The Verticillium dahliae Small Cysteine-Rich Protein VdSCP23 Manipulates Host Immunity
Abstract
:1. Introduction
2. Results
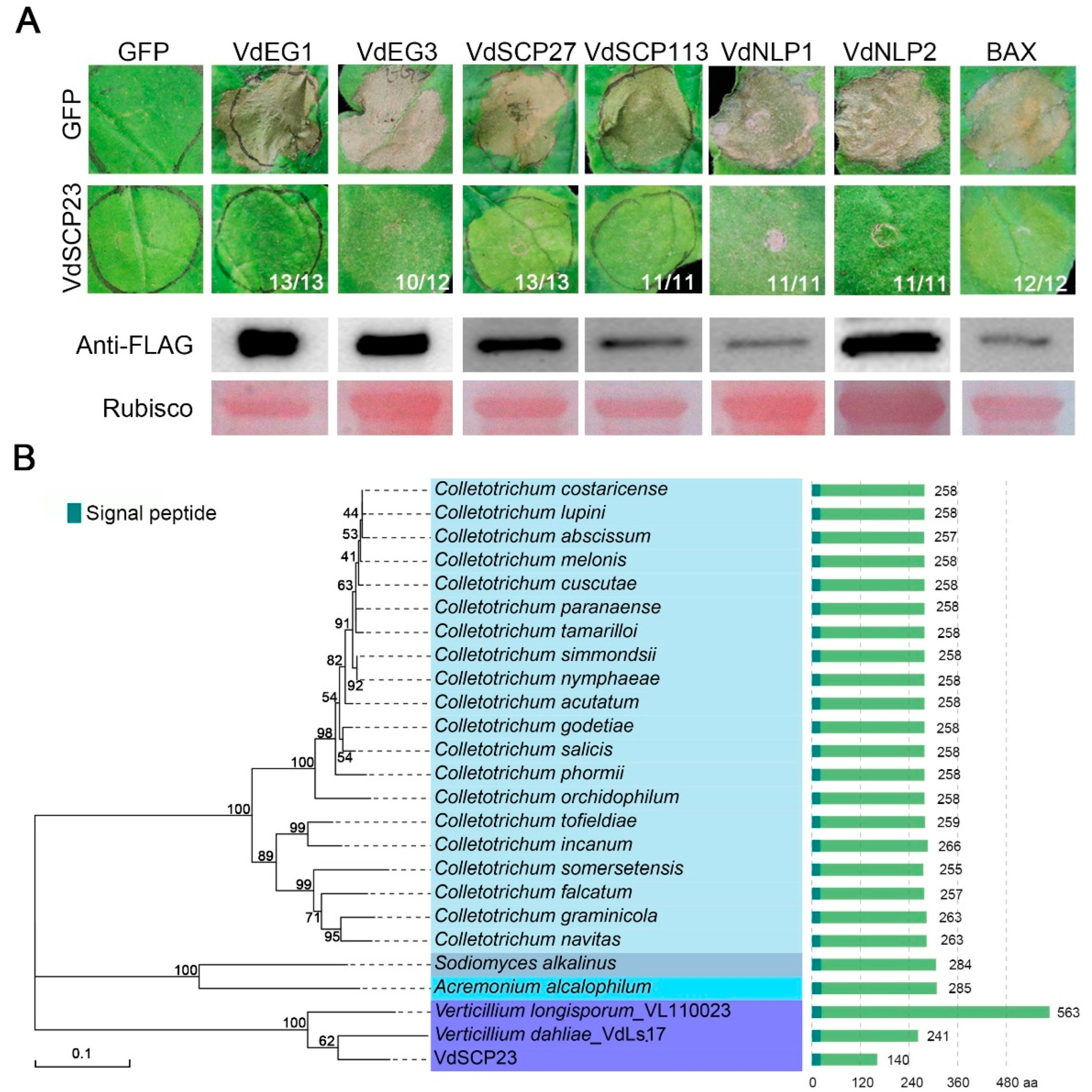
2.1. VdSCP23 Displays Broad-Spectrum Cell Death Suppression
2.2. VdSCP23 Is Conserved among Strains of Verticillium dahliae
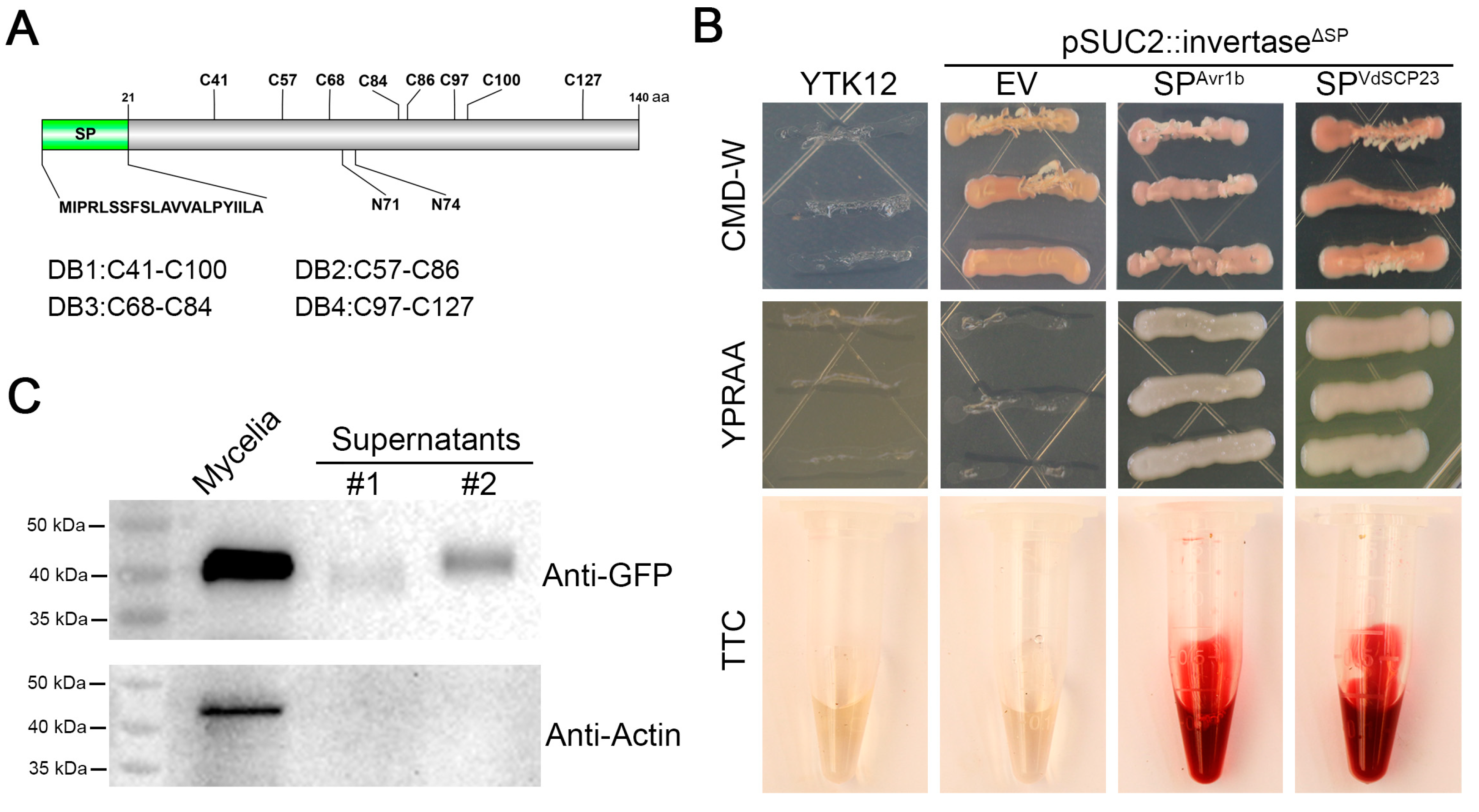
2.3. VdSCP23 Is Secreted to the Extracellular Region of Verticillium dahliae
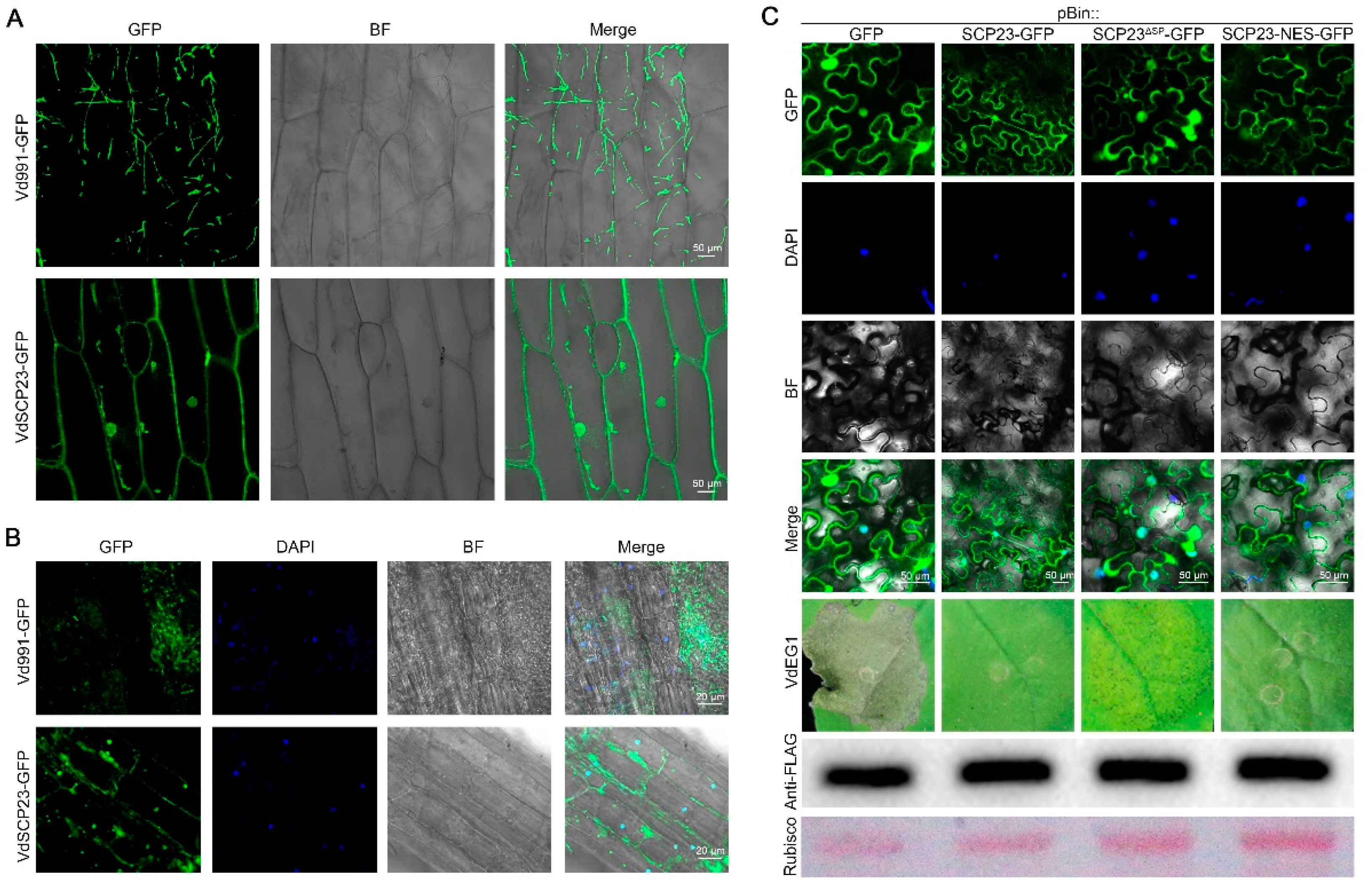
2.4. VdSCP23 Is Mainly Localized on Plasma Membrane or Cytoplasm for the Exertion of Its Cell Death-Suppression Function
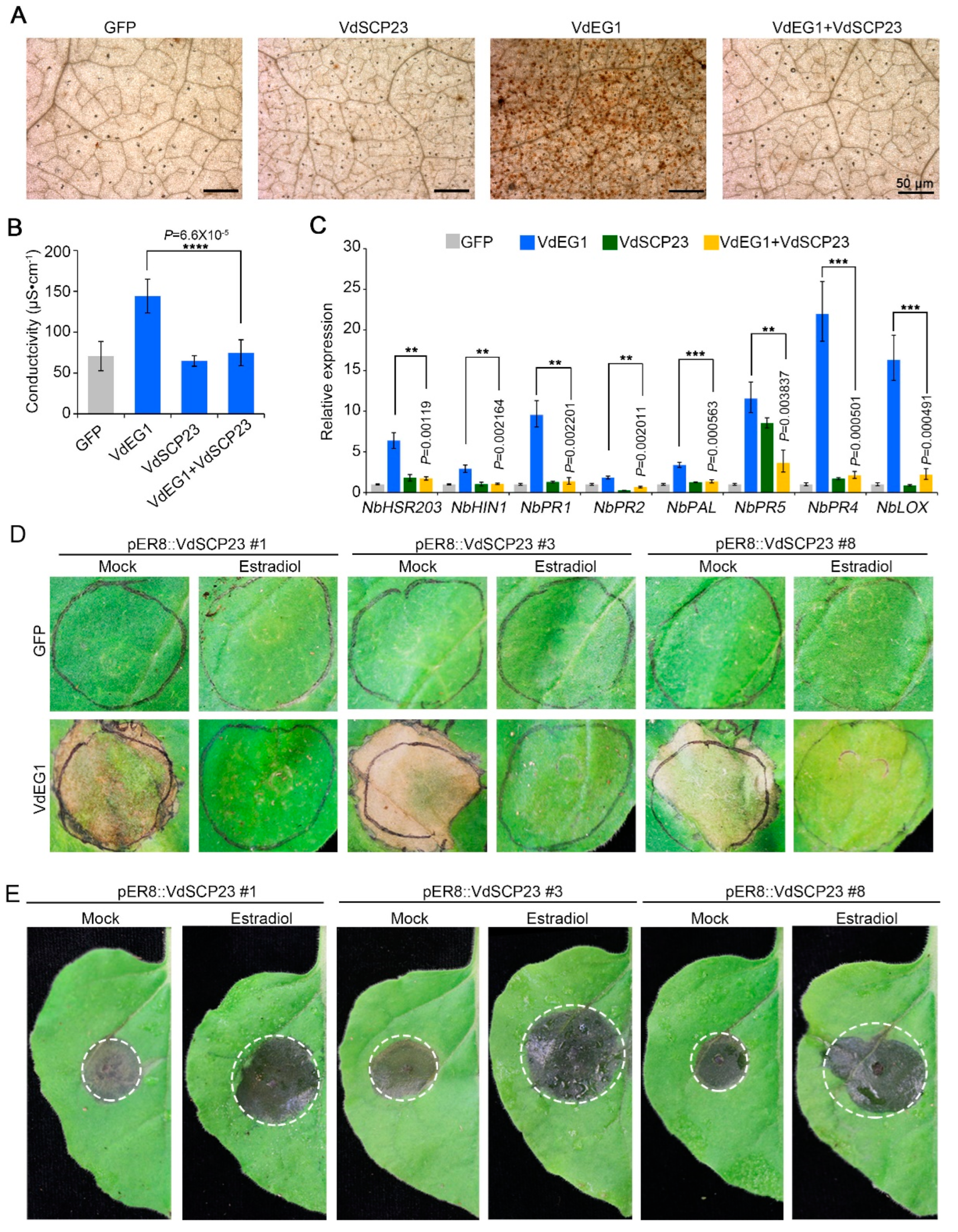
2.5. VdSCP23 Significantly Enhanced Host Susceptibility by Suppressing Immunity
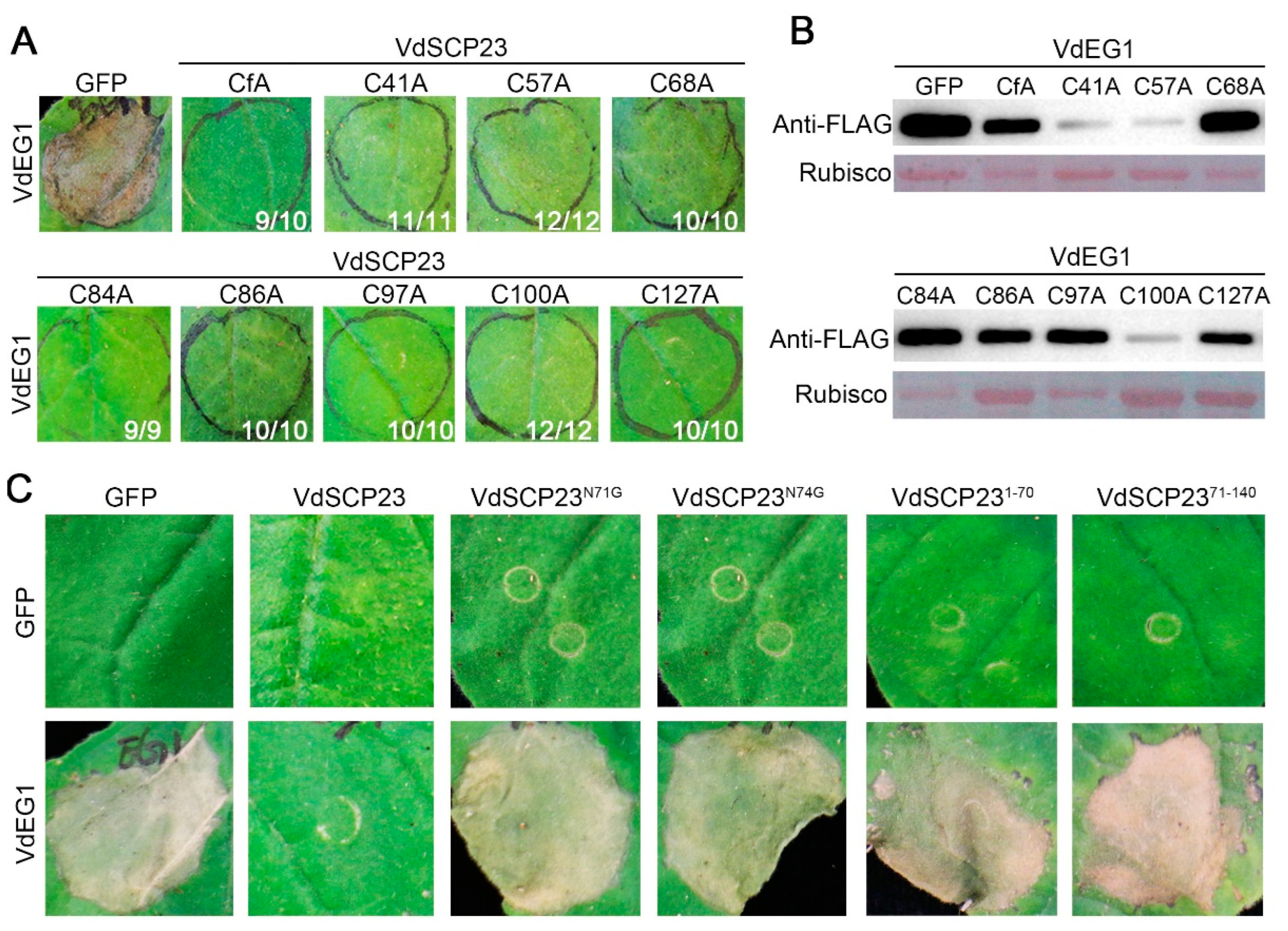
2.6. N-glycosylation Sites and Protein Structural Integrity, but Not Cysteine Residues, Are Necessary for the Immunity-Inhibiting Function of VdSCP23
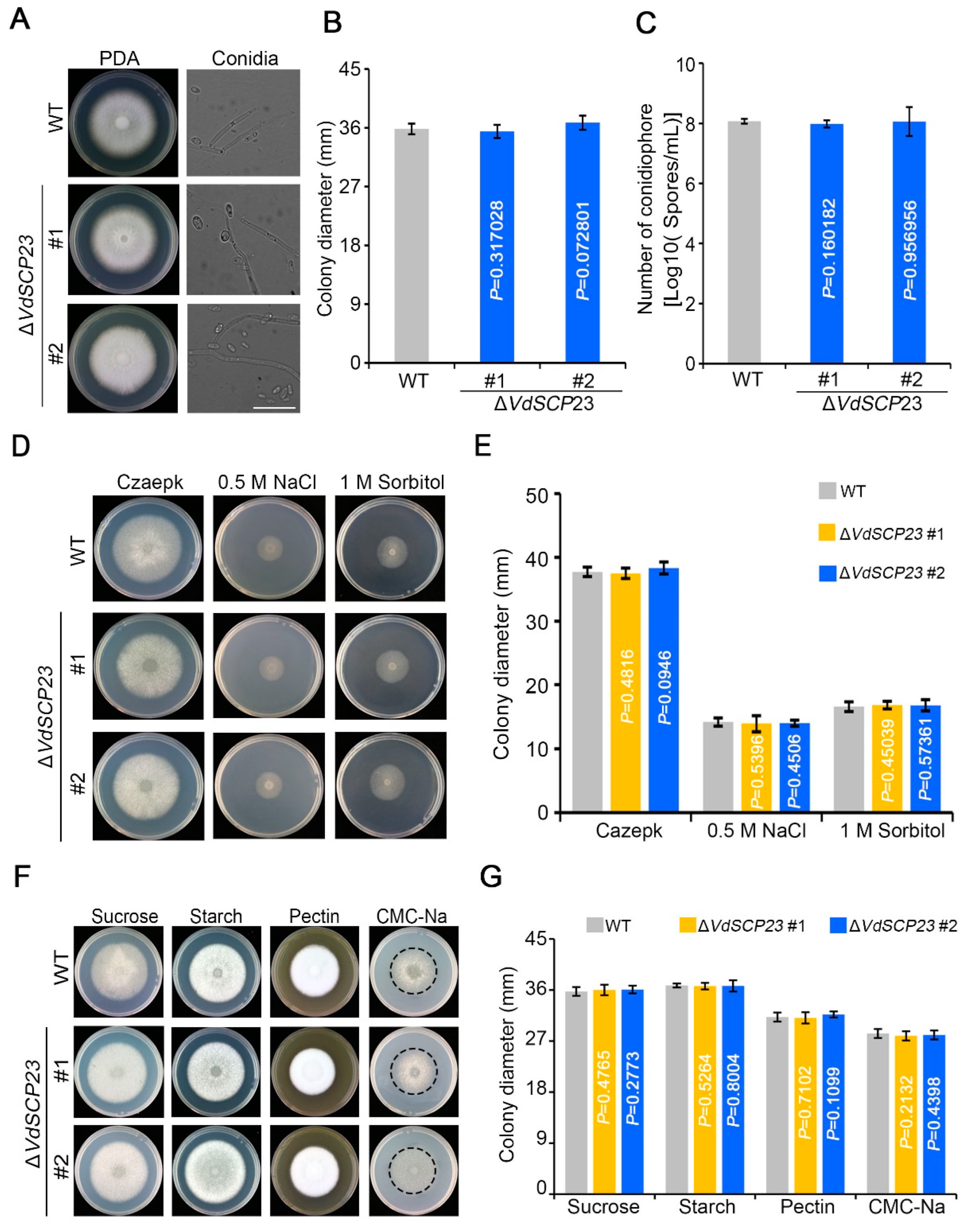
2.7. VdSCP23 Was Not Required for Growth, Normal Morphology, Stress Tolerance, or Carbon Source Utilization
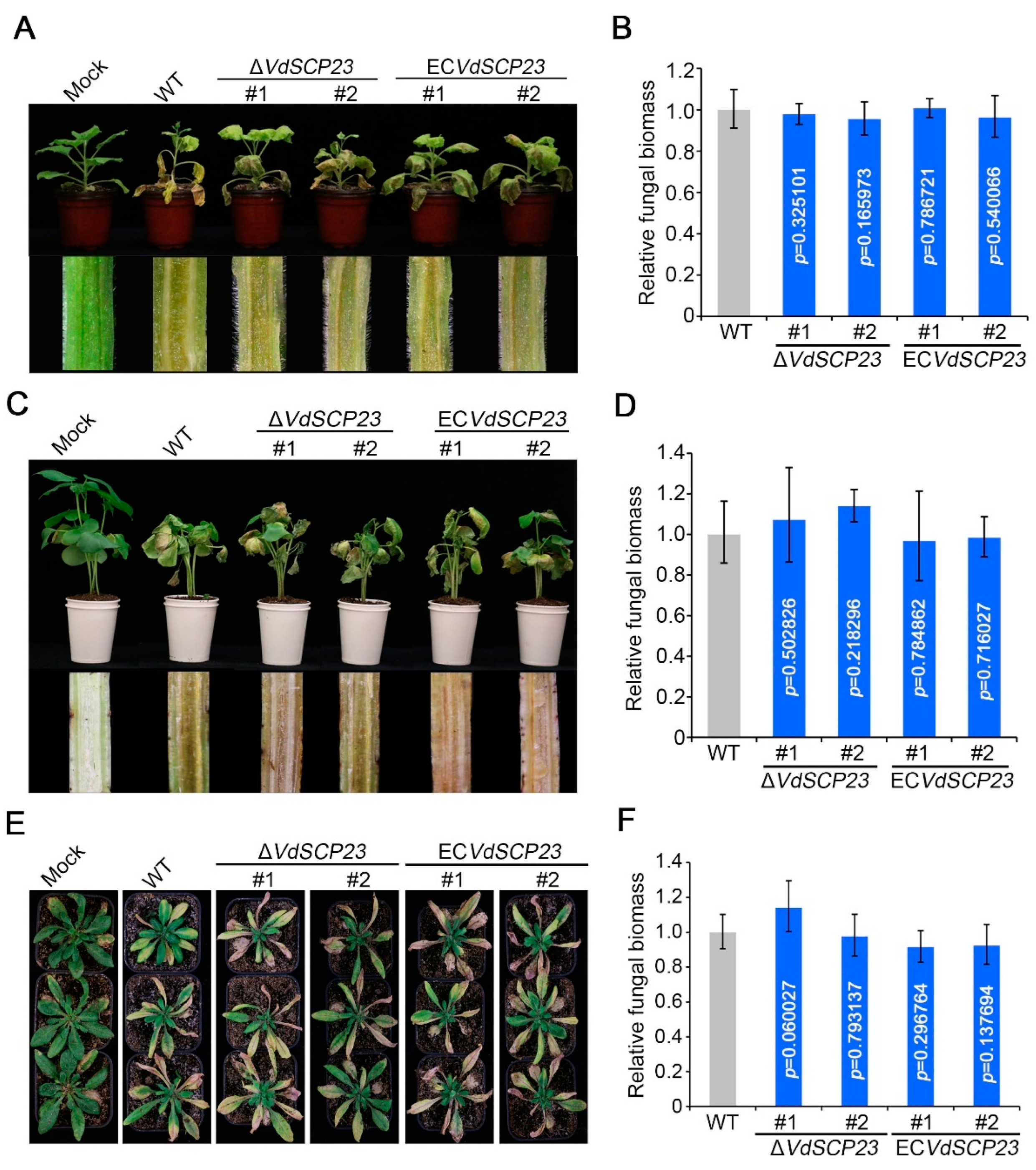
2.8. VdSCP23 Is Not Required for Virulence
3. Discussion
4. Materials and Methods
4.1. Cultivation of Microbial Strains and Plant Materials
4.2. Identification and Phylogenetic Analysis of VdSCP23
4.3. Transient Expression in Nicotiana benthamiana
4.4. Yeast Signal Sequence Trap System
4.5. In Vitro Secretion Activity Assay
4.6. Subcellular Localization Assays
4.7. ROS and Electrolyte Leakage Detection
4.8. Analyses of Gene Expression Levels
4.9. Fungal Transformations for Gene Deletion and Complementation
4.10. Analyses of Growth and Morphology of the VdSCP23 Deletion Mutants
4.11. Virulence Assays
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cook, D.E.; Mesarich, C.H.; Thomma, B.P. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015, 53, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, D.; Robatzek, S. Pattern recognition receptors: From the cell surface to intracellular dynamics. Mol. Plant-Microbe Interact. MPMI 2007, 20, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- van Kan, J.A.; van den Ackerveken, G.F.; de Wit, P.J. Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. MPMI 1991, 4, 52–59. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Wang, J.; Li, C.; Revote, J.; Zhang, Y.; Naderer, T.; Hayashida, M.; Akutsu, T.; Webb, G.I.; Lithgow, T.; et al. SecretEPDB: A comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems. Sci. Rep. 2017, 7, 41031. [Google Scholar] [CrossRef]
- Sharpee, W.C.; Dean, R.A. Form and function of fungal and oomycete effectors. Fungal Biol. Rev. 2016, 30, 62–73. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Bélanger, R.R. Computational prediction of effector proteins in fungi: Opportunities and challenges. Front. Plant Sci. 2016, 7, 126. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; de Wit, P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Liu, C.; Gui, Y.J.; Si, K.W.; Zhang, D.D.; Wang, J.; Short, D.P.G.; Huang, J.Q.; Li, N.Y.; Liang, Y.; et al. Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.T.; Sorensen, E.S.; Stachowiak, D.; Boldt, H.B.; Kristensen, L.; Sottrup-Jensen, L.; Oxvig, C. Complex of pregnancy-associated plasma protein-A and the proform of eosinophil major basic protein: Disulfide structure and carbohydrate attachment sites. J. Biol. Chem. 2003, 278, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef]
- Chen, X.L.; Shi, T.; Yang, J.; Shi, W.; Gao, X.; Chen, D.; Xu, X.; Xu, J.R.; Talbot, N.J.; Peng, Y.L. N-glycosylation of effector proteins by an α-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell 2014, 26, 1360–1376. [Google Scholar] [CrossRef]
- Fang, A.; Han, Y.; Zhang, N.; Zhang, M.; Liu, L.; Li, S.; Lu, F.; Sun, W. Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol. Plant-Microbe Interact. MPMI 2016, 29, 405–416. [Google Scholar] [CrossRef]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Guo, X.; Zhang, X.; Chen, Y.; Liu, Y.; et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 2015, 11, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Mueller, O.; Kahmann, R.; Aguilar, G.; Trejo-Aguilar, B.; Wu, A.; de Vries, R.P. The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. FG B 2008, 45 (Suppl. 1), S63–S70. [Google Scholar] [CrossRef]
- Wang, D.; Tian, L.; Zhang, D.D.; Song, J.; Song, S.S.; Yin, C.M.; Zhou, L.; Liu, Y.; Wang, B.L.; Kong, Z.Q.; et al. Functional analyses of small secreted cysteine-rich proteins identified candidate effectors in Verticillium dahliae. Mol. Plant Pathol. 2020, 21, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Seitner, D.; Uhse, S.; Gallei, M.; Djamei, A. The core effector Cce1 is required for early infection of maize by Ustilago maydis. Mol. Plant Pathol. 2018, 19, 2277–2287. [Google Scholar] [CrossRef]
- Qi, M.; Link, T.I.; Müller, M.; Hirschburger, D.; Pudake, R.N.; Pedley, K.F.; Braun, E.; Voegele, R.T.; Baum, T.J.; Whitham, S.A. A small cysteine-rich protein from the Asian soybean rust fungus, Phakopsora pachyrhizi, suppresses plant immunity. PLoS Pathog. 2016, 12, e1005827. [Google Scholar] [CrossRef] [PubMed]
- Inderbitzin, P.; Subbarao, K.V. Verticillium systematics and evolution: How confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Zhang, D.D.; Dai, X.F.; Klosterman, S.J.; Subbarao, K.V.; Chen, J.Y. The secretome of Verticillium dahliae in collusion with plant defence responses modulates Verticillium wilt symptoms. Biol. Rev. Camb. Philos. Soc. 2022, 97, 1810–1822. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Zeng, H.; Liu, H.; Zhou, T.; Tan, B.; Yuan, J.; Guo, L.; Qiu, D. The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl. Microbiol. Biotechnol. 2012, 93, 191–201. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, B.S.; Zhao, J.H.; Huang, J.F.; Jia, P.S.; Wang, S.; Zhang, J.; Zhou, J.M.; Guo, H.S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Kombrink, A.; Rovenich, H.; Shi-Kunne, X.; Rojas-Padilla, E.; van den Berg, G.C.; Domazakis, E.; de Jonge, R.; Valkenburg, D.J.; Sánchez-Vallet, A.; Seidl, M.F.; et al. Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol. Plant Pathol. 2017, 18, 596–608. [Google Scholar] [CrossRef]
- Gui, Y.J.; Chen, J.Y.; Zhang, D.D.; Li, N.Y.; Li, T.G.; Zhang, W.Q.; Wang, X.Y.; Short, D.P.G.; Li, L.; Guo, W.; et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 2017, 19, 1914–1932. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.J.; Zhang, W.Q.; Zhang, D.D.; Zhou, L.; Short, D.P.G.; Wang, J.; Ma, X.F.; Li, T.G.; Kong, Z.Q.; Wang, B.L.; et al. A Verticillium dahliae extracellular cutinase modulates plant immune responses. Mol. Plant-Microbe Interact. MPMI 2018, 31, 260–273. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Maruthachalam, K.; Bolton, M.D.; Santhanam, P.; Saber, M.K.; Zhang, Z.; Usami, T.; Lievens, B.; Subbarao, K.V.; et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 5110–5115. [Google Scholar] [CrossRef]
- Zhang, L.; Ni, H.; Du, X.; Wang, S.; Ma, X.W.; Nürnberger, T.; Guo, H.S.; Hua, C. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 2017, 215, 368–381. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, D.D.; Song, J.; Li, J.J.; Wang, J.; Li, R.; Klosterman, S.J.; Kong, Z.Q.; Lin, F.Z.; Dai, X.F.; et al. Verticillium dahliae CFEM proteins manipulate host immunity and differentially contribute to virulence. BMC Biol. 2022, 20, 55. [Google Scholar] [CrossRef]
- Qin, J.; Wang, K.; Sun, L.; Xing, H.; Wang, S.; Li, L.; Chen, S.; Guo, H.S.; Zhang, J. The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife 2018, 7, 34902. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.; Chen, Z.; Henrissat, B.; Lee, Y.H.; Park, J.; et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef]
- Zhou, B.J.; Jia, P.S.; Gao, F.; Guo, H.S. Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant-Microbe Interact. MPMI 2012, 25, 964–975. [Google Scholar] [CrossRef]
- Lacomme, C.; Santa Cruz, S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 1999, 96, 7956–7961. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Young, C.; Lee, M.; Oliva, R.; Bozkurt, T.O.; Cano, L.M.; Win, J.; Bos, J.I.; Liu, H.Y.; van Damme, M.; et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 2009, 21, 2928–2947. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef]
- Kubo, M.; Imai, A.; Nishiyama, T.; Ishikawa, M.; Sato, Y.; Kurata, T.; Hiwatashi, Y.; Reski, R.; Hasebe, M. System for stable β-estradiol-inducible gene expression in the moss Physcomitrella patens. PLoS ONE 2013, 8, e77356. [Google Scholar] [CrossRef]
- Ferrè, F.; Clote, P. DiANNA 1.1: An extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res. 2006, 34, W182–W185. [Google Scholar] [CrossRef]
- Liu, C.; Talbot, N.J.; Chen, X.L. Protein glycosylation during infection by plant pathogenic fungi. New Phytol. 2021, 230, 1329–1335. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Kim, K.T.; Jeon, J.; Choi, J.; Cheong, K.; Song, H.; Choi, G.; Kang, S.; Lee, Y.H. Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 2016, 7, 186. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef]
- Sevier, C.S.; Kaiser, C.A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002, 3, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Luderer, R.; Takken, F.L.; de Wit, P.J.; Joosten, M.H. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 2002, 45, 875–884. [Google Scholar] [CrossRef]
- van den Burg, H.A.; Westerink, N.; Francoijs, K.J.; Roth, R.; Woestenenk, E.; Boeren, S.; de Wit, P.J.; Joosten, M.H.; Vervoort, J. Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 2003, 278, 27340–27346. [Google Scholar] [CrossRef]
- van den Hooven, H.W.; van den Burg, H.A.; Vossen, P.; Boeren, S.; de Wit, P.J.; Vervoort, J. Disulfide bond structure of the AVR9 elicitor of the fungal tomato pathogen Cladosporium fulvum: Evidence for a cystine knot. Biochemistry 2001, 40, 3458–3466. [Google Scholar] [CrossRef] [PubMed]
- Van’t Klooster, J.W.; Van der Kamp, M.W.; Vervoort, J.; Beekwilder, J.; Boeren, S.; Joosten, M.H.; Thomma, B.P.; De Wit, P.J. Affinity of Avr2 for tomato cysteine protease Rcr3 correlates with the Avr2-triggered Cf-2-mediated hypersensitive response. Mol. Plant Pathol. 2011, 12, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 2016, 12, e1005435. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Z.; Faris, J.D.; Oliver, R.P.; Syme, R.; McDonald, M.C.; McDonald, B.A.; Solomon, P.S.; Lu, S.; Shelver, W.L.; et al. The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 2012, 8, e1002467. [Google Scholar] [CrossRef]
- Chen, X.R.; Li, Y.P.; Li, Q.Y.; Xing, Y.P.; Liu, B.B.; Tong, Y.H.; Xu, J.Y. SCR96, a small cysteine-rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 2016, 17, 577–587. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, J.; Fang, A.; Wang, J.; Li, D.; Li, Y.; Wang, S.; Cui, F.; Yu, J.; Liu, Y.; et al. The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region. Mol. Plant Pathol. 2020, 21, 445–459. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wang, T.; Zhou, S.; Liang, X.; Xie, C.; Kang, Z.; Chen, D.; Zheng, L. The small secreted protein FoSsp1 elicits plant defenses and negatively regulates pathogenesis in Fusarium oxysporum f. sp. cubense (Foc4). Front. Plant Sci. 2022, 13, 873451. [Google Scholar] [CrossRef]
- Wang, W.; An, B.; Feng, L.; He, C.; Luo, H. A Colletotrichum gloeosporioides cerato-platanin protein, CgCP1, contributes to conidiation and plays roles in the interaction with rubber tree. Can. J. Microbiol. 2018, 64, 826–834. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Birney, E.; Biswas, M.; Bucher, P.; Cerutti, L.; Corpet, F.; Croning, M.D.; et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001, 29, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, J.; Guo, W.; Zhang, T. Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular wilt fungus Verticillium dahliae. J. Genet. Genom. 2013, 40, 421–431. [Google Scholar] [CrossRef]
- Bindschedler, L.V.; Dewdney, J.; Blee, K.A.; Stone, J.M.; Asai, T.; Plotnikov, J.; Denoux, C.; Hayes, T.; Gerrish, C.; Davies, D.R.; et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. Cell Mol. Biol. 2006, 47, 851–863. [Google Scholar] [CrossRef]
- Oh, C.S.; Pedley, K.F.; Martin, G.B. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 2010, 22, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Chen, J.Y.; Wang, J.L.; Li, L.; Xiao, H.L.; Adam, S.M.; Dai, X.F. Molecular characterization and functional analysis of a specific secreted protein from highly virulent defoliating Verticillium dahliae. Gene 2013, 529, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, P.; van Esse, H.P.; Albert, I.; Faino, L.; Nürnberger, T.; Thomma, B.P. Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant-Microbe Interact. MPMI 2013, 26, 278–286. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wang, D.; Ji, X.; Wang, J.; Klosterman, S.J.; Dai, X.; Chen, J.; Subbarao, K.V.; Hao, X.; Zhang, D. The Verticillium dahliae Small Cysteine-Rich Protein VdSCP23 Manipulates Host Immunity. Int. J. Mol. Sci. 2023, 24, 9403. https://doi.org/10.3390/ijms24119403
Wang J, Wang D, Ji X, Wang J, Klosterman SJ, Dai X, Chen J, Subbarao KV, Hao X, Zhang D. The Verticillium dahliae Small Cysteine-Rich Protein VdSCP23 Manipulates Host Immunity. International Journal of Molecular Sciences. 2023; 24(11):9403. https://doi.org/10.3390/ijms24119403
Chicago/Turabian StyleWang, Jie, Dan Wang, Xiaobin Ji, Jun Wang, Steven J. Klosterman, Xiaofeng Dai, Jieyin Chen, Krishna V. Subbarao, Xiaojuan Hao, and Dandan Zhang. 2023. "The Verticillium dahliae Small Cysteine-Rich Protein VdSCP23 Manipulates Host Immunity" International Journal of Molecular Sciences 24, no. 11: 9403. https://doi.org/10.3390/ijms24119403




