Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin
Abstract
:1. Introduction
2. Results
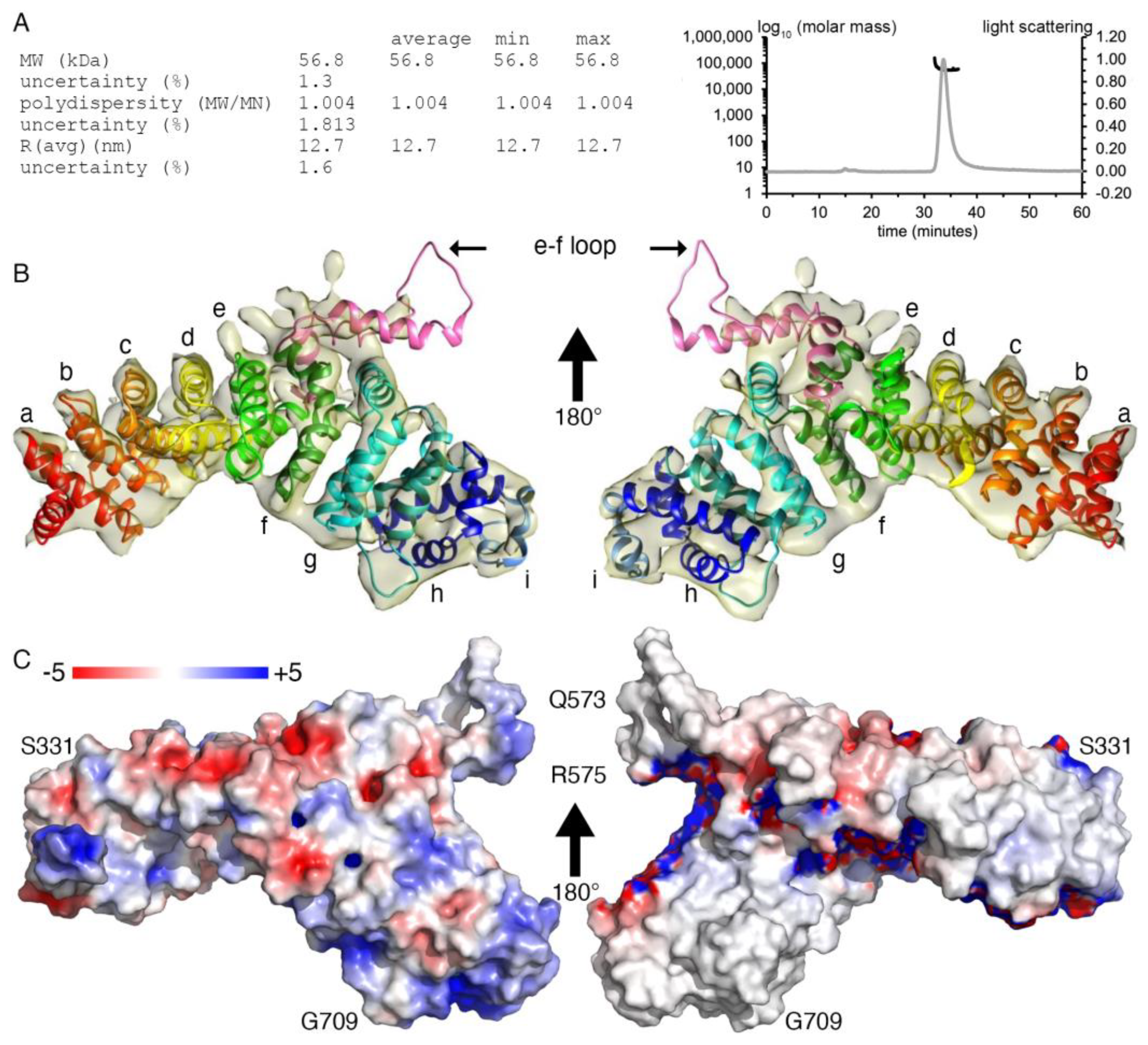
2.1. Plakophilin-3 cryoEM Structure
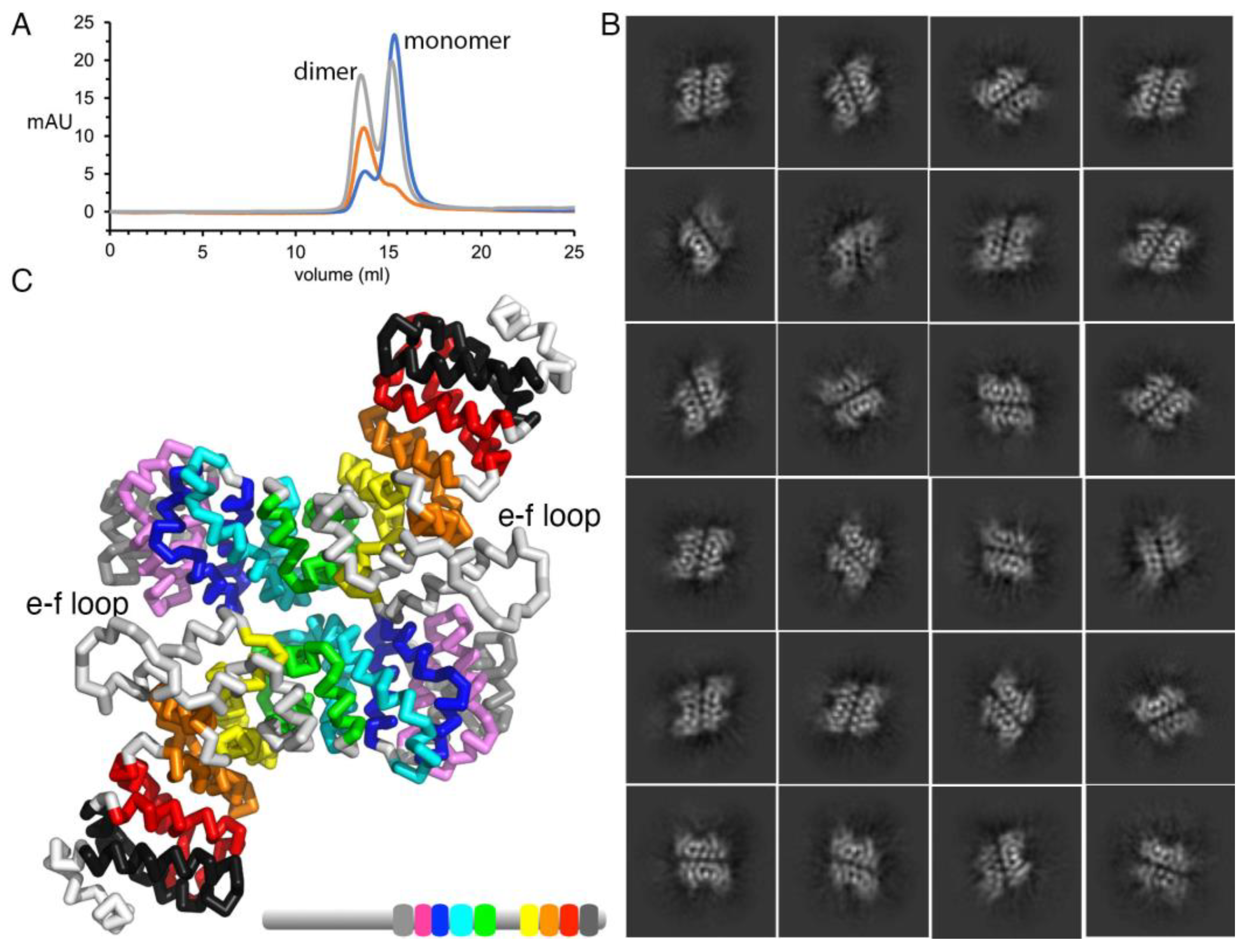
2.2. Plakophilin-3 Forms a Dimer and Monomer
2.3. Plakophilin-3 Binds to the Membrane
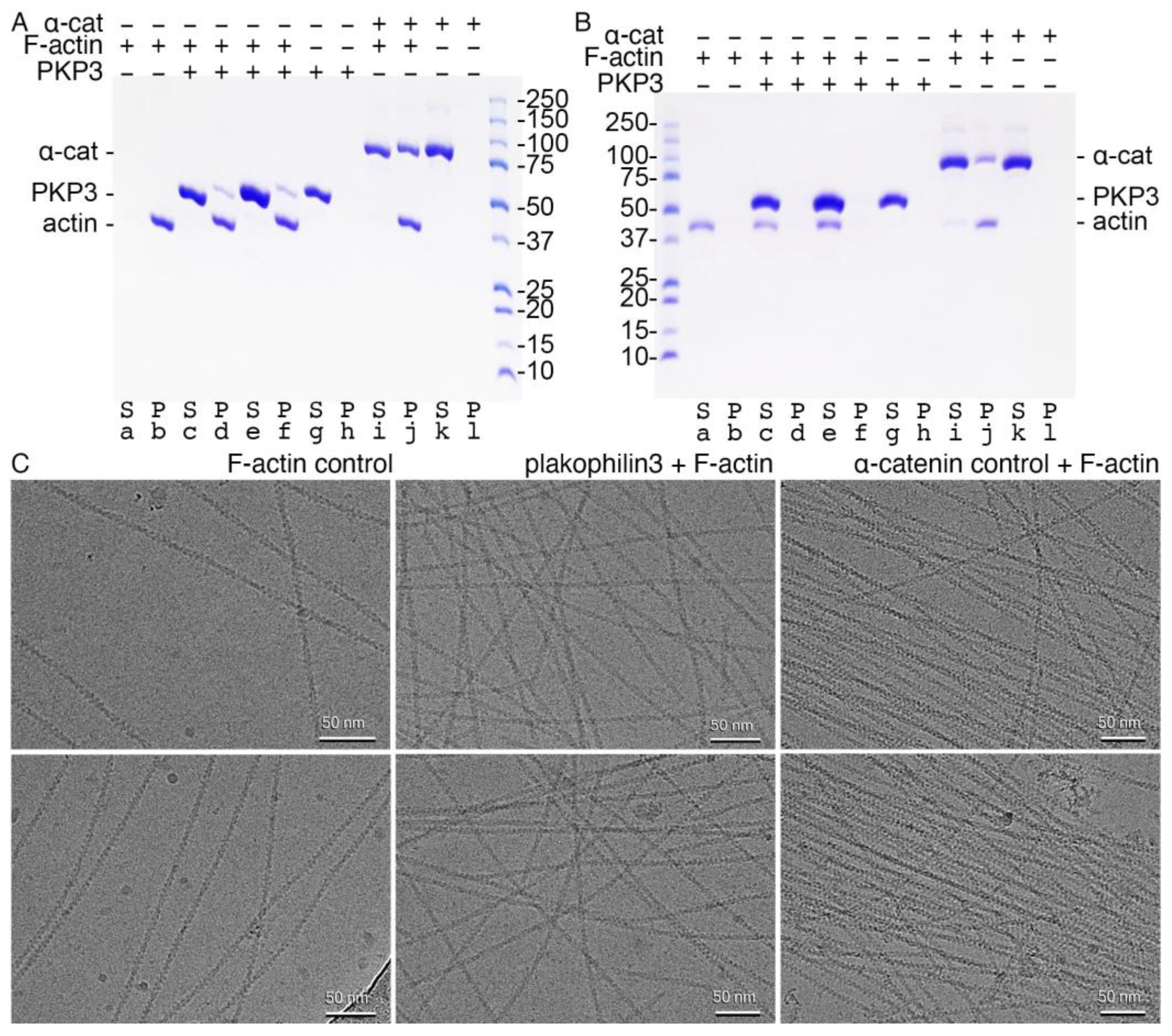
2.4. Plakophilin-3 Binds Actin Filaments but Does Not Bundle F-Actin
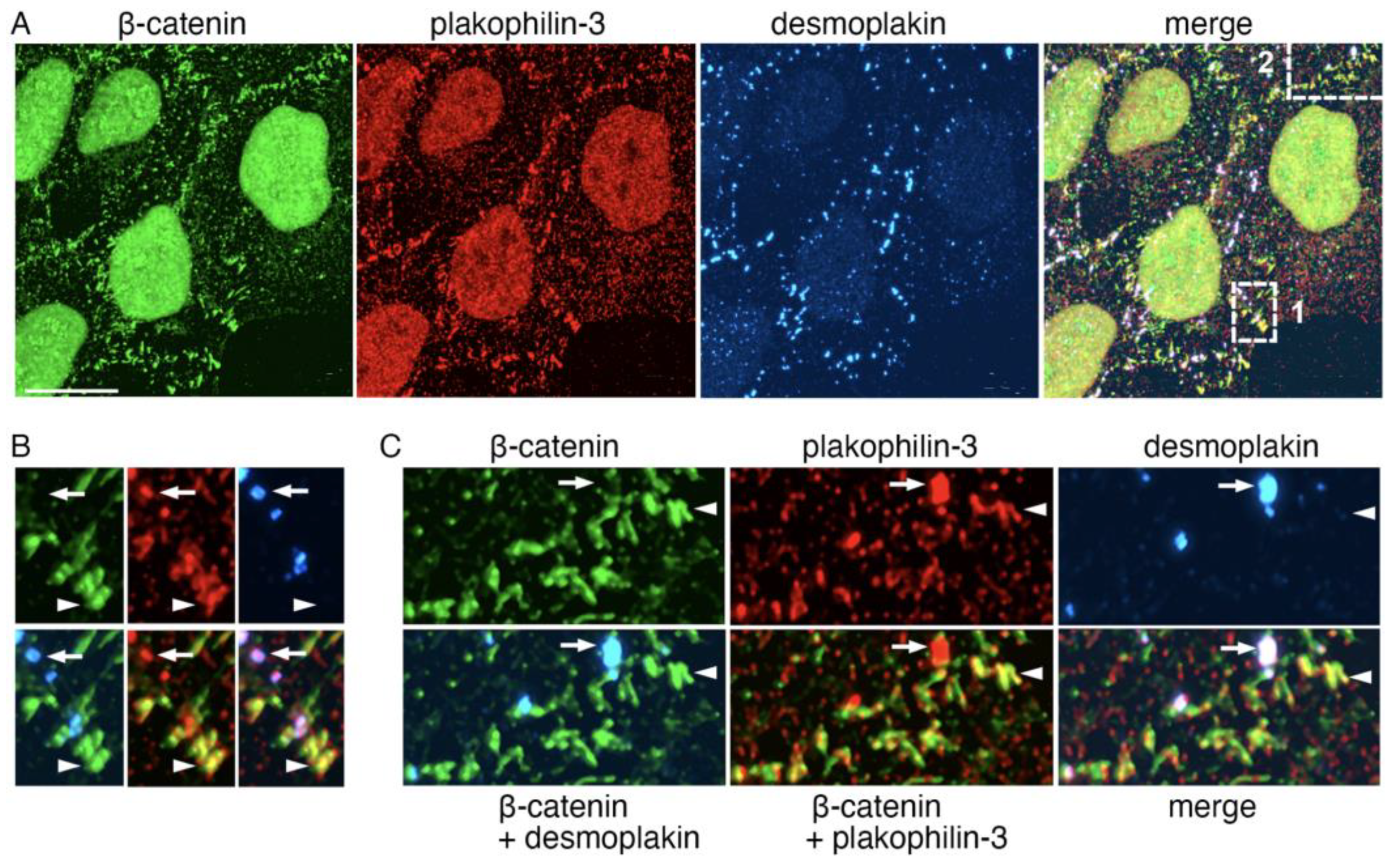
2.5. Plakophilin-3 Forms Actin-Associated Clusters in the Vicinity of Adherens Junctions
3. Discussion
4. Materials and Methods
4.1. Cloning
4.2. Protein Purification
4.3. Size Exclusion Chromatography and Multi-Angle Light Scattering
4.4. Lipid Vesicle Co-Sedimentation
4.5. Preparation of Nanodiscs
4.6. Nanodisc-Plakophilin-3 Binding Assay
4.7. Actin Polymerization
4.8. Actin Binding Assays
4.9. Cryogenic Electron Microscopy Grid Preparation
4.10. Cryogenic Electron Microscopy Data Collection
4.11. CryoEM Data Processing
4.12. Structure Determination
4.13. Immunofluorescence Microscopy
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bass-Zubek, A.E.; Godsel, L.M.; Delmar, M.; Green, K.J. Plakophilins: Multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 2009, 21, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Neuber, S.; Muhmer, M.; Wratten, D.; Koch, P.J.; Moll, R.; Schmidt, A. The desmosomal plaque proteins of the plakophilin family. Dermatol. Res. Pract. 2010, 2010, 101452. [Google Scholar] [CrossRef]
- Hatzfeld, M.; Wolf, A.; Keil, R. Plakophilins in desmosomal adhesion and signaling. Cell Commun. Adhes. 2014, 21, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, I. Plakophilins and their roles in diseased states. Cell Tissue Res. 2020, 379, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Nagatomo, A.; Tsuda, M.; Obata, S.; Sakuma, T.; Yamamoto, T.; Suzuki, S.T. Desmocollin-2 alone forms functional desmosomal plaques, with the plaque formation requiring the juxtamembrane region and plakophilins. J. Biochem. 2015, 158, 339–353. [Google Scholar] [CrossRef]
- Todorovic, V.; Koetsier, J.L.; Godsel, L.M.; Green, K.J. Plakophilin 3 mediates Rap1-dependent desmosome assembly and adherens junction maturation. Mol. Biol. Cell 2014, 25, 3749–3764. [Google Scholar] [CrossRef]
- Indra, I.; Troyanovsky, R.B.; Green, K.J.; Troyanovsky, S.M. Plakophilin 3 and Par3 facilitate desmosomes’ association with the apical junctional complex. Mol. Biol. Cell 2021, 32, 1824–1837. [Google Scholar] [CrossRef]
- Schmelz, M.; Way, D.L.; Borgs, P.; Peitsch, W.K.; Schmidt, H.; Witte, M.H.; Witte, C.L.; Franke, W.W.; Moll, R. A novel type of adhering junction in an epithelioid tumorigenic rat cell culture line. Cell Tissue Res. 1998, 294, 11–25. [Google Scholar] [CrossRef]
- Franke, W.W.; Rickelt, S.; Barth, M.; Pieperhoff, S. The junctions that don’t fit the scheme: Special symmetrical cell-cell junctions of their own kind. Cell Tissue Res. 2009, 338, 1–17. [Google Scholar] [CrossRef]
- Pieperhoff, S.; Barth, M.; Rickelt, S.; Franke, W.W. Desmosomal molecules in and out of adhering junctions: Normal and diseased States of epidermal, cardiac and mesenchymally derived cells. Dermatol. Res. Pract. 2010, 2010, 139167. [Google Scholar] [CrossRef]
- Mohammed, F.; Chidgey, M. Desmosomal protein structure and function and the impact of disease-causing mutations. J. Struct. Biol. 2021, 213, 107749. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.P.; Hatzfeld, M.; Bornslaeger, E.A.; Kopp, D.S.; Borgwardt, J.E.; Corcoran, C.M.; Settler, A.; Green, K.J. The head domain of plakophilin-1 binds to desmoplakin and enhances its recruitment to desmosomes—Implications for cutaneous disease. J. Biol. Chem. 1999, 274, 18145–18148. [Google Scholar] [CrossRef]
- Hatzfeld, M.; Haffner, C.; Schulze, K.; Vinzens, U. The function of plakophilin 1 in desmosome assembly and actin filament organization. J. Cell Biol. 2000, 149, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Bonne, S.; Gilbert, B.; Hatzfeld, M.; Chen, X.; Green, K.J.; van Roy, F. Defining desmosomal plakophilin-3 interactions. J. Cell Biol. 2003, 161, 403–416. [Google Scholar] [CrossRef]
- Fulle, J.B.; Huppert, H.; Liebl, D.; Liu, J.; Alves de Almeida, R.; Yanes, B.; Wright, G.D.; Lane, E.B.; Garrod, D.R.; Ballestrem, C. Desmosome dualism—Most of the junction is stable, but a plakophilin moiety is persistently dynamic. J. Cell Sci. 2021, 134, jcs258906. [Google Scholar] [CrossRef] [PubMed]
- Hatzfeld, M. The p120 family of cell adhesion molecules. Eur. J. Cell Biol. 2005, 84, 205–214. [Google Scholar] [CrossRef] [PubMed]
- McCrea, P.D.; Park, J.I. Developmental functions of the P120-catenin sub-family. Biochim. Biophys. Acta 2007, 1773, 17–33. [Google Scholar] [CrossRef]
- Tewari, R.; Bailes, E.; Bunting, K.A.; Coates, J.C. Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 2010, 20, 470–481. [Google Scholar] [CrossRef]
- Choi, H.J.; Weis, W.I. Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 2005, 346, 367–376. [Google Scholar] [CrossRef]
- Ishiyama, N.; Lee, S.H.; Liu, S.; Li, G.Y.; Smith, M.J.; Reichardt, L.F.; Ikura, M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 2010, 141, 117–128. [Google Scholar] [CrossRef]
- Sobolik-Delmaire, T.; Katafiasz, D.; Wahl, J.K., 3rd. Carboxyl terminus of Plakophilin-1 recruits it to plasma membrane, whereas amino terminus recruits desmoplakin and promotes desmosome assembly. J. Biol. Chem. 2006, 281, 16962–16970. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.J.; Reddy, R.; Wahl, J.K., 3rd. Stratifin (14-3-3 sigma) limits plakophilin-3 exchange with the desmosomal plaque. PLoS ONE 2013, 8, e77012. [Google Scholar] [CrossRef] [PubMed]
- Dries, A.M.; Kirillova, A.; Reuter, C.M.; Garcia, J.; Zouk, H.; Hawley, M.; Murray, B.; Tichnell, C.; Pilichou, K.; Protonotarios, A.; et al. The genetic architecture of Plakophilin 2 cardiomyopathy. Genet. Med. 2021, 23, 1961–1968. [Google Scholar] [CrossRef]
- Wu, M.; Lander, G.C. How low can we go? Structure determination of small biological complexes using single-particle cryo-EM. Curr. Opin. Struct Biol. 2020, 64, 9–16. [Google Scholar] [CrossRef]
- Aiyer, S.; Zhang, C.; Baldwin, P.R.; Lyumkis, D. Evaluating Local and Directional Resolution of Cryo-EM Density Maps. Methods Mol. Biol. 2021, 2215, 161–187. [Google Scholar] [CrossRef]
- Tan, Y.Z.; Baldwin, P.R.; Davis, J.H.; Williamson, J.R.; Potter, C.S.; Carragher, B.; Lyumkis, D. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 2017, 14, 793–796. [Google Scholar] [CrossRef]
- Kirchner, F.; Schuetz, A.; Boldt, L.H.; Martens, K.; Dittmar, G.; Haverkamp, W.; Thierfelder, L.; Heinemann, U.; Gerull, B. Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations. Circ. Cardiovasc. Genet. 2012, 5, 400–411. [Google Scholar] [CrossRef]
- Munoz, W.A.; Kloc, M.; Cho, K.; Lee, M.; Hofmann, I.; Sater, A.; Vleminckx, K.; McCrea, P.D. Plakophilin-3 is required for late embryonic amphibian development, exhibiting roles in ectodermal and neural tissues. PLoS ONE 2012, 7, e34342. [Google Scholar] [CrossRef] [PubMed]
- Bonne, S.; van Hengel, J.; Nollet, F.; Kools, P.; van Roy, F. Plakophilin-3, a novel armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 1999, 112, 2265–2276. [Google Scholar] [CrossRef]
- Madhurantakam, C.; Varadamsetty, G.; Grutter, M.G.; Pluckthun, A.; Mittl, P.R. Structure-based optimization of designed Armadillo-repeat proteins. Protein Sci. 2012, 21, 1015–1028. [Google Scholar] [CrossRef]
- Wood, M.N.; Ishiyama, N.; Singaram, I.; Chung, C.M.; Flozak, A.S.; Yemelyanov, A.; Ikura, M.; Cho, W.; Gottardi, C.J. α-Catenin homodimers are recruited to phosphoinositide-activated membranes to promote adhesion. J. Cell Biol. 2017, 216, 3767–3783. [Google Scholar] [CrossRef] [PubMed]
- Indra, I.; Troyanovsky, R.B.; Shapiro, L.; Honig, B.; Troyanovsky, S.M. Sensing Actin Dynamics through Adherens Junctions. Cell Rep. 2020, 30, 2820–2833.e2823. [Google Scholar] [CrossRef]
- Roberts, B.J.; Pashaj, A.; Johnson, K.R.; Wahl, J.K., 3rd. Desmosome dynamics in migrating epithelial cells requires the actin cytoskeleton. Exp. Cell Res. 2011, 317, 2814–2822. [Google Scholar] [CrossRef] [PubMed]
- Beggs, R.R.; Rao, T.C.; Dean, W.F.; Kowalczyk, A.P.; Mattheyses, A.L. Desmosomes undergo dynamic architectural changes during assembly and maturation. Tissue Barriers 2022, 10, 2017225. [Google Scholar] [CrossRef] [PubMed]
- Moch, M.; Schieren, J.; Leube, R.E. Cortical tension regulates desmosomal morphogenesis. Front. Cell Dev. Biol. 2022, 10, 946190. [Google Scholar] [CrossRef]
- Fuchs, M.; Radeva, M.Y.; Spindler, V.; Vielmuth, F.; Kugelmann, D.; Waschke, J. Cytoskeletal anchorage of different Dsg3 pools revealed by combination of hybrid STED/SMFS-AFM. Cell Mol. Life Sci. 2023, 80, 25. [Google Scholar] [CrossRef]
- Koeser, J.; Troyanovsky, S.M.; Grund, C.; Franke, W.W. De novo formation of desmosomes in cultured cells upon transfection of genes encoding specific desmosomal components. Exp. Cell Res. 2003, 285, 114–130. [Google Scholar] [CrossRef]
- Nekrasova, O.; Green, K.J. Desmosome assembly and dynamics. Trends Cell Biol. 2013, 23, 537–546. [Google Scholar] [CrossRef]
- Bianchini, J.M.; Kitt, K.N.; Gloerich, M.; Pokutta, S.; Weis, W.I.; Nelson, W.J. Reevaluating αE-catenin monomer and homodimer functions by characterizing E-cadherin/αE-catenin chimeras. J. Cell Biol. 2015, 210, 1065–1074. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Izard, T. Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 2013, 20, 188–193. [Google Scholar] [CrossRef]
- Wen, Y.; Vogt, V.M.; Feigenson, G.W. PI(4,5)P(2) Clustering and Its Impact on Biological Functions. Annu. Rev. Biochem. 2021, 90, 681–707. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, S.; Vazquez Nunez, R.; Biertumpfel, C.; Mizuno, N. Bottom-up reconstitution of focal adhesion complexes. FEBS J. 2022, 289, 3360–3373. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.M.; Ramachandran, S.; Case, L.B.; Tolbert, C.E.; Tandon, A.; Pershad, M.; Dokholyan, N.V.; Waterman, C.M.; Campbell, S.L. A Structural Model for Vinculin Insertion into PIP(2)-Containing Membranes and the Effect of Insertion on Vinculin Activation and Localization. Structure 2017, 25, 264–275. [Google Scholar] [CrossRef]
- Chinthalapudi, K.; Rangarajan, E.S.; Patil, D.N.; George, E.M.; Brown, D.T.; Izard, T. Lipid binding promotes oligomerization and focal adhesion activity of vinculin. J. Cell Biol. 2014, 207, 643–656. [Google Scholar] [CrossRef]
- Chinthalapudi, K.; Patil, D.; Rangarajan, E.; Rader, C.; Izard, T. Lipid-directed vinculin dimerization. Biochemistry 2015, 54, 2758–2768. [Google Scholar] [CrossRef]
- Chinthalapudi, K.; Rangarajan, E.S.; Brown, D.T.; Izard, T. Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization. Proc. Natl. Acad. Sci. USA 2016, 113, 9539–9544. [Google Scholar] [CrossRef] [PubMed]
- Chinthalapudi, K.; Mandati, V.; Zheng, J.; Sharff, A.J.; Bricogne, G.; Griffin, P.R.; Kissil, J.; Izard, T. Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2. Nat. Commun. 2018, 9, 1338. [Google Scholar] [CrossRef]
- Chinthalapudi, K.; Rangarajan, E.S.; Izard, T. The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10339–10344. [Google Scholar] [CrossRef]
- Roberts, B.J.; Johnson, K.E.; McGuinn, K.P.; Saowapa, J.; Svoboda, R.A.; Mahoney, M.G.; Johnson, K.R.; Wahl, J.K., 3rd. Palmitoylation of plakophilin is required for desmosome assembly. J. Cell Sci. 2014, 127, 3782–3793. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Izard, T. α-Catenin unfurls upon binding to vinculin. J. Biol. Chem. 2012, 287, 18492–18499. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Smith, E.W.; Izard, T. Distinct inter-domain interactions of dimeric versus monomeric α-catenin link cell junctions to filaments. Commun. Biol. 2023, 6, 276. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, E.S.; Primi, M.C.; Colgan, L.A.; Chinthalapudi, K.; Yasuda, R.; Izard, T. A distinct talin2 structure directs isoform specificity in cell adhesion. J. Biol. Chem. 2020, 295, 12885–12899. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, K.; Alder, N.N. Reconstitution of Mitochondrial Membrane Proteins into Nanodiscs by Cell-Free Expression. Methods Mol. Biol. 2017, 1567, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Troyanovsky, R.B.; Sergeeva, A.P.; Indra, I.; Chen, C.S.; Kato, R.; Shapiro, L.; Honig, B.; Troyanovsky, S.M. Sorting of cadherin-catenin-associated proteins into individual clusters. Proc. Natl. Acad. Sci. USA 2021, 118, e2105550118. [Google Scholar] [CrossRef]
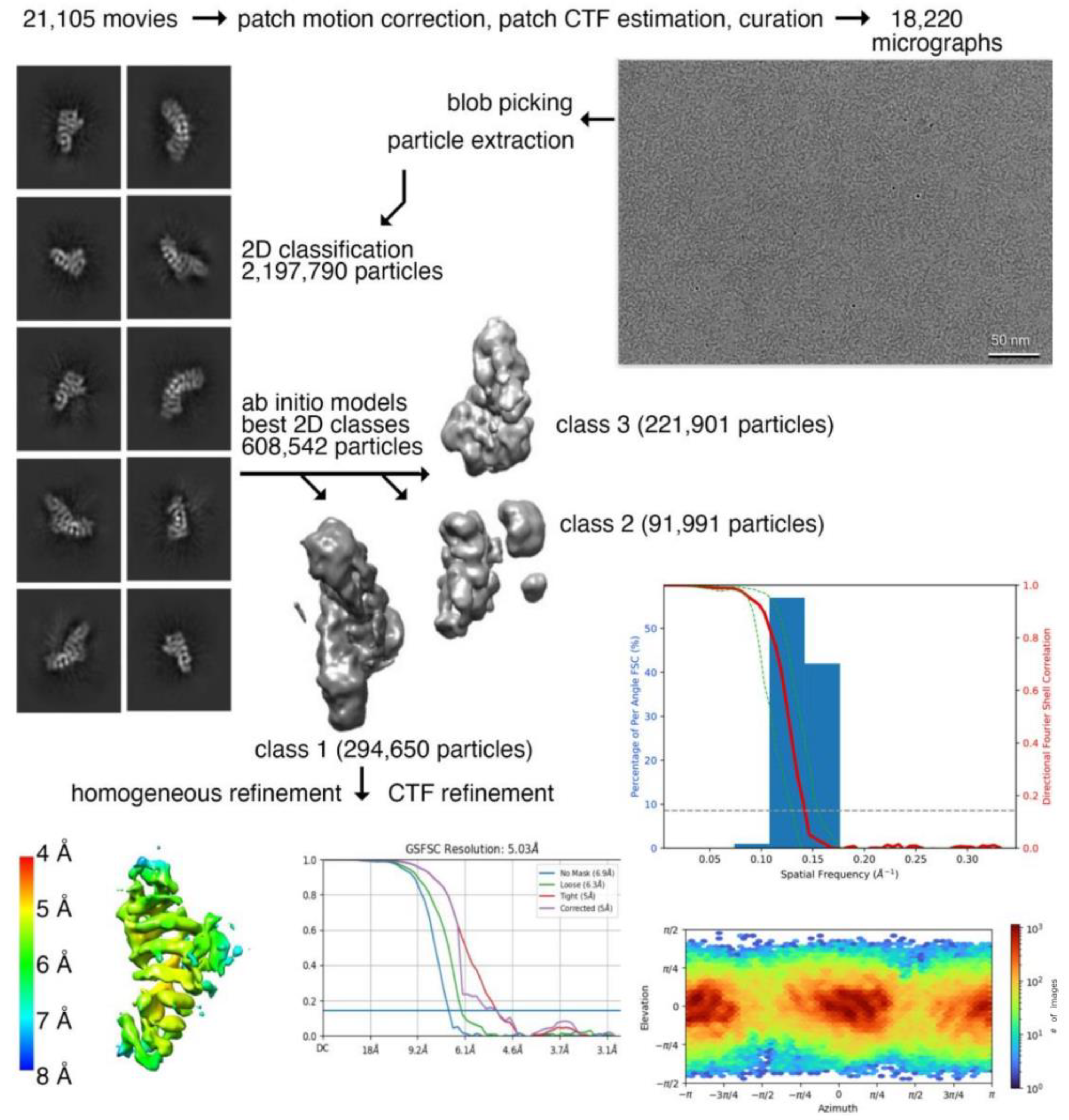



| Data Collection and Processing | Plakophilin-3 (Residues 305-797) |
|---|---|
| magnification | 60,000 |
| total dose | 60 e− per Å2 |
| frames | 50 |
| pixel size | 0.72 Å per pixel |
| defocus range | −0.8 to −2.4 μm |
| number of micrographs | 18,220 |
| number of initial particles | 608,542 |
| number of final particles | 294,650 |
| map resolution | 5.03 Å |
| Fourier shell correlation threshold | 0.143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, J.; Rangarajan, E.S.; Troyanovsky, R.B.; Indra, I.; Troyanovsky, S.M.; Izard, T. Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin. Int. J. Mol. Sci. 2023, 24, 9458. https://doi.org/10.3390/ijms24119458
Gupta J, Rangarajan ES, Troyanovsky RB, Indra I, Troyanovsky SM, Izard T. Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin. International Journal of Molecular Sciences. 2023; 24(11):9458. https://doi.org/10.3390/ijms24119458
Chicago/Turabian StyleGupta, Jyoti, Erumbi S. Rangarajan, Regina B. Troyanovsky, Indrajyoti Indra, Sergey M. Troyanovsky, and Tina Izard. 2023. "Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin" International Journal of Molecular Sciences 24, no. 11: 9458. https://doi.org/10.3390/ijms24119458
APA StyleGupta, J., Rangarajan, E. S., Troyanovsky, R. B., Indra, I., Troyanovsky, S. M., & Izard, T. (2023). Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin. International Journal of Molecular Sciences, 24(11), 9458. https://doi.org/10.3390/ijms24119458







