Exogenously Induced Silencing of Four MYB Transcription Repressor Genes and Activation of Anthocyanin Accumulation in Solanum lycopersicum
Abstract
1. Introduction
2. Results
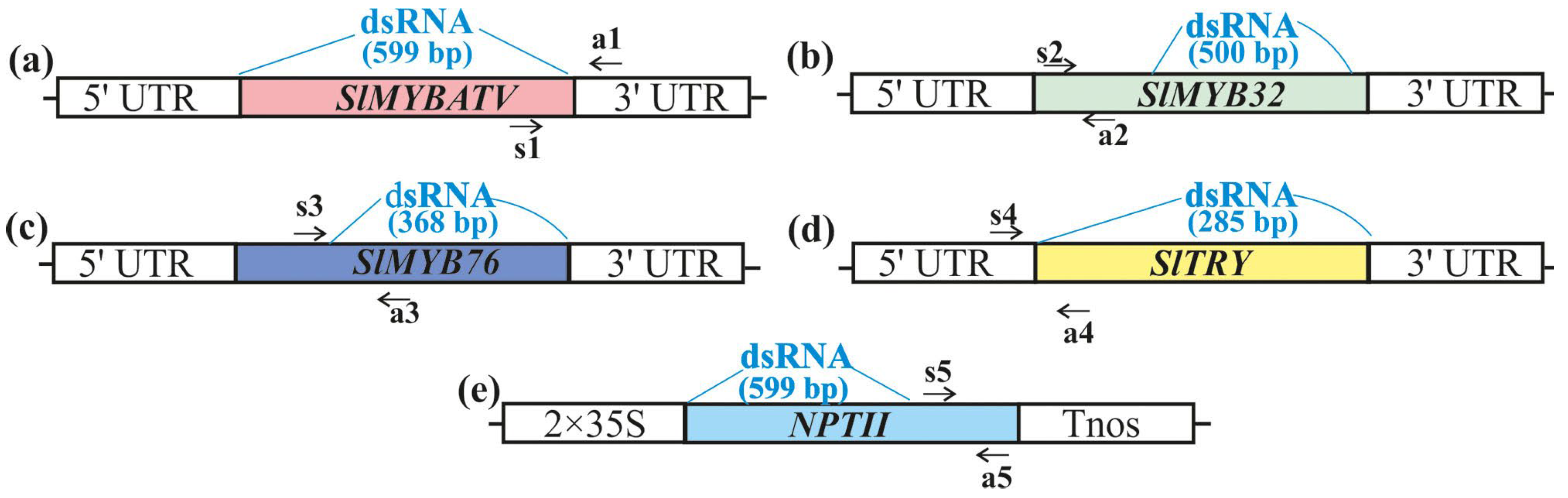
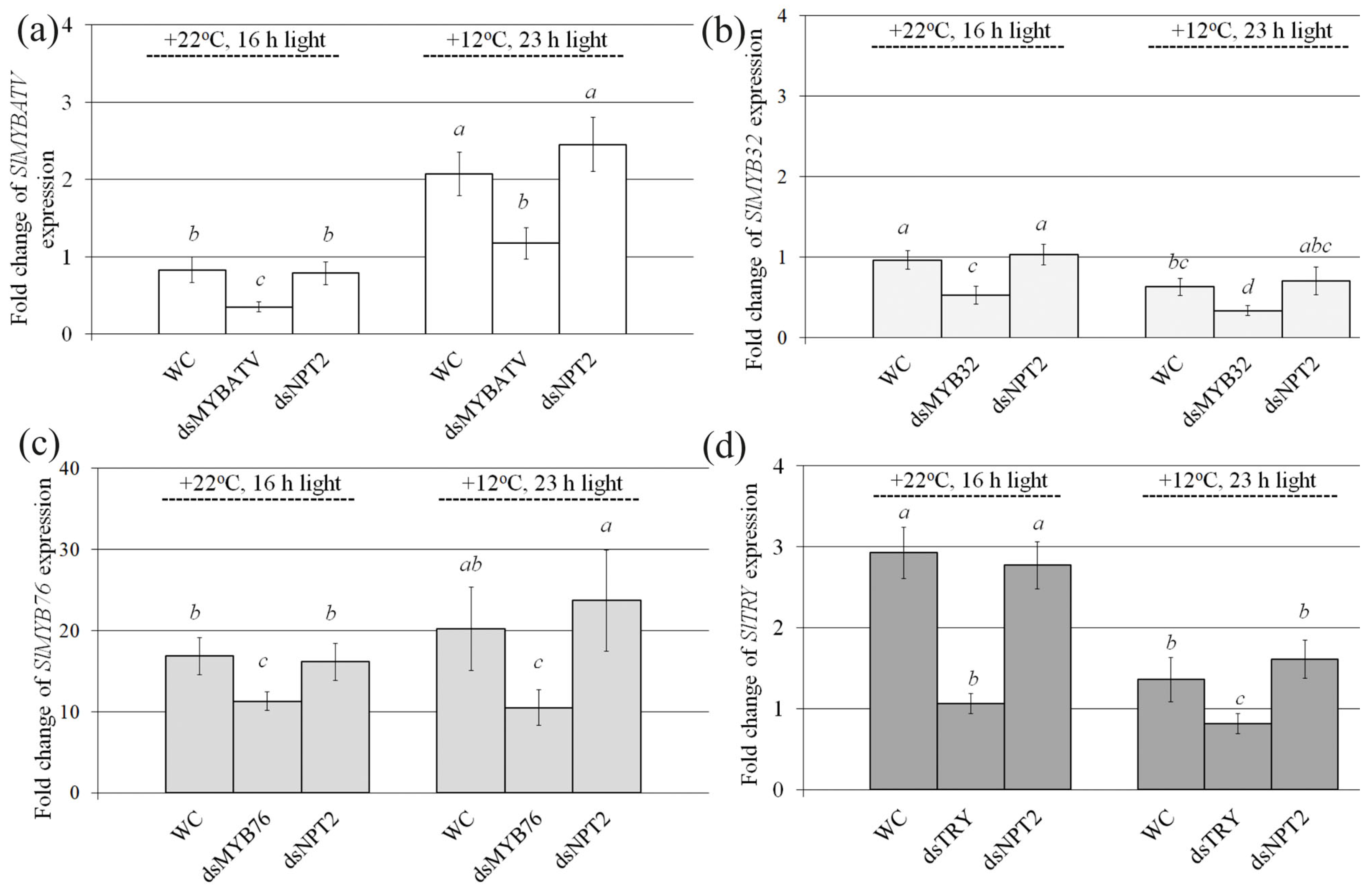
2.1. Exogenous dsRNAs Downregulate mRNA Levels of SlMYBATV, SlMYB32, SlMYB76, and SlTRY Transcription Factors
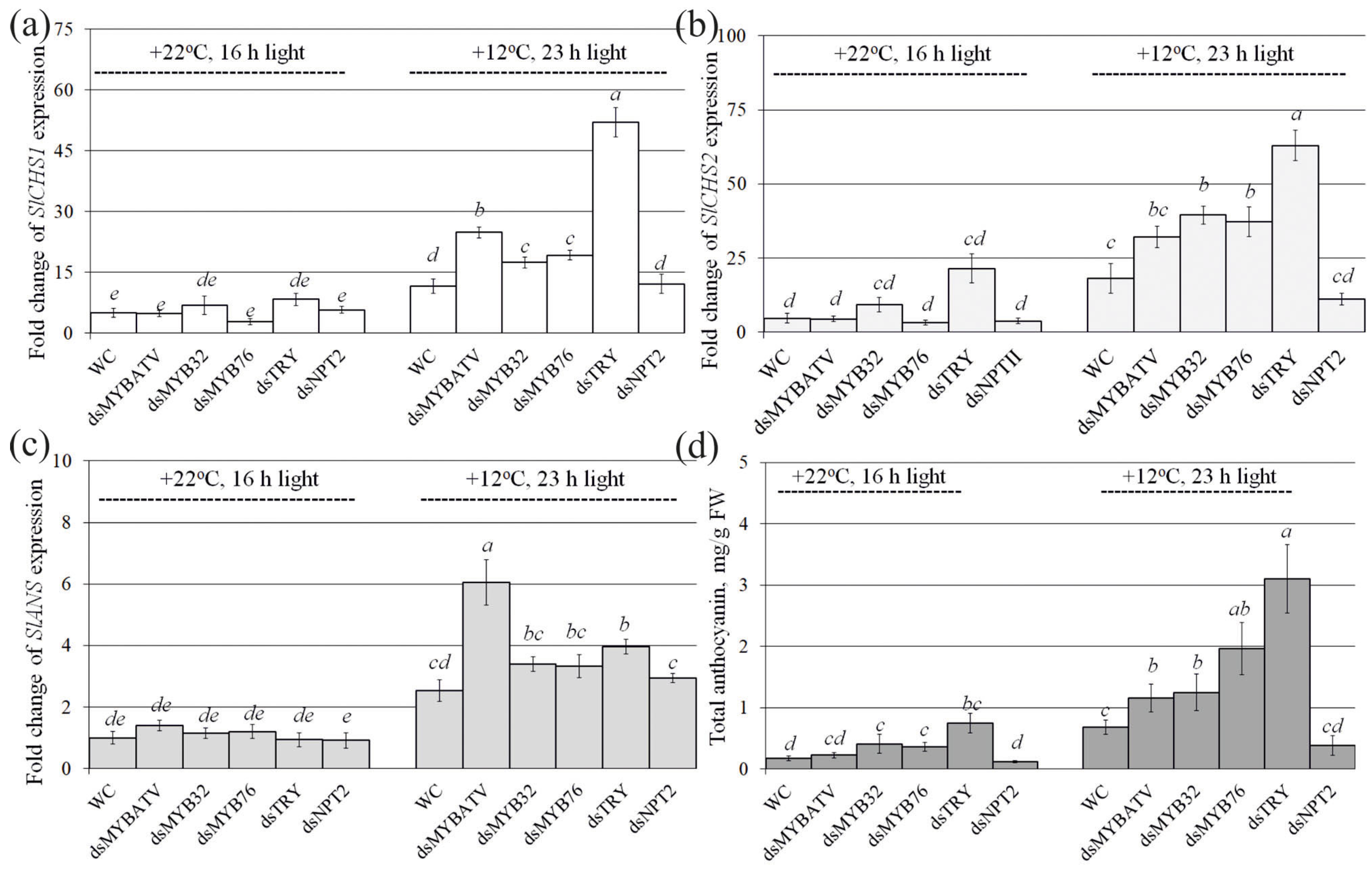
2.2. Exogenous dsRNAs Upregulate mRNA Levels of Anthocyanin Biosynthesis-Related Genes and Anthocyanin Content
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Isolation and Sequencing of SlMYBATV, SlMYB32, SlMYB76, and SlTRY Transcripts
4.3. dsRNA Synthesis and Application
4.4. RNA Isolation and Reverse Transcription
4.5. Gene Expression Analysis by qRT–PCR
4.6. Quantification of Anthocyanins
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 507, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Guo, H.S. RNA silencing: From discovery and elucidation to application and perspectives. J. Integr. Plant Biol. 2022, 64, 476–498. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Guleria, P.; Mahajan, M.; Bhardwaj, J.; Yadav, S.K. Plant small RNAs: Biogenesis, mode of action and their roles in abiotic stresses. Genom. Proteom. Bioinform. 2011, 6, 183–199. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA interference: A natural immune system of plants to counteract biotic stressors. Cells 2019, 8, 38. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-stranded RNAs in plant protection against pathogenic organisms and viruses in agriculture. Acta Nat. 2019, 11, 13–21. [Google Scholar] [CrossRef]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef]
- Koch, A.; Wassenegger, M. Host-induced gene silencing-mechanisms and applications. New Phytol. 2021, 231, 54–59. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 3, 487–501. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a foliar spray: Efficiency and challenges to field applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 2019, 20, 2282. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; McMillan, J.N.; Cardoza, V.; Maegele, I.; Dadami, E.; Runne, M.; Krczal, G.; Wassenegger, M. Induction of silencing in plants by high-pressure spraying of in vitro-synthesized small RNAs. Front. Plant Sci. 2016, 7, 1327. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 6749. [Google Scholar] [CrossRef] [PubMed]
- Konakalla, N.C.; Kaldis, A.; Berbati, M.; Masarapu, H.; Voloudakis, A.E. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 2016, 244, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Song, X.S.; Gu, K.X.; Duan, X.X.; Xiao, X.M.; Hou, Y.P.; Duan, Y.B.; Wang, J.X.; Zhou, M.G. A myosin5 dsRNA that reduces the fungicide resistance and pathogenicity of Fusarium asiaticum. Pest. Biochem. Physiol. 2018, 150, 1–9. [Google Scholar] [CrossRef]
- Song, X.S.; Gu, K.X.; Duan, X.X.; Xiao, X.M.; Hou, Y.P.; Duan, Y.B.; Wang, J.X.; Yu, N.; Zhou, M.G. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 2018, 19, 2543–2560. [Google Scholar] [CrossRef]
- Kaldis, A.; Berbati, M.; Melita, O.; Reppa, C.; Holeva, M.; Otten, P.; Voloudakis, A. Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol. Plant Pathol. 2008, 19, 883–895. [Google Scholar] [CrossRef]
- Gu, K.X.; Song, X.S.; Xiao, X.M.; Duan, X.X.; Wang, J.X.; Duan, Y.B.; Hou, Y.P.; Zhou, M.G. A β2-tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pest. Biochem. Physiol. 2019, 153, 36–46. [Google Scholar] [CrossRef]
- Werner, B.T.; Gaffar, F.Y.; Schuemann, J.; Biedenkopf, D.; Koch, A.M. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 2020, 11, 476. [Google Scholar] [CrossRef]
- Qiao, L.; Lan, C.; Capriotti, L.; Ah-Fong, A.; Nino Sanchez, J.; Hamby, R.; Heller, J.; Zhao, H.; Glass, N.L.; Judelson, H.S.; et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 2021, 19, 1756–1768. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H. Spray-induced gene silencing: A powerful innovative strategy for crop protection. Trends Microbiol. 2017, 25, 4–6. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of exogenous dsRNAs-induced RNAi in agriculture: Challenges and triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Akbar, S.; Wei, Y.; Zhang, M.-Q. RNA interference: Promising approach to combat plant viruses. Int. J. Mol. Sci. 2022, 23, 5312. [Google Scholar] [CrossRef]
- Lau, S.E.; Schwarzacher, T.; Othman, R.Y.; Harikrishna, J.A. dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol. 2015, 15, 194. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef] [PubMed]
- Marcianò, D.; Ricciardi, V.; Fassolo, E.M.; Passera, A.; BIANCO, P.A.; Failla, O.; Casati, P.; Maddalena, G.; De Lorenzis, G.; Toffolatti, S.L. RNAi of a putative grapevine susceptibility gene as a possible downy mildew control strategy. Front. Plant Sci. 2021, 12, 667319. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Guaschino, M.; Pagliarani, C.; De Rosso, M.; Lovisolo, C.; Chitarra, W. Spray-induced gene silencing targeting a glutathione S-transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 2022, 45, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Sammons, R.; Ivashuta, S.; Liu, H.; Wang, D.; Feng, P.; Kouranov, A.; Andersen, S. Polynucleotide molecules for gene regulation in plants. U.S. Patent 20110296556, 1 September 2015. [Google Scholar]
- Jiang, L.; Ding, L.; He, B.; Shen, J.; Xu, Z.; Yin, M.; Zhang, X. Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 2014, 6, 9965–9969. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Pennisi, F.; Cressoni, C.; Vitulo, N.; Dusi, V.; Speghini, A.; Pandolfini, T. Nanovector-mediated exogenous delivery of dsRNA induces silencing of target genes in very young tomato flower buds. Nanoscale Adv. 2022, 4, 4542–4553. [Google Scholar] [CrossRef] [PubMed]
- Killiny, N.; Gonzalez-Blanco, P.; Gowda, S.; Martini, X.; Etxeberria, E. Plant functional genomics in a few days: Laser-assisted delivery of double-stranded RNA to higher plants. Plants 2021, 10, 93. [Google Scholar] [CrossRef]
- Nityagovsky, N.N.; Kiselev, K.V.; Suprun, A.R.; Dubrovina, A.S. Exogenous dsRNA induces RNA interference of a chalcone synthase gene in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 5325. [Google Scholar] [CrossRef]
- Kong, J.M.; Chia, L.S.; Goh, N.K.; Chia, T.F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef]
- Konczak, I.; Zhang, W. Anthocyanins—More than nature’s colours. J. Biomed. Biotech. 2004, 5, 239. [Google Scholar] [CrossRef]
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023, 361, 12–29. [Google Scholar] [CrossRef]
- Verhoeyen, M.E.; Bovy, A.; Collins, G.; Muir, S.; Robinson, S.; De Vos CH, R.; Colliver, S. Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J. Exp. Bot. 2002, 53, 2099–2106. [Google Scholar] [CrossRef]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef]
- Chaves-Silva, S.; Dos Santos, A.L.; Chalfun-Júnior, A.; Zhao, J.; Peres, L.E.P.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27. [Google Scholar] [CrossRef]
- Cao, X.; Qiu, Z.; Wang, X.; Van Giang, T.; Liu, X.; Wang, J.; Wang, X.; Gao, J.; Guo, Y.; Du, Y.; et al. A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp Bot. 2017, 68, 5745–5758. [Google Scholar] [CrossRef]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 2018, 9, 830. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Gen. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef]
- Nukumizu, Y.; Wada, T.; Tominaga-Wada, R. Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal Behav. 2013, 8, e24575. [Google Scholar]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kostetsky, E.Y.; Dubrovina, A.S. The specificity of transgene suppression in plants by exogenous dsRNA. Plants 2022, 11, 715. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Dubrovina, A.S. Physiological conditions and dsRNA application approaches for exogenously induced RNA interference in Arabidopsis thaliana. Plants 2021, 10, 264. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Tudor-Radu, M.; Vijan, L.E.; Tudor-Radu, C.M.; Tita, I.; Sima, R.; Mitrea, R. Assessment of Ascorbic Acid, Polyphenols, Flavonoids, Anthocyanins and Carotenoids Content in Tomato Fruits. Not. Bot. Horti Agrobot. Cluj-Napoca 2016, 44, 477–483. [Google Scholar] [CrossRef]
- Mazzucato, A.; Willems, D.; Bernini, R.; Picarella, M.E.; Santangelo, E.; Ruiu, F.; Tilesi, F.; Soressi, G.P. Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation. Plant Physiol. Biochem. 2013, 72, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 11, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Shen, J.; Liu, H.; Li, F.; Ding, N.; Gao, C.; Pattanaik, S.; Patra, B.; Li, R.; Yuan, L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 2015, 242, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Singh, A.K.; Behera, T.K. CRISPR/Cas genome editing in tomato improvement: Advances and applications. Front Plant Sci. 2023, 14, 1121209. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Faqir Napar, W.P.; Kaleri, A.R.; Ahmed, A.; Nabi, F.; Sajid, S.; Ćosić, T.; Yao, Y.; Liu, J.; Raspor, M.; Gao, Y. The anthocyanin-rich tomato genotype LA-1996 displays superior efficiency of mechanisms of tolerance to salinity and drought. J. Plant Physiol. 2022, 271, 153662. [Google Scholar] [CrossRef]
- Numata, K.; Ohtani, M.; Yoshizumi, T.; Demura, T.; Kodama, Y. Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 2014, 12, 1027–1034. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Aleynova, O.A.; Kalachev, A.V.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Induction of transgene suppression in plants via external application of synthetic dsRNA. Int. J. Mol. Sci. 2019, 20, 1585. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Aleynova, O.A.; Suprun, A.R.; Ogneva, Z.V.; Kiselev, K.V. Transgene suppression in plants by foliar application of in vitro-synthesized small interfering RNAs. Appl. Microbiol. Biotechnol. 2020, 104, 2125–2135. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Aleynova, O.A.; Ogneva, Z.V.; Suprun, A.R.; Ananev, A.A.; Kiselev, K.V. The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis. Plants 2019, 8, 602. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Dubrovina, A.S.; Shumakova, O.A.; Karetin, Y.A.; Manyakhin, A.Y. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013, 32, 431–442. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. The role of calcium-dependent protein kinase genes VaCPK1 and VaCPK26 in the response of Vitis amurensis (in vitro) and Arabidopsis thaliana (in vivo) to abiotic stresses. Russ. J. Genet. 2019, 55, 319–329. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Abdullin, S.R.; Nikulin, V.Y.; Nikulin, A.Y.; Manyakhin, A.Y.; Bagmet, V.B.; Suprun, A.R.; Gontcharov, A.A. Roholtiella mixta sp. nov. (Nostocales, Cyanobacteria): Morphology, molecular phylogeny, and carotenoid content. Phycologia 2021, 60, 73–82. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Zhou, Z.; Qiu, Z.; Cui, X. Rapid analysis of anthocyanin and its structural modifications in fresh tomato fruit. Food Chem. 2020, 333, 127439. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suprun, A.R.; Kiselev, K.V.; Dubrovina, A.S. Exogenously Induced Silencing of Four MYB Transcription Repressor Genes and Activation of Anthocyanin Accumulation in Solanum lycopersicum. Int. J. Mol. Sci. 2023, 24, 9344. https://doi.org/10.3390/ijms24119344
Suprun AR, Kiselev KV, Dubrovina AS. Exogenously Induced Silencing of Four MYB Transcription Repressor Genes and Activation of Anthocyanin Accumulation in Solanum lycopersicum. International Journal of Molecular Sciences. 2023; 24(11):9344. https://doi.org/10.3390/ijms24119344
Chicago/Turabian StyleSuprun, Andrey R., Konstantin V. Kiselev, and Alexandra S. Dubrovina. 2023. "Exogenously Induced Silencing of Four MYB Transcription Repressor Genes and Activation of Anthocyanin Accumulation in Solanum lycopersicum" International Journal of Molecular Sciences 24, no. 11: 9344. https://doi.org/10.3390/ijms24119344
APA StyleSuprun, A. R., Kiselev, K. V., & Dubrovina, A. S. (2023). Exogenously Induced Silencing of Four MYB Transcription Repressor Genes and Activation of Anthocyanin Accumulation in Solanum lycopersicum. International Journal of Molecular Sciences, 24(11), 9344. https://doi.org/10.3390/ijms24119344






