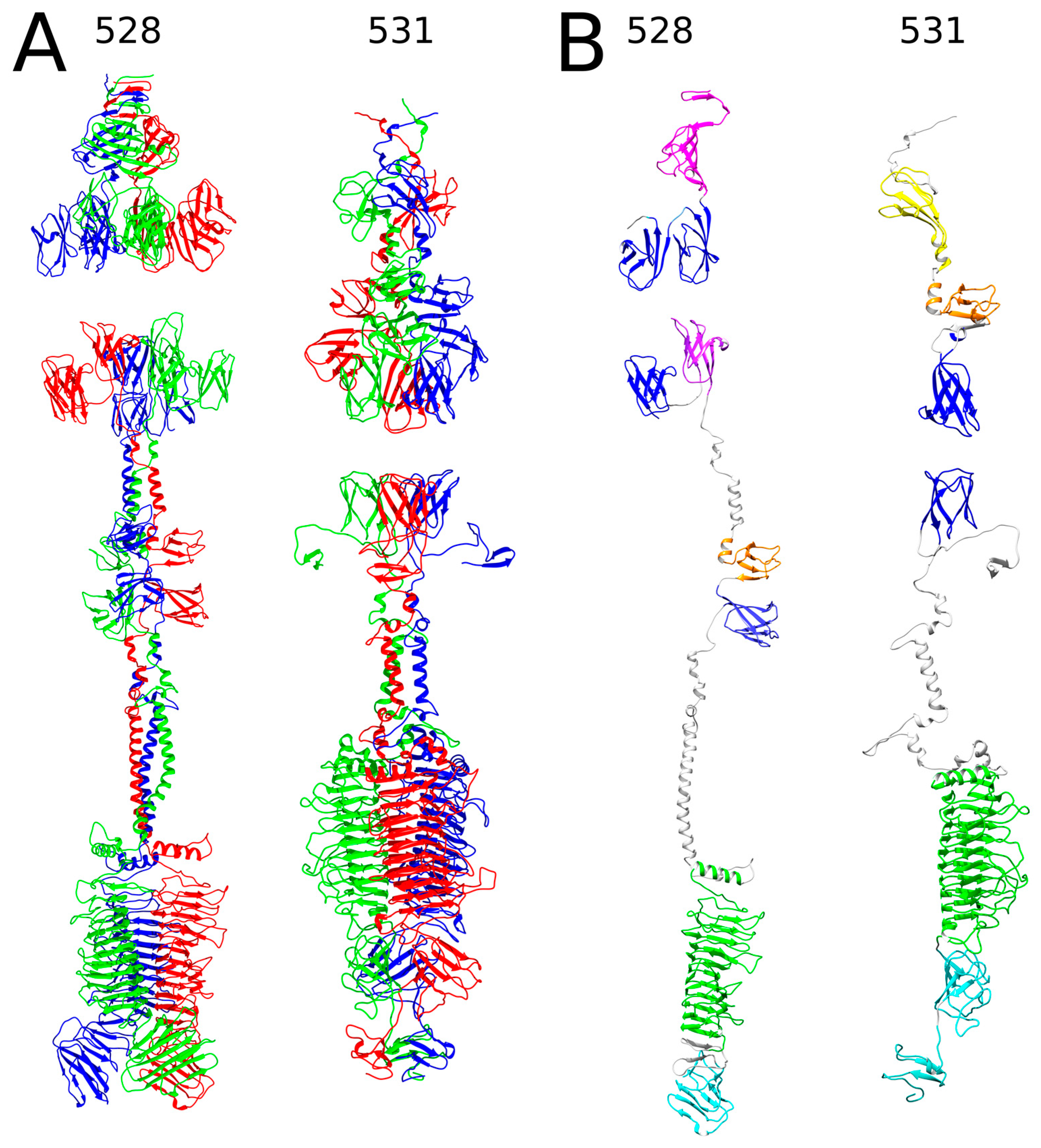
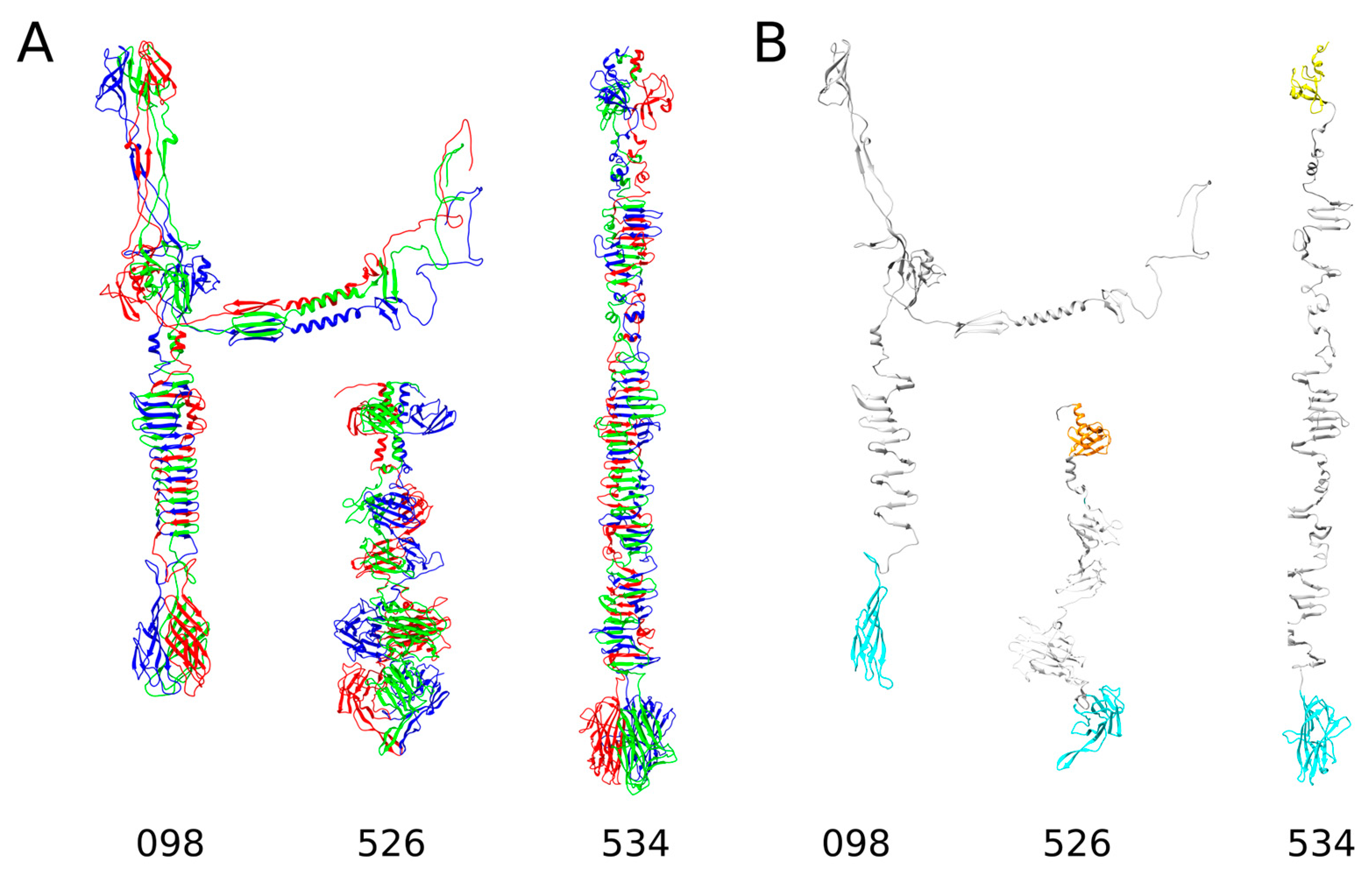
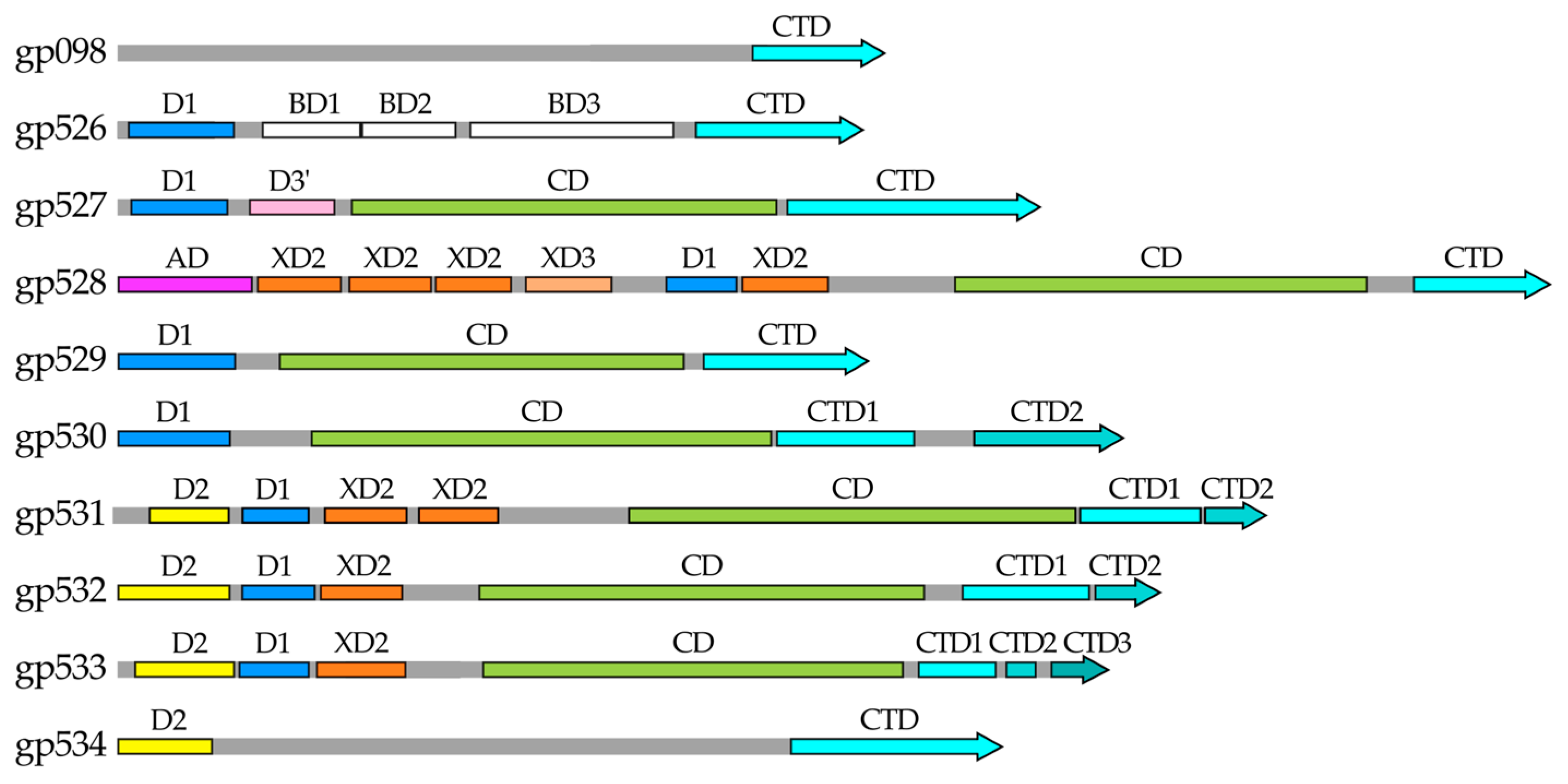
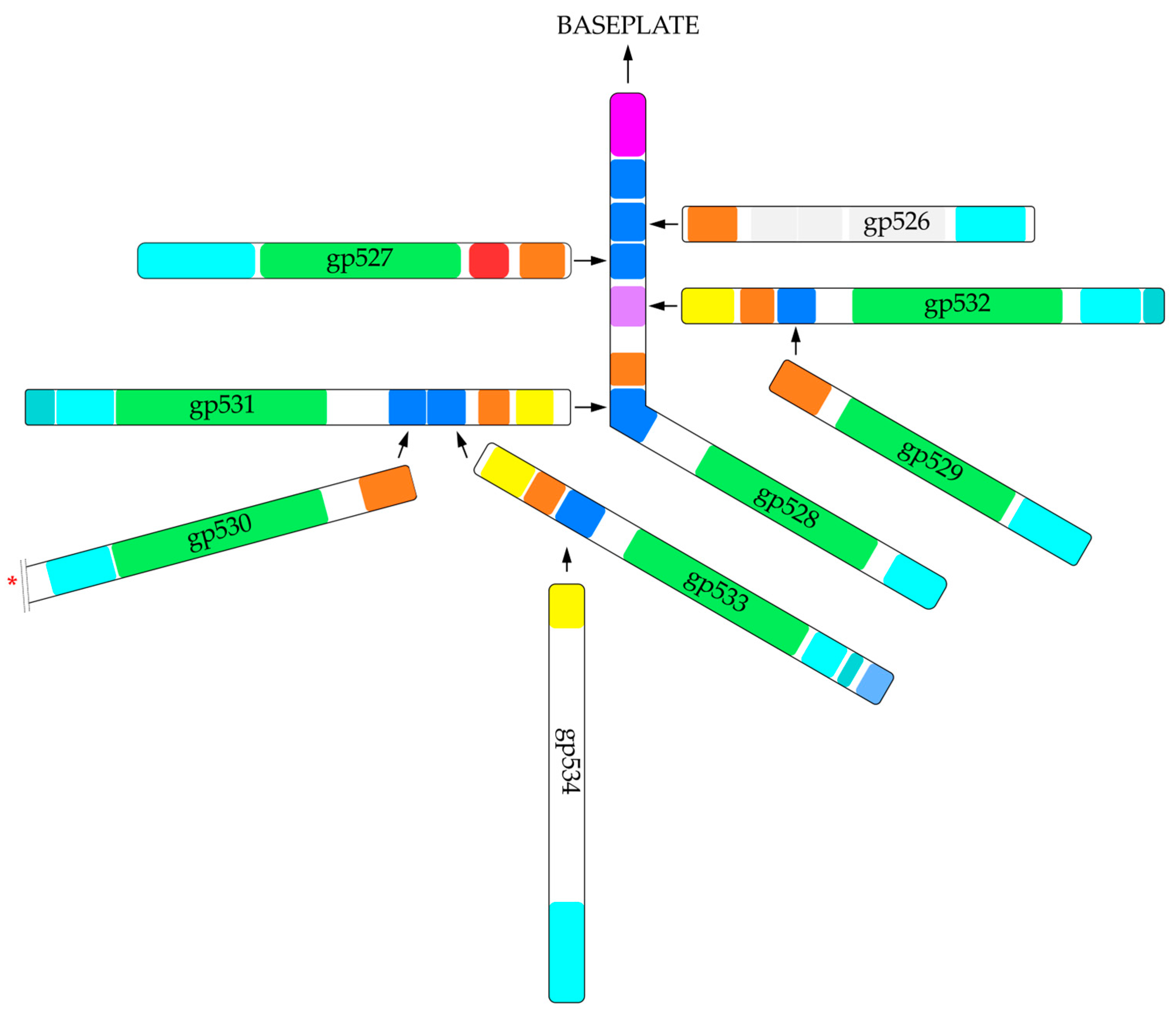
2.1. In Silico Analysis of RaK2 TFPs
2.2. Production of Recombinant RaK2 TFPs
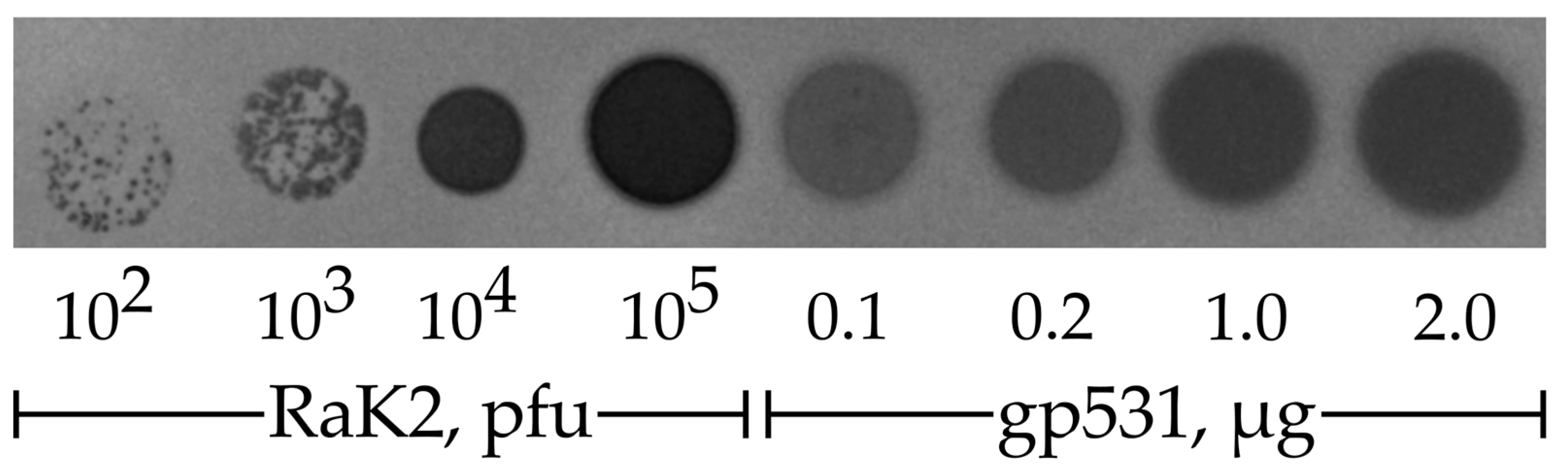
2.3. Depolymerase Activity Test
2.4. Western Blot Analysis of Antisera Raised against Recombinant RaK2 TFPs
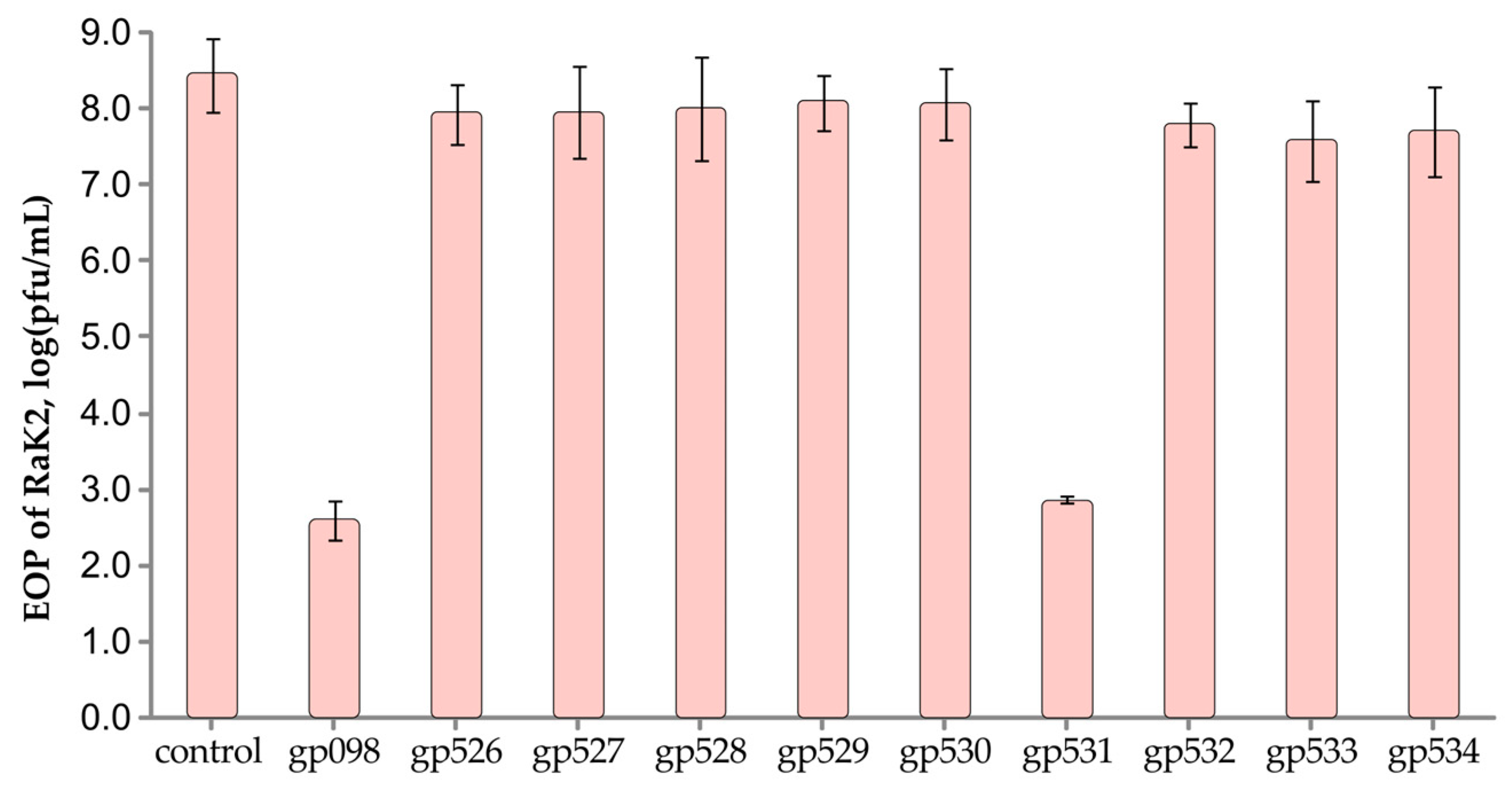
2.5. RaK2 Inhibition with Antisera
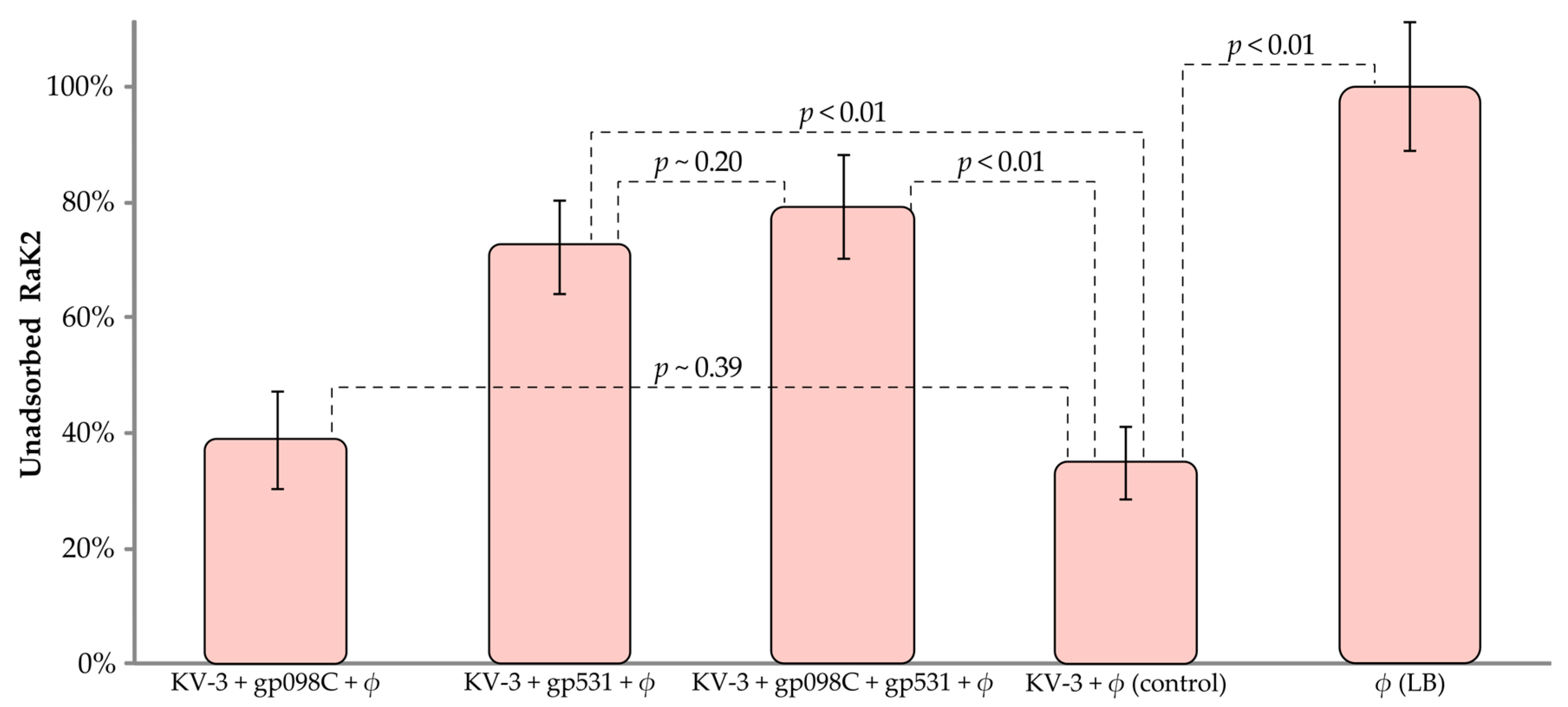
2.6. Adsorption Inhibition Assay
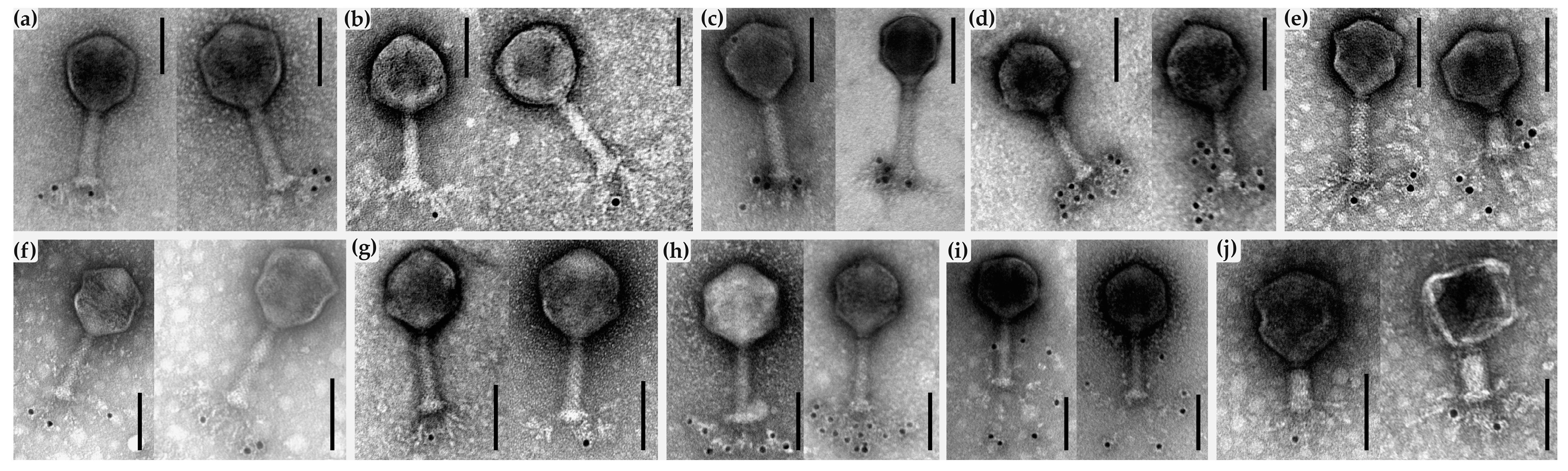
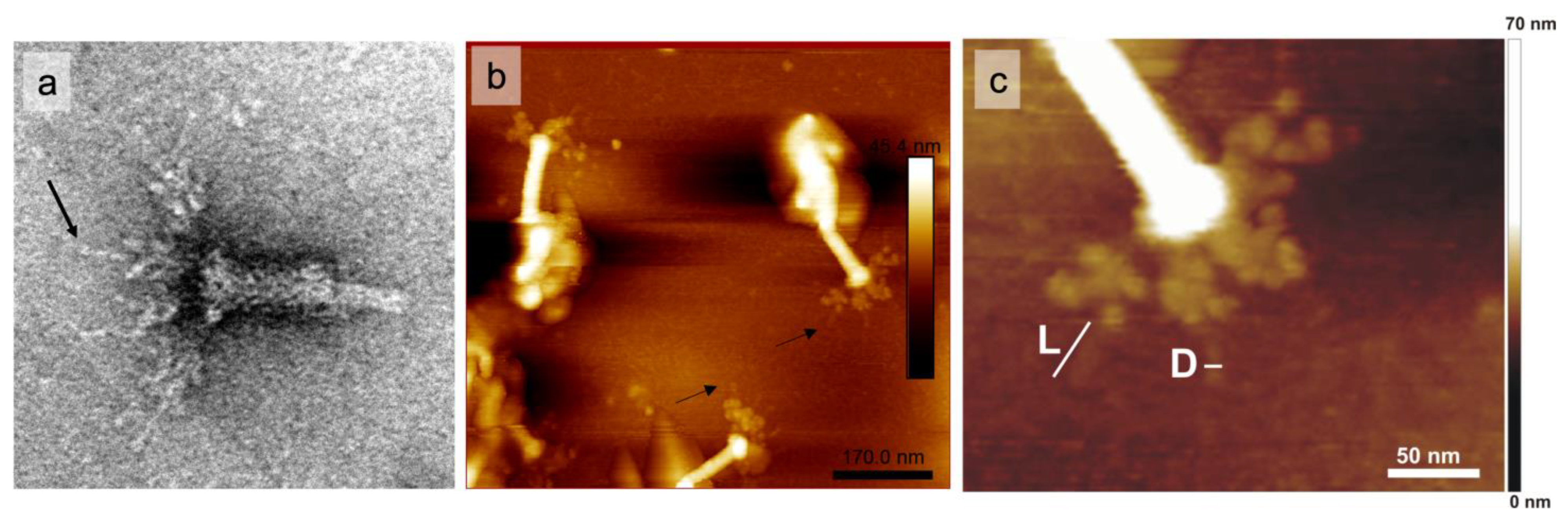
2.7. AFM and Immunogold-TEM Analyses
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis
4.2. Phage, Bacterial Strains, and Growth Conditions
4.3. Plasmid Construction
4.4. Protein Production and Purification
4.5. Depolymerase Activity Assay
4.6. RaK2 Adsorption Inhibition with Recombinant Proteins
4.7. Antisera Production
4.8. RaK2 Adsorption Inhibition with Policlonal Antiserum
4.9. Immunogold Labeling and TEM Analysis
4.10. AFM Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses 2018, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Philipson, L. Attachment of viruses to cell receptors and penetration of viruses into cells. In Textbook of Medical Virology; Elsevier: Amsterdam, The Netherlands, 1983; pp. 51–56. [Google Scholar] [CrossRef]
- Veesler, D.; Cambillau, C. A Common Evolutionary Origin for Tailed-Bacteriophage Functional Modules and Bacterial Machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Dennehy, J.J.; Abedon, S.T. Adsorption: Phage Acquisition of Bacteria. In Bacteriophages: Biology, Technology, Therapy; Springer International Publishing: Cham, Switzerland, 2021; pp. 93–117. [Google Scholar]
- Leprince, A.; Mahillon, J. Phage Adsorption to Gram-Positive Bacteria. Viruses 2023, 15, 196. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Bacteriophage Classifcation. In Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2004; pp. 67–89. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Sanz-Gaitero, M.; Seoane-Blanco, M.; van Raaij, M.J. Structure and Function of Bacteriophages. In Bacteriophages: Biology, Technology, Therapy; Springer International Publishing: Cham, Switzerland, 2021; pp. 19–91. [Google Scholar]
- Taylor, N.M.I.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
- Yap, M.L.; Rossmann, M.G. Structure and function of bacteriophage T4. Futur. Microbiol. 2014, 9, 1319–1327. [Google Scholar] [CrossRef]
- Gil, J.; Paulson, J.; Brown, M.; Zahn, H.; Nguyen, M.M.; Eisenberg, M.; Erickson, S. Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins. Viruses 2023, 15, 286. [Google Scholar] [CrossRef]
- Sørensen, A.N.; Woudstra, C.; Sørensen, M.C.H.; Brøndsted, L. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput. Struct. Biotechnol. J. 2021, 19, 4854–4867. [Google Scholar] [CrossRef]
- Walter, M.; Fiedler, C.; Grassl, R.; Biebl, M.; Rachel, R.; Hermo-Parrado, X.L.; Llamas-Saiz, A.L.; Seckler, R.; Miller, S.; van Raaij, M.J. Structure of the Receptor-Binding Protein of Bacteriophage Det7: A Podoviral Tail Spike in a Myovirus. J. Virol. 2008, 82, 2265–2273. [Google Scholar] [CrossRef]
- Broeker, N.K.; Roske, Y.; Valleriani, A.; Stephan, M.S.; Andres, D.; Koetz, J.; Heinemann, U.; Barbirz, S. Time-resolved DNA release from an O-antigen–specific Salmonella bacteriophage with a contractile tail. J. Biol. Chem. 2019, 294, 11751–11761. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Lemire, S.; Grimon, D.; Dams, D.; Maciejewska, B.; Lu, T.; Drulis-Kawa, Z.; Briers, Y. Engineering the Modular Receptor-Binding Proteins of Klebsiella Phages Switches Their Capsule Serotype Specificity. mBio 2021, 12, e00455-21. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2019, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Leiman, P.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front. Microbiol. 2019, 10, 2649. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Sycheva, L.V.; Shneider, M.M.; Popova, A.V.; Ziganshin, R.H.; Volozhantsev, N.V.; Miroshnikov, K.A.; Leiman, P.G. Crystal Structure of the Putative Tail Fiber Protein Gp53 from the Acinetobacter Baumannii Bacteriophage AP22. bioRxiv 2019, 518761. [Google Scholar] [CrossRef]
- Rostøl, J.T.; Marraffini, L. (Ph)ighting Phages: How Bacteria Resist Their Parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Blundell-Hunter, G.; Enright, M.C.; Negus, D.; Dorman, M.J.; Beecham, G.E.; Pickard, D.J.; Wintachai, P.; Voravuthikunchai, S.P.; Thomson, N.R.; Taylor, P.W. Characterisation of Bacteriophage-Encoded Depolymerases Selective for Key Klebsiella pneumoniae Capsular Exopolysaccharides. Front. Cell. Infect. Microbiol. 2021, 11, 686090. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Leiman, P.G.; Battisti, A.J.; Bowman, V.D.; Stummeyer, K.; Mühlenhoff, M.; Gerardy-Schahn, R.; Scholl, D.; Molineux, I.J. The Structures of Bacteriophages K1E and K1-5 Explain Processive Degradation of Polysaccharide Capsules and Evolution of New Host Specificities. J. Mol. Biol. 2007, 371, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.; Taylor, N.M.I.; Buth, S.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, R.; Costa, A.R.; Cassidy, C.K.; Otwinowska, A.; Williams, V.C.J.; Latka, A.; Stansfeld, P.J.; Drulis-Kawa, Z.; Briers, Y.; Pelt, D.M.; et al. High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. Nat. Commun. 2022, 13, 7241. [Google Scholar] [CrossRef]
- Greenfield, J.; Shang, X.; Luo, H.; Zhou, Y.; Heselpoth, R.D.; Nelson, D.C.; Herzberg, O. Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3). Sci. Rep. 2019, 9, 7349. [Google Scholar] [CrossRef]
- Chen, C.; Bales, P.; Greenfield, J.; Heselpoth, R.D.; Nelson, D.C.; Herzberg, O. Crystal Structure of ORF210 from E. coli O157:H1 Phage CBA120 (TSP1), a Putative Tailspike Protein. PLoS ONE 2014, 9, e93156. [Google Scholar] [CrossRef]
- Greenfield, J.; Shang, X.; Luo, H.; Zhou, Y.; Linden, S.B.; Heselpoth, R.D.; Leiman, P.G.; Nelson, D.C.; Herzberg, O. Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7. Sci. Rep. 2020, 10, 15402. [Google Scholar] [CrossRef]
- Chao, K.L.; Shang, X.; Greenfield, J.; Linden, S.B.; Alreja, A.B.; Nelson, D.C.; Herzberg, O. Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N). Sci. Rep. 2022, 12, 2061. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef]
- Šimoliūnas, E.; Kaliniene, L.; Truncaite, L.; Klausa, V.; Zajančkauskaite, A.; Meškys, R. Genome of Klebsiella sp.-Infecting Bacteriophage vB_KleM_RaK2. J. Virol. 2012, 86, 5406. [Google Scholar] [CrossRef]
- Šimoliūnas, E.; Kaliniene, L.; Truncaite, L.; Zajančkauskaite, A.; Staniulis, J.; Kaupinis, A.; Ger, M.; Valius, M.; Meškys, R. Klebsiella Phage vB_KleM-RaK2—A Giant Singleton Virus of the Family Myoviridae. PLoS ONE 2013, 8, e60717. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-J.; Lin, T.-L.; Chen, C.-C.; Tsai, Y.-T.; Cheng, Y.-H.; Chen, Y.-Y.; Hsieh, P.-F.; Lin, Y.-T.; Wang, J.-T. Klebsiella Phage ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91, e02457-16. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-J.; Lin, T.-L.; Lin, Y.-T.; Su, P.-A.; Chen, C.-T.; Hsieh, P.-F.; Hsu, C.-R.; Chen, C.-C.; Hsieh, Y.-C.; Wang, J.-T. Identification of Capsular Types in Carbapenem-Resistant Klebsiella pneumoniae Strains by wzc Sequencing and Implications for Capsule Depolymerase Treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Taxon Details|ICTV. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202106653 (accessed on 30 March 2023).
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Goulet, A.; Cambillau, C. Structure and Topology Prediction of Phage Adhesion Devices Using AlphaFold2: The Case of Two Oenococcus oeni Phages. Microorganisms 2021, 9, 2151. [Google Scholar] [CrossRef] [PubMed]
- Goulet, A.; Joos, R.; Lavelle, K.; Van Sinderen, D.; Mahony, J.; Cambillau, C. A structural discovery journey of streptococcal phages adhesion devices by AlphaFold2. Front. Mol. Biosci. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Holm, L. Using Dali for Protein Structure Comparison. Methods Mol. Biol. 2020, 2112, 29–42. [Google Scholar] [CrossRef]
- Holm, L.; Rosenström, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef]
- Squeglia, F.; Maciejewska, B.; Łątka, A.; Ruggiero, A.; Briers, Y.; Drulis-Kawa, Z.; Berisio, R. Structural and Functional Studies of a Klebsiella Phage Capsule Depolymerase Tailspike: Mechanistic Insights into Capsular Degradation. Structure 2020, 28, 613–624.e4. [Google Scholar] [CrossRef]
- Müller, J.J.; Barbirz, S.; Heinle, K.; Freiberg, A.; Seckler, R.; Heinemann, U. An Intersubunit Active Site between Supercoiled Parallel β Helices in the Trimeric Tailspike Endorhamnosidase of Shigella flexneri Phage Sf6. Structure 2008, 16, 766–775. [Google Scholar] [CrossRef]
- Kostyuchenko, V.A.; Navruzbekov, G.A.; Kurochkina, L.P.; Strelkov, S.V.; Mesyanzhinov, V.V.; Rossmann, M.G. The structure of bacteriophage T4 gene product 9: The trigger for tail contraction. Structure 1999, 7, 1213–1222. [Google Scholar] [CrossRef]
- Kurochkina, L.P.; Vishnevskiy, A.Y.; Zhemaeva, L.V.; Sykilinda, N.N.; Strelkov, S.V.; Mesyanzhinov, V.V. Structure, stability, and biological activity of bacteriophage T4 gene product 9 probed with mutagenesis and monoclonal antibodies. J. Struct. Biol. 2006, 154, 122–129. [Google Scholar] [CrossRef]
- Kostyuchenko, V.A.; Leiman, P.G.; Chipman, P.R.; Kanamaru, S.; van Raaij, M.J.; Arisaka, F.; Mesyanzhinov, V.V.; Rossmann, M.G. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Mol. Biol. 2003, 10, 688–693. [Google Scholar] [CrossRef]
- Garcia-Doval, C.; Castón, J.R.; Luque, D.; Granell, M.; Otero, J.M.; Llamas-Saiz, A.L.; Renouard, M.; Boulanger, P.; Van Raaij, M.J. Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone. Viruses 2015, 7, 6424–6440. [Google Scholar] [CrossRef]
- Xiang, Y.; Leiman, P.G.; Li, L.; Grimes, S.; Anderson, D.L.; Rossmann, M.G. Crystallographic Insights into the Autocatalytic Assembly Mechanism of a Bacteriophage Tail Spike. Mol. Cell 2009, 34, 375–386. [Google Scholar] [CrossRef]
- Seul, A.; Müller, J.J.; Andres, D.; Stettner, E.; Heinemann, U.; Seckler, R. Bacteriophage P22 tailspike: Structure of the complete protein and function of the interdomain linker. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1336–1345. [Google Scholar] [CrossRef]
- Granell, M.; Namura, M.; Alvira, S.; Kanamaru, S.; Van Raaij, M.J. Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34. Viruses 2017, 9, 168. [Google Scholar] [CrossRef]
- Buth, S.A.; Shneider, M.M.; Scholl, D.; Leiman, P.G. Structure and Analysis of R1 and R2 Pyocin Receptor-Binding Fibers. Viruses 2018, 10, 427. [Google Scholar] [CrossRef]
- Wilson, J.J.; Matsushita, O.; Okabe, A.; Sakon, J. A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 2003, 22, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Rozeboom, H.J.; Bjerkan, T.M.; Kalk, K.H.; Ertesvåg, H.; Holtan, S.; Aachmann, F.L.; Valla, S.; Dijkstra, B.W. Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii. J. Biol. Chem. 2008, 283, 23819–23828. [Google Scholar] [CrossRef] [PubMed]
- Urusova, D.V.; Kinsella, R.L.; Salinas, N.; Haurat, M.F.; Feldman, M.F.; Tolia, N.H. The structure of Acinetobacter-secreted protease CpaA complexed with its chaperone CpaB reveals a novel mode of a T2SS chaperone–substrate interaction. J. Biol. Chem. 2019, 294, 13344–13354. [Google Scholar] [CrossRef] [PubMed]
- Irmscher, T.; Roske, Y.; Gayk, I.; Dunsing, V.; Chiantia, S.; Heinemann, U.; Barbirz, S. Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold. J. Biol. Chem. 2021, 296, 100286. [Google Scholar] [CrossRef]
- Lee, I.-M.; Tu, I.-F.; Yang, F.-L.; Ko, T.-P.; Liao, J.-H.; Lin, N.-T.; Wu, C.-Y.; Ren, C.-T.; Wang, A.H.-J.; Chang, C.-M.; et al. Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage ΦAB6 tailspike protein. Sci. Rep. 2017, 7, srep42711. [Google Scholar] [CrossRef]
- Hymowitz, S.G.; Compaan, D.M.; Yan, M.; Wallweber, H.J.; Dixit, V.M.; Starovasnik, M.A.; de Vos, A.M. The Crystal Structures of EDA-A1 and EDA-A2: Splice Variants with Distinct Receptor Specificity. Structure 2003, 11, 1513–1520. [Google Scholar] [CrossRef]
- McCabe, O.; Spinelli, S.; Farenc, C.; Labbé, M.; Tremblay, D.; Blangy, S.; Oscarson, S.; Moineau, S.; Cambillau, C. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol. Microbiol. 2015, 96, 875–886. [Google Scholar] [CrossRef]
- Hyman, P.; van Raaij, M. Bacteriophage T4 long tail fiber domains. Biophys. Rev. 2018, 10, 463–471. [Google Scholar] [CrossRef]
- North, O.I.; Davidson, A.R. Phage Proteins Required for Tail Fiber Assembly Also Bind Specifically to the Surface of Host Bacterial Strains. J. Bacteriol. 2021, 203, e00406-20. [Google Scholar] [CrossRef]
- Pacheco, B.; Crombet, L.; Loppnau, P.; Cossar, D. A screening strategy for heterologous protein expression in Escherichia coli with the highest return of investment. Protein Expr. Purif. 2012, 81, 33–41. [Google Scholar] [CrossRef]
- Gopal, G.J.; Kumar, A. Strategies for the Production of Recombinant Protein in Escherichia coli. Protein J. 2013, 32, 419–425. [Google Scholar] [CrossRef]
- Tang, S.R.; Somasundaram, B.; Lua, L.H.L. Protein Expression Optimization Strategies in E. coli: A Tailored Approach in Strain Selection and Parallelizing Expression Conditions. In Insoluble Proteins: Methods and Protocols; Springer: New York, NY, USA, 2022; pp. 93–111. [Google Scholar] [CrossRef]
- Ortega, C.; Oppezzo, P.; Correa, A. Overcoming the Solubility Problem in E. coli: Available Approaches for Recombinant Protein Production. In Insoluble Proteins: Methods and Protocols; Springer: New York, NY, USA, 2022; pp. 35–64. [Google Scholar] [CrossRef]
- Hayes, S.; Duhoo, Y.; Neve, H.; Murphy, J.; Noben, J.-P.; Franz, C.M.A.P.; Cambillau, C.; Mahony, J.; Nauta, A.; Van Sinderen, D. Identification of Dual Receptor Binding Protein Systems in Lactococcal 936 Group Phages. Viruses 2018, 10, 668. [Google Scholar] [CrossRef]
- Miernikiewicz, P.; Kłopot, A.; Soluch, R.; Szkuta, P.; Kęska, W.; Hodyra-Stefaniak, K.; Konopka, A.; Nowak, M.; Lecion, D.; Kaźmierczak, Z.; et al. T4 Phage Tail Adhesin Gp12 Counteracts LPS-Induced Inflammation In Vivo. Front. Microbiol. 2016, 7, 1112. [Google Scholar] [CrossRef]
- Vinga, I.; Baptista, C.; Auzat, I.; Petipas, I.; Lurz, R.; Tavares, P.; Santos, M.A.; São-José, C. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 2011, 83, 289–303. [Google Scholar] [CrossRef]
- Boulanger, P.; Jacquot, P.; Plançon, L.; Chami, M.; Engel, A.; Parquet, C.; Herbeuval, C.; Letellier, L. Phage T5 Straight Tail Fiber Is a Multifunctional Protein Acting as a Tape Measure and Carrying Fusogenic and Muralytic Activities. J. Biol. Chem. 2008, 283, 13556–13564. [Google Scholar] [CrossRef]
- Braun, P.; Wolfschläger, I.; Reetz, L.; Bachstein, L.; Jacinto, A.C.; Tocantins, C.; Poppe, J.; Grass, G. Rapid Microscopic Detection of Bacillus anthracis by Fluorescent Receptor Binding Proteins of Bacteriophages. Microorganisms 2020, 8, 934. [Google Scholar] [CrossRef]
- Akuta, T.; Maruyama, T.; Sakuma, C.; Nakagawa, M.; Tomioka, Y.; Entzminger, K.; Fleming, J.K.; Sato, R.; Shibata, T.; Kurosawa, Y.; et al. A New Method to Characterize Conformation-Specific Antibody by a Combination of Agarose Native Gel Electrophoresis and Contact Blotting. Antibodies 2022, 11, 36. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hendrix, R.W. Jumbo Bacteriophages. Poxviruses 2009, 328, 229–240. [Google Scholar] [CrossRef]
- Buttimer, C.; Hendrix, H.; Oliveira, H.; Casey, A.; Neve, H.; McAuliffe, O.; Ross, R.P.; Hill, C.; Noben, J.-P.; O’Mahony, J.; et al. Things Are Getting Hairy: Enterobacteria Bacteriophage vB_PcaM_CBB. Front. Microbiol. 2017, 8, 44. [Google Scholar] [CrossRef]
- Van Raaij, M.J.; Schoehn, G.; Burda, M.R.; Miller, S. Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 2001, 314, 1137–1146. [Google Scholar] [CrossRef]
- Santos, S.B.; Carvalho, C.M.; Sillankorva, S.; Nicolau, A.; Ferreira, E.C.; Azeredo, J. The use of antibiotics to improve phage detection and enumeration by the double-layer agar technique. BMC Microbiol. 2009, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Dupont, K.; Vogensen, F.K.; Neve, H.; Bresciani, J.; Josephsen, J. Identification of the Receptor-Binding Protein in 936-Species Lactococcal Bacteriophages. Appl. Environ. Microbiol. 2004, 70, 5818–5824. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Klausa, V.; Truncaitė, L. Low-temperature T4-like coliphages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20. Arch. Virol. 2010, 155, 871–880. [Google Scholar] [CrossRef] [PubMed]
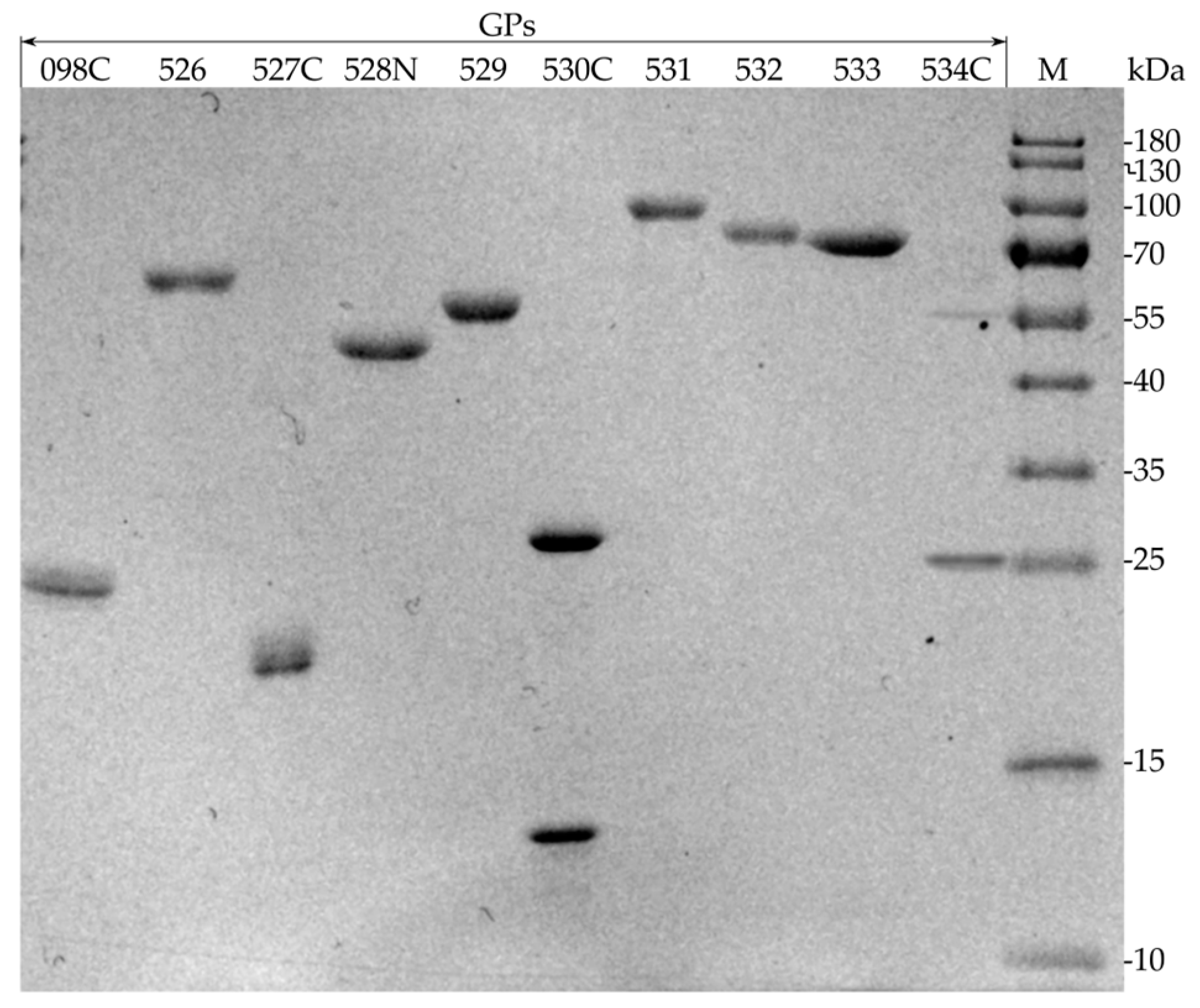
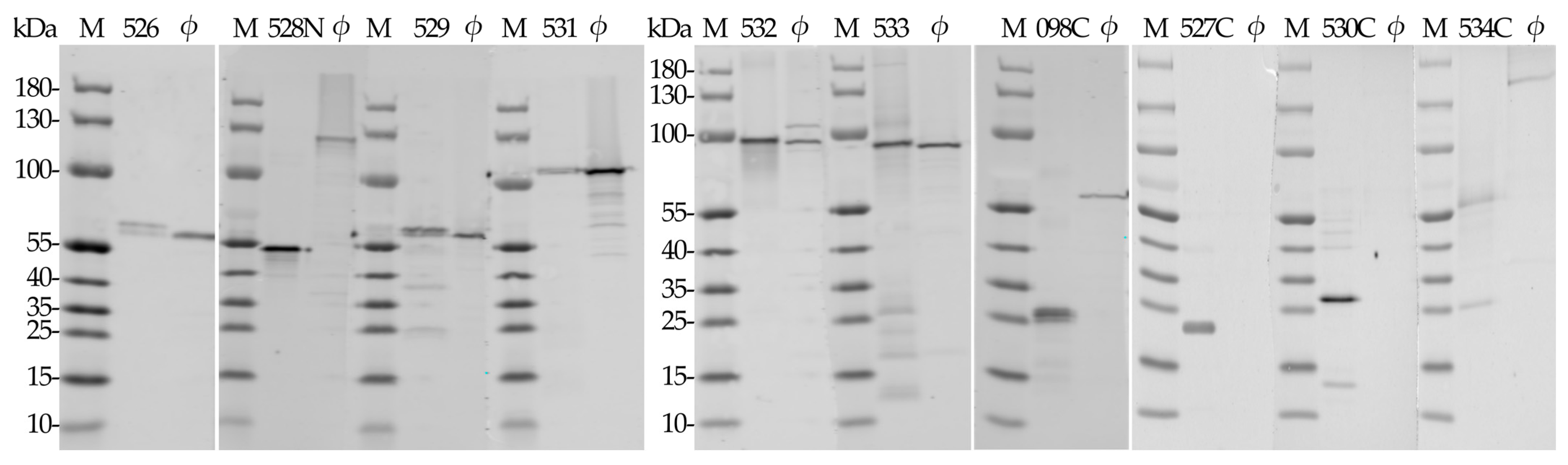
| Recombinant Protein | MW, kDa 1 | MW, kDa 2 | IPTG, mM | Temperature, °C | Time, h | E. coli Strain |
|---|---|---|---|---|---|---|
| 098C | 62.9 | 26.3 * | 1.0 | 22 | 18 | Rosetta DE3 |
| 526 | 63.1 | 66.1 | 1.0 | 20 | 18 | Rosetta DE3 |
| 527C | 79.5 | 21.6 * | 0.5 | 30 | 18 | Rosetta DE3 |
| 528N | 121.8 | 46.3 * | 0.8 | 25 | 18 | HMS174 DE3 |
| 529 | 62.5 | 64.9 | 0.5 | 17 | 17 | Rosetta DE3 |
| 530C | 86.1 | 29.7 * | 0.5 | 30 | 18 | Rosetta DE3 |
| 531 | 97.8 | 100.3 | 0.5 | 17 | 19 | Rosetta DE3 |
| 532 | 87.3 | 89.8 | 1.0 | 18 | 17 | HMS174 DE3 |
| 533 | 82.8 | 85.4 | 0.5 | 30 | 21 | Rosetta DE3 |
| 534C | 70.8 | 28.4 * | 0.5 | 30 | 18 | Rosetta DE3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noreika, A.; Rutkiene, R.; Dumalakienė, I.; Vilienė, R.; Laurynėnas, A.; Povilonienė, S.; Skapas, M.; Meškys, R.; Kaliniene, L. Insights into the Alcyoneusvirus Adsorption Complex. Int. J. Mol. Sci. 2023, 24, 9320. https://doi.org/10.3390/ijms24119320
Noreika A, Rutkiene R, Dumalakienė I, Vilienė R, Laurynėnas A, Povilonienė S, Skapas M, Meškys R, Kaliniene L. Insights into the Alcyoneusvirus Adsorption Complex. International Journal of Molecular Sciences. 2023; 24(11):9320. https://doi.org/10.3390/ijms24119320
Chicago/Turabian StyleNoreika, Algirdas, Rasa Rutkiene, Irena Dumalakienė, Rita Vilienė, Audrius Laurynėnas, Simona Povilonienė, Martynas Skapas, Rolandas Meškys, and Laura Kaliniene. 2023. "Insights into the Alcyoneusvirus Adsorption Complex" International Journal of Molecular Sciences 24, no. 11: 9320. https://doi.org/10.3390/ijms24119320
APA StyleNoreika, A., Rutkiene, R., Dumalakienė, I., Vilienė, R., Laurynėnas, A., Povilonienė, S., Skapas, M., Meškys, R., & Kaliniene, L. (2023). Insights into the Alcyoneusvirus Adsorption Complex. International Journal of Molecular Sciences, 24(11), 9320. https://doi.org/10.3390/ijms24119320







