Abstract
Diaphorina citri, a vector of citrus huanglongbing (HLB) disease, frequently leads to HLB outbreaks and reduces Rutaceae crop production. Recent studies have investigated the effects of RNA interference (RNAi) targeting the Vitellogenin (Vg4) and Vitellogenin receptor (VgR) genes, which are involved in egg formation in this pest, providing a theoretical foundation for developing new strategies to manage D. citri populations. This study presents RNAi methods for Vg4 and VgR gene expression interference and reveals that dsVgR is more effective than dsVg4 against D. citri. We demonstrated that dsVg4 and dsVgR persisted for 3–6 days in Murraya odorifera shoots when delivered via the in-plant system (IPS) and effectively interfered with Vg4 and VgR gene expression. Following Vg4 and VgR gene expression interference, egg length and width in the interference group were significantly smaller than those in the negative control group during the 10–30-day development stages. Additionally, the proportion of mature ovarian eggs in the interference group was significantly lower than that in the negative control group at the 10, 15, 20, 25, and 30-day developmental stages. DsVgR notably suppresses oviposition in D. citri, with fecundity decreasing by 60–70%. These results provide a theoretical basis for controlling D. citri using RNAi to mitigate the spread of HLB disease.
1. Introduction
Diaphorina citri (Kuwayama) (Hemiptera: Psyllidae), a vector of citrus huanglongbing (HLB), frequently causes disease outbreaks [1,2]. Research has shown that D. citri adults can disperse over long distances under high wind speeds in autumn, and the widespread cultivation of Rutaceae crops globally has exacerbated the growth of individual D. citri populations and the resulting damage [3,4]. Consequently, controlling the D. citri population is crucial for suppressing the spread of HLB disease [5,6,7]. Currently, many orchards predominantly rely on pesticides for D. citri management. However, the long-term use of chemical pesticides increases the proportion of insecticide-resistant individuals within the D. citri population, leading to elevated control costs. Hence, it is essential to explore new methods for the integrated management of D. citri.
Advancements in molecular biology technology have enabled the application of sophisticated genetic engineering techniques, such as RNA interference (RNAi) and CRISPR-associated protein 9 (CRISPR/Cas9), in pest management. Recent studies have focused on the interference and knockout of oviposition genes closely associated with pest population development, including the phospholipase A2, sugar gustatory receptor, odorant receptor coreceptor, diapause hormone, dopa decarboxylase, and sucrose hydrolase gene families [8,9,10,11,12,13]. The Vitellogenin (Vg) and Vitellogenin receptor (VgR) gene families have been shown to play a direct role in egg formation [14]. For instance, knocking out the Tudor gene in Bactrocera dorsalis (Diptera: Trypetidae) resulted in reduced ovarian development and mating rates, with dsTudor interference efficiency reaching 95%; interfering with the expression of Vg and VgR genes effectively hindered Vitellogenin accumulation and mature egg formation in the ovaries [15,16,17]. A significant reduction in the next generation’s population size will occur if the pest cannot accumulate sufficient Vitellogenin.
It is well established that Vg protein is critical to insect egg production and embryonic development. In order to mitigate the damage caused by D. citri and suppress the spread of HLB disease, RNAi has been employed to inhibit Vg and VgR gene expression in the ovaries, thus controlling egg formation and representing a novel approach to managing D. citri populations. A study revealed that female egg production requires the mobilization of nutrients for egg cells in the ovary to synthesize Vitellogenin [18]. Vitellogenin is produced in the fat body, secreted into the hemolymph, and subsequently incorporated into developing oocytes via VgR-mediated endocytosis [14]. Consequently, RNAi was utilized to interfere with the expression of female Vg and VgR genes and egg formation as a viable method for controlling pest damage. Previous research indicated that Vg gene expression can directly or indirectly affect insect ovary development, while VgR gene expression can influence egg formation, oviposition behavior, and other physiological activities related to oviposition [15,18].
Vitellogenin-1-like-1 (Vg1) (Gene id: LOC103,523,873; gene full length: 1858 bp), Vitellogenin-1-like-2 (Vg2) (Gene id: LOC103,523,874; gene full length: 1116 bp), Vitellogenin-2-like (Vg3) (Gene id: LOC103,513,507; gene full length: 1197 bp), Vitellogenin-3-like (Vg4) (Gene id: LOC103,523,199; gene full length: 4808 bp), Vitellogenin-like (Vg5) (Gene id: LOC113,469,177; gene full length: 1002 bp), and VgR (Gene id: LOC103,524,089; gene full length: 5950 bp) were identified. Bioinformatics information was comprehensively described based on the D. citri female transcriptome and proteome we previously reported (NCBI: PRJNA669507; ProteomeXchange: PDX022359) [19]. However, the developmental characteristics of the ovary were not documented after interference with the Vg and VgR gene expression in D. citri. Currently, RNAi has been widely applied in insect gene function studies, primarily employing the delivery of double-stranded ribonucleic acid (dsRNA) into insects via quantitative injection. The advantage of the quantitative injection method is the control over the amount of dsRNA administered to insects. However, its disadvantages include causing injury to insects, resulting in a reduced lifespan of research subjects and the inability to perform multiple injections. Small insects such as D. citri and aphids are not suitable for using the injection method to deliver dsRNA. Instead, the direct oral feeding of dsRNA on D. citri was mainly applied to verify the effect or function of relative genes for a period of time [20,21]. The indirect delivery of dsRNA to D. citri through plants was less reported and the observation time was usually short, with the longest observation time not exceeding 25 days [22]. The effects of dsRNA long-term (more than 30 days) feeding on the gene interference expression of D. citri are almost unknown.
We further discovered that the Vg4 protein, corresponding to the Vg4 gene, had the most complex amino acid structure in the Vg gene family and was highly expressed in the female abdomen. This finding suggests that the Vg4 gene may play a crucial role in egg formation physiology. There was only one VgR gene, which was directly selected as an interference target. In this study, we improved and innovated Andrade’s method of feeding dsRNA through plants (in-plant system: IPS) to observe the long-term effects of feeding dsVg4 and dsVgR on the oviposition activities of D. citri [22]. We tested whether dsVg4 and dsVgR could effectively reduce the expression levels of Vg4 and VgR genes in D. citri, thereby interfering with egg formation. To address our research question, we established two treatments (i.e., dsVg4 and dsVgR) and two controls (i.e., double-stranded green fluorescent protein (dsGFP) and blank control) to examine the following aspects: (1) the stability of dsRNA in Murraya odorifera shoots using IPS delivery; (2) Vg4 and VgR gene expression levels and ovarian development; (3) differences in egg formation and the number of malformed eggs and nymphs; (4) variations in fecundity and egg hatchability; and (5) suppression effects of dsVg4 and dsVgR on female oviposition.
2. Results
2.1. Stability of dsRNA in M. odorifera Shoots
Gel electrophoresis was employed to assess the stability of dsVg4, dsVgR, and dsGFP in treated M. odorifera shoots over six days. Clear gel electrophoresis bands were observed at 1–6 days in the M. odorifera shoots treated with dsGFP, indicating the feasibility of IPS for delivering dsRNA (Figure 1). DsVg4 and dsVgR displayed clear bands at 1–2 days, suggesting that dsVg4 and dsVgR maintained a relatively complete double-stranded structure during this period. Once D. citri females fed on the M. odorifera shoots for 1–2 days, dsVg4 and dsVgR readily entered the female body. The bands became blurred after 3–6 days, signifying that dsVg4 and dsVgR started to decompose at this stage. However, traces of bands in dsVg4 gel electrophoresis indicated that the structures of some dsVg4 and dsVgR remained intact at 3–6 days (Figure 1).
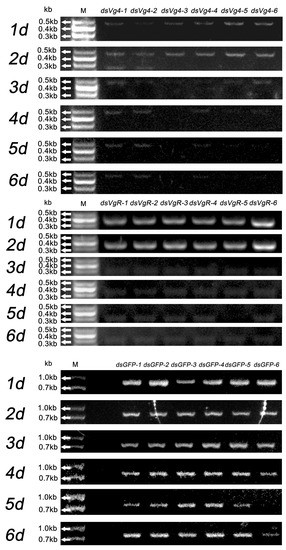
Figure 1.
DsVg4, dsVgR, and dsGFP were absorbed by tender M. odorifera shoots for six days, and gel electrophoresis was used to detect the persistence of dsVg4, dsVgR, and dsGFP after each day. The gel electrophoresis bands of dsVg4 were relatively clear at 1–6 d; the gel electrophoresis bands of dsVgR were relatively clear at 1–2 d, but blurry at 3–6 d; the gel electrophoresis bands of dsGFP were relatively clear at 1–6 d. The dsVg4-1, 2, 3, 4, 5, 6/dsVgR-1, 2, 3, 4, 5, 6/dsGFP-1, 2, 3, 4, 5, 6 represent the six replicates for each treatment.
Chromas analysis of gene sequencing results for dsVg4 and dsVgR was consistent with the template gene used to synthesize dsRNA (Figure S1). Gel electrophoresis was also performed on the dsGFP group and blank control group using Vg4 and VgR PCR primers to demonstrate the reliability of the PCR results. No clear bands were observed in the gel electrophoresis when Vg4 and VgR PCR primers were used in the PCR process for the dsGFP group and blank control group, proving the specificity of Vg4 and VgR PCR primers (Figure S2). The original gel electrophoresis photos of these PCR tests are available in the Supplementary Materials (Figures S3–S5).
2.2. Gene Expressions of Vg4 and VgR and Ovarian Development
After D. citri females were fed dsVg4, the expression of the Vg4 gene decreased during the 1–30 d period. The expression of the Vg4 gene was most significantly decreased at 15 d, and the relative expression declined by 99.06% (p < 0.05). However, the expression of the Vg4 gene abnormally increased at 20 d, with the relative expression rising by 143.65% (p < 0.05) (Figure 2). This finding indicates that, at the 20 d developmental stage, females exhibited the strong immune regulation of dsVg4. By dissecting the internal reproductive organs of females with interference in Vg4 gene expression, we observed that the number of eggs in the ovary of the Vg4 interference group was lower than that in the negative control group at 1–30 d. Ovaries from females in the negative control group began to form a large number of eggs at 15 d, and a certain number of eggs could be observed in the ovaries at 15–30 d, which was similar to the normal development rate (Supplementary Materials). Fewer mature and immature eggs were observed in the ovaries of the interference group, and a large number of eggs had not yet formed in the ovaries.
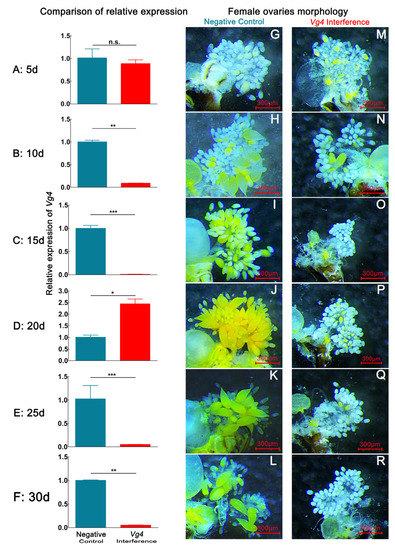
Figure 2.
Interference effect of dsGFP and dsVg4 via continuous dsVg4 feeding over 30 d on the Vg4 expression and ovary developmental morphological characteristics in D. citri. (A,G,M), (B,H,N), (C,I,O), (D,J,P), (E,K,Q), (F,L,R) represent the relative expression level of Vg4 and ovarian morphology of Vg4 interference-treated and negative-control groups on the 5th, 10th, 20th, 25th, and 30th days, respectively. Data are shown as the mean ± SD. Values of P were based on the independent samples t test: *** p < 0.001; ** p < 0.01; * p < 0.05; ns, p ≥ 0.05. The eggs have a yellow color. The blue color in the bar chart represents the negative control group, and the red color represents the Vg4 interference group. (G–L) correspond to the ovaries of negative control group females. (M–R) correspond to ovaries of Vg4 interference group female.
After D. citri females were fed dsVgR, the expression of the VgR gene decreased during the 1–30 d period. The expression of the VgR gene decreased most significantly at 25 d, with the relative expression declining by 99.57% (p < 0.05). The expression of the VgR gene abnormally increased at 20 d, and the relative expression rose by 256.93% (p < 0.05) (Figure 3). By dissecting the internal reproductive organs of females with interference in VgR gene expression, we observed that the number of eggs in the ovary of the interference group was lower than that of the negative control group at 1–30 d, similar to the findings for the Vg4 interference group (Table 1).
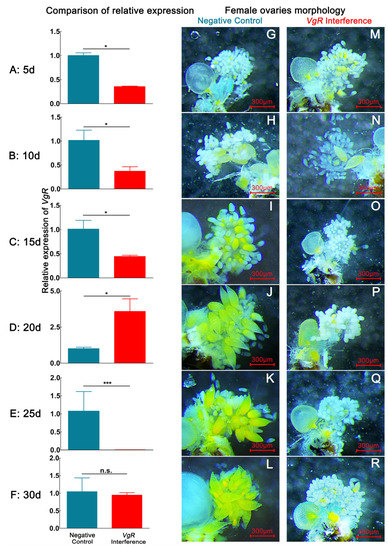
Figure 3.
The interference effect of dsGFP and dsVgR on VgR expression and ovary developmental morphological characteristics in D. citri via continuous dsVgR feeding within 30 d. (A,G,M), (B,H,N), (C,I,O), (D,J,P), (E,K,Q), and (F,L,R) represent the relative expression level of VgR and ovarian morphology of VgR interference-treated and negative-control groups on the 5th, 10th, 20th, 25th, and 30th days, respectively. Data are shown as the mean ± SD. P values are from the independent samples t test: *** p < 0.001; * p < 0.05; ns, p ≥ 0.05. The eggs have a yellow color. The blue color in the bar chart represents the negative control group, the red color represents the VgR interference group. (G–L) correspond to the ovaries of negative control group female. (M–R) correspond to the ovaries of VgR interference group female.

Table 1.
Comparison of egg size and number of mature eggs of D. citri in different treatments. The egg length and width, number of eggs, and proportion of mature eggs were compared between 1 and 30 d. Data from the same day were compared vertically.
2.3. Effects of dsVg4 and dsVgR on the Morphology of Eggs and Nymphs
Interference with Vg4 and VgR gene expression in D. citri females resulted in a relatively small number of mature ovarian eggs at the 15–30 d stage (Figure 2 and Figure 3, Table 1). At the 5 d developmental stage, the ovaries of both the interference group and the negative control group had not formed any eggs. Mature eggs were only found in the dissected ovaries of the Vg4 and VgR interference groups at the 10 d developmental stage. At this stage, eggs in the Vg4 interference group exhibited an average length of 136.30 ± 15.55 µm, an average width of 62.58 ± 6.30 µm, a maximum number of mature eggs of 1.03 ± 0.37 eggs, and a maximum percentage of mature eggs of 8.33 ± 1.67%. In the VgR interference group at the 10 d developmental stage, eggs exhibited an average length of 123.32 ± 30.03 µm, average width of 61.62 ± 6.59 µm, maximum number of mature eggs of 0.75 ± 0.25 eggs, and maximum mature egg percentage of 7.75 ± 0.50%. The egg size and mature egg proportion of the Vg4 and VgR interference groups were lower than those of the negative control group at the 10 d developmental stage. At the 15–30 d developmental stage, mature eggs were not observed in the Vg4 and VgR interference groups, and only a few immature eggs were observed in the ovarioles.
After interfering with Vg4 and VgR gene expression in D. citri females, abnormal eggs and nymphs were found on the M. odorifera shoots (Figure 4). Females in the negative control group produced normal eggs, which were shaped like droplets and had smooth, yellow surfaces. In contrast, the egg surfaces of the Vg4 and VgR interference groups were abnormal, appearing shriveled and brown compared to normal eggs. Normal nymphs in the negative control group had smooth backs and yellow bodies, while abnormal nymphs in the Vg4 and VgR interference groups exhibited wrinkled backs, brownish-yellow bodies, and some translucent nymph bodies in the Vg4 interference group. None of the abnormal eggs hatched, and the abnormal nymphs displayed abnormal characteristics at the 1–2 instar stage.
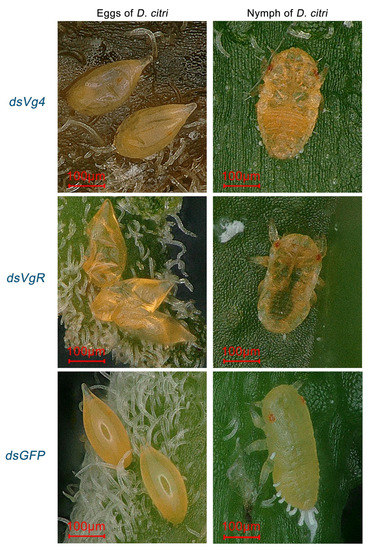
Figure 4.
Typical abnormal shriveled eggs and wrinkled nymphs of the Vg4 and VgR interference groups in D. citri. Eggs in dsGFP have a water droplet-like shape with a smooth, yellow surface. Eggs in the Vg4 and VgR interference groups appeared shrunken and brown compared to normal eggs, with an abnormal surface. Nymphs in dsGFP have a yellow body with a smooth back, while abnormal nymphs in the Vg4 and VgR interference groups showed wrinkled backs with a brown-yellow body color, and some nymphs in the Vg4 interference group had a semi-transparent body. Abnormal eggs did not hatch, and abnormalities were observed in the nymphs at the 1–2 instar stage.
2.4. Comparison of Oviposition Suppression between dsVg4 and dsVgR
In the Vg4 interference group, a total of 1379 eggs were collected, with an average of 45.97 ± 7.31 eggs per female. Out of these, 1190 eggs hatched into nymphs, resulting in an average hatchability of 84.41 ± 6.50%. The total deformity rates for eggs and nymphs were 4.42% and 4.12%, respectively (Table 2). In the VgR interference group, a total of 638 eggs were collected, with an average of 20.80 ± 9.37 eggs per female. Among these, 594 eggs hatched into nymphs, yielding an average hatchability of 81.11 ± 7.30%. The total deformation rates for eggs and nymphs were 6.80% and 1.35%, respectively. In the dsGFP and blank control groups, 2804 and 2457 eggs were collected, with an average of 93.47 ± 10.20 and 84.97 ± 12.28 eggs per female, respectively. In these groups, 2570 and 2380 eggs hatched into nymphs, with average hatchability rates of 94.89 ± 2.76% and 96.67 ± 1.94%, respectively. No deformed eggs or nymphs were observed in these groups.

Table 2.
Comparison of D. citri oviposition and nymph formation in interference group and control group. The items compared were the total number of eggs deposited, number of eggs deposited per female, total number of nymphs, egg hatchability, egg deformity, and nymph deformity. Units of comparison items are provided in parentheses.
3. Discussion
Our findings indicate that RNAi through the IPS method could successfully suppress egg formation in D. citri. This suppression is attributed to the interference of dsVg4 and dsVgR, which disturbed ovarian development and subsequently decreased fecundity and egg hatchability [23,24,25]. This was especially true for dsVgR, which proved to be more effective than dsVg4 in terms of suppressing egg formation.
Based on the results of dsRNA stability in M. odorifera shoots, our study demonstrated that delivering dsRNA to an insect is feasible through plant absorption (Figure 1). Since the dsRNA solution could be directly analyzed by RT-PCR even after plant absorption, the total plant RNA could be extracted for RT-PCR to verify the presence of dsRNA (Supplementary Materials). This detection method is widely used in research on dsRNA viruses and dsRNA transgenic crops [26,27,28]. DsRNA can persist for a period of time in liquid form under certain environmental conditions. For instance, a study found that after leaf tissue absorbed dsGFP, the tissue tested positive for dsGFP at 24 h post-treatment and remained positive for up to 40 days post-treatment [22]. DsRNA can be detected by reverse transcription reaction after being absorbed by plants, as dsRNA is not stable in the environment. It has been reported that the half-life of naked siRNA in serum ranges from several minutes to about an hour [26]. This indicates that dsRNA has difficulty maintaining the stability of its double-stranded structure for extended periods in the complex internal environment of plants. Consequently, if dsRNA persists in plants for more than 24 h and no suitable amplification site can be found, dsRNA will transition to a single strand and further decompose. The cDNA and PCR reagents can be used to detect the presence of dsRNA when the dsRNA in plants becomes single-stranded. It is worth noting that, compared to the Vg4 and VgR interference treatment groups, M. odorifera shoots treated with dsGFP (726 bp) displayed relatively clear bands within 1–6 days, suggesting that the half-life of dsRNA may be related to the length of its base sequence (Figure 1 and Figure S5). A study found that the 1454 bp dsRNA virus of Trichomonas vaginalis requires boiling water treatment for qRT-PCR analysis, which also supports the idea that a longer dsRNA base sequence provides greater stability and a longer half-life [27]. This finding indicates that designing target gene dsRNA interference fragments and extending the base length of dsRNA within a controllable range may increase dsRNA’s half-life, although more experiments are needed for verification. In this study, the gel electrophoresis of dsVg4 and dsVgR demonstrated that dsVg4 and dsVgR could persist for 2–5 days (Figure 1), indicating that dsVg4 and dsVgR could be absorbed by M. odorifera shoots and accumulate in the leaves, effectively delivering dsVg4 and dsVgR to females feeding on M. odorifera shoots. Consequently, dsRNA formulated into an appropriate molecular dosage form may be suitable for the integrated management of D. citri populations and/or can be used in combination with certain chemical pesticides [29].
Gene expression data demonstrated that the Vg4 and VgR expression levels were significantly affected by dsVg4 and dsVgR in D. citri. At the 1–30-day developmental stage, the relative expression of the Vg4 and VgR genes was detected at six time points, revealing an overall decrease in their expression. However, the relative expression of the VgR gene abnormally increased by 256.93% at the 20-day developmental stage (Figure 3). This phenomenon may be attributed to cellular immune regulation by dsRNA. Previous studies have indicated that dsRNA-degrading enzymes (dsRNases) are important factors in reducing RNA interference efficiency in different insect species and can lead to an increase in gene expression [30,31].
The gene expression results show that after D. citri females were fed dsRNA for an extended period, they were likely to exhibit a set of immune physiological effects against dsRNA through an immune response, which may result in a decline in the interference effect of dsRNA on the target gene expression. Typically, the secretion of dsRNA-degrading enzymes is part of the insect immune response [32]. Whether the long-term feeding of dsRNA leads to sustained immune response activation remains unclear, and research on pest resistance to dsRNA pesticides is not comprehensive.
The large-scale application of dsRNA for managing D. citri would begin by bypassing the immune response physiology of D. citri and enhancing the integrated management effect of dsRNA without stimulating the immune response of D. citri. However, further research is required to understand the immune response in D. citri induced by dsRNA and the related mechanisms.
DsVg4 and dsVgR significantly suppress egg formation in D. citri, demonstrating that the Vg and VgR gene families have a crucial impact on ovarian development [19]. This study’s results also revealed that the number of mature eggs in the ovary and the number of eggs/female significantly decreased after dsVg4 and VgR interference with the Vg4 and VgR genes. The length and width of female eggs appeared intermittently in the 10–30-day VgR interference group (Table 1) because dsVgR disrupted the formation of the VgR receptor, preventing the normal transport of Vg proteins to the ovary [33].
Although the ovary could not accumulate enough Vg proteins to form eggs, dsVgR only interfered with, rather than blocked, Vg protein accumulation. Females could still carry out oviposition activities to deposit eggs on plants. After oviposition activities, the ovary needed to re-accumulate Vg protein in case of insufficient VgR receptor. Consequently, the length and width of eggs appeared intermittently in the 10–30 day VgR interference group. The suppressive effect of dsVgR on egg formation was stronger than that of dsVg4, suggesting that the effect of VgR gene expression was more influential than the Vg4 gene on mature egg formation [34]. However, the deletion of the VgR receptor can lead to the insufficient accumulation of Vg4 protein in the ovary. Thus, after interference with Vg4 and VgR gene expression, the ovarian development characteristics between the Vg4 and VgR interference groups were similar.
From the experimental results, there may be an upstream and downstream relationship between the Vg4 gene and VgR gene expression, with the VgR gene likely being the upstream gene of the Vg4 gene. In reality, the gene expression network involved in ovarian Vg protein synthesis and transport may be more complex and requires a comprehensive and systematic study of the Vg protein synthesis and transport gene family. Constructing a gene bank of D. citri egg formation-related genes would be beneficial for the future integrated management of D. citri populations using molecular biology technology.
Although the RNAi of the expression of the Vg4 and VgR genes could not entirely block female oviposition, if combined with other integrated pest management (IPM) control methods and strictly following the IPM principle, the occurrence rate of D. citri outbreaks could be reduced to very low levels. In this study, an interference system for the long-term feeding of dsVg4 and dsVgR to knock down the expression of female Vg4 and VgR genes was established. It can continuously feed adults with dsRNA from the emergence to death stages, allowing for multiple oviposition phenotype and behavior studies. In the context of the successful RNAi of dsVg4 and dsVgR, the functions of more oviposition-related genes can be evaluated in D. citri, such as upstream and downstream gene families associated with Vg4 and VgR.
The functions of regulatory genes of Vg and VgR have been reported in some insects, while relative reports in D. citri are limited. For example, interference of the Krüppel homolog 1 (Kr-h1) gene in Bombyx mori female pupae severely inhibited VgR transcription, leading to reduced Vg deposition in oocytes [35]; Vg synthesis is directly affected by doublesex in Agrotis ipsilon, mucin genes in Nilaparvata lugens [36,37]; as well as DOPA decarboxylase in Drosophila sechellia (Diptera: Drosophilidae) and Plutella xylostella (Lepidoptera: Plutellidae) [38,39], etc. Therefore, further studies on the regulatory genes associated with the expression of the Vg and VgR gene family can help provide theoretical support for the systematic control of D. citri populations using RNAi. In addition to the results in the current study, further experiments are required to elucidate the impact of the Vg4 and VgR gene families on the oviposition physiology and behavior of D. citri females. These research efforts can provide a gene data theoretical basis for controlling the damage caused by D. citri populations using RNAi technology in the future [40].
Moreover, in terms of pest management, the application of RNAi not only depends on the suppressive effect of dsRNA on oviposition but also requires the establishment of proper methods for utilizing dsRNA. To minimize insect resistance and the environmental impact of insecticides, it is crucial to develop appropriate molecular pesticide formulations with dsRNA. Previous studies have reported that the effectiveness of dsRNA combined with specific nanomaterials may be superior to some chemical pesticides against insect pests, such as Anopheles gambiae, B. dorsalis, and Apolygus lucorum [41,42,43,44,45,46,47]. If dsRNA can be combined with nanomaterials or formulated into corresponding agents, it can be employed for the large-scale integrated management of D. citri populations. However, it is also necessary to evaluate the interference effect of dsRNA on different day-ages in D. citri and determine the optimal time for suppressing female oviposition.
4. Materials and Methods
4.1. Insect Rearing and Collection
Adult D. citri were collected from M. odorifera plants in the citrus orchard of Guangxi University, China (108.290° E, 22.849° N), and raised in five indoor cages (90 × 90 × 100 cm3). M. odorifera plants with heights ranging from 80 to 100 cm were transferred to insect cages as food. Six hundred adults that emerged after 8–11 days were paired in the indoor cages mentioned above, and tender shoots of M. odorifera seedlings were used as oviposition substrates. Newly emerged adults of the next generation were used as the experimental insects. All experiments were conducted indoors at a temperature of 25 ± 1 °C, relative humidity of 75 ± 5%, and a light: dark cycle of 14:10 h.
Males and females that emerged on the same day were used in the following experiments, and all were reared in plastic containers. The plastic container consisted of a cup (148 mm high × 60 mm wide at the bottom), a cover (95 mm in diameter), and a plastic platform (70 mm in diameter) and contained 1–2 M. odorifera shoots and a 1.5 mL Eppendorff (ep) tube. A cover could be opened and closed to facilitate the replacement of dsRNA solution and M. odorifera shoots (Figure 5). For the RNAi assay, dsRNA was dissolved in water, added to the tube, and then transported to the tender shoot through the xylem of M. odorifera for feeding the females. Mating and oviposition among the adults in the rearing container were observed and recorded.
Figure 5.
Insect rearing device. (A) Plastic container with M. odorifera; and (B) Mating of male and female adults on M. odorifera.
4.2. Ovary Anatomy and Measurement
Female D. citri were rendered immobile by dipping in liquid nitrogen prior to dissection and quickly transferred to alcohol-treated wax tablets for ovary dissection. The specimens were soaked in precooled 1X PBS (37 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4) and immediately dissected under a stereoscope (SMZ800N, Nikon, Japan) using fine insect anatomical needles. In brief, a complete ovary was obtained by cutting the side of the female abdomen and removing the abdominal exoskeleton and other tissues. Subsequently, a typical ovary was photographed, and a hand drawing of the ovary was created by tracing on sulfuric acid paper (Figure 6).
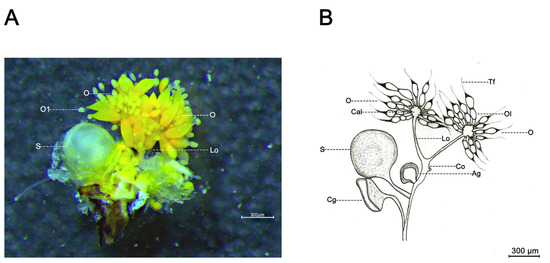
Figure 6.
Ovarian morphology of D. citri female. (A) Ovarian morphology; and (B) Hand drawing of ovarian morphology. Ag, accessory glands; Cal, calyx; Cg, colleterial gland; Co, common oviduct; Lo, lateral oviduct; O, ovary; Ol, ovariole; S, spermatheca; Tf, terminal filament.
4.3. DsRNA Synthesis
Ovaries of D. citri were dissected as previously described and immediately frozen in liquid nitrogen in a 1.5 mL Eppendorff tube for total RNA extraction using TransZol Up (TransGen Biotech, Beijing, China). Then, cDNA synthesis was carried out using the PrimescriptTM RT reagent kit with gDNA eraser (perfect real time) (TaKaRa, Tokyo, Japan) following the manufacturer’s instructions: (1) prepare the following mixture in an RNase-free Eppendorff tube, RNase-free ddH2O to 16 µL, 4 µL 4 × 2-Step Gdna Erase-Out Mix, total RNA 1 pg-1 µg, 42 °C reaction for 2 min; (2) add 4 µL 5× ToloScript qRT EasyMix directly to the reaction solution in step 1, conduct reverse transcription reaction, 37 °C for 15 min, 85 °C for 5 s. Vg4 and VgR gene interference fragments were designed using E-RNAi Webservice https://www.dkfz.de/signaling/e-rnai3/idseq.php (accessed on 15 February 2023) (E-RNAi team, Thomas Horn and Michael Boutros). Using the nucleotide sequences of the Vg4 and VgR genes, primers were designed for PCR, quantitative real-time PCR (qRT–PCR), and dsRNA synthetic primers in NCBI (National Center for Biotechnology Information, Bethesda, Maryland, USA) (Table 3). Vg4 (477 bp) cDNA fragments were generated with the primer pair Vg4 F3/R3 using 2 × FastPfu Premix (TOLOBIO, Shanghai, China). The purified PCR Vg4 fragments were inserted into the plasmid of the pMD18T vector (DH5α, Thermo Fisher Scientific, Waltham, Massachusetts, USA). The resulting Vg4 plasmids were used as templates to generate dsVg4 with the dsRNA synthesis kit (RNAsyn Biottech Co., Ltd., Jiangsu, Suzhou, China; Supplementary Materials), following the manufacturer’s instructions: the preparation of dsRNA synthesis (DEPC water 51.6 μL, 5× transcription buffer 24 μL, NTP 20.4 μL, transcriptional template 6 μL, RNase enzyme inhibitor 3.6 μL, DTT 2.4 μL, RNA polymerase 12 μL), transcript at 37 °C for 1 h, add 12 μL DNaseI and react at 37 °C for 30 min, remove the template, and purify using NEB’s RNA purification kit (NEB, Ipswich, Massachusetts, USA) to obtain positive and negative Vg4 RNA; mix positive and negative Vg4 RNA in equal proportions and anneal to obtain dsVg4, with detailed synthesis methods provided in the Supplementary Materials. Synthesized dsRNA was quantitated by an N80 Touch nanophotometer (Implen, Munich, Bavaria, Germany) at 260 nm, and the integrity was analyzed by agarose gel electrophoresis. The syntheses of dsVgR (346 bp) and dsGPF (726 bp) were carried out as described for dsVg4 (Table 3).

Table 3.
Oligonucleotide primer pairs used in this study.
4.4. Delivery and Stability of dsVg4 and dsVgR in M. odorifera Shoots
Six M. odorifera shoots were immersed in a plastic container containing dsVg4 solution (1 mL) at a concentration of 10 ng/µL. The dsVg4 solution in the Eppendorff tubes was maintained at 1 mL by adding water. Individual M. odorifera shoot and leaf samples were collected once a day for six days, immediately frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. Total RNA and cDNA from shoots and leaves of M. odorifera were sequentially prepared as previously described. The cDNA, used as a template, was employed to clone Vg4, and the PCR product was visualized by gel electrophoresis as described previously (repeated at least six times). Clear bands indicated that dsVg4 was stable and transferred to shoots and leaves as expected. The stability of dsVgR was the same as that of dsVg4. The clear bands of Vg4 and VgR during gel electrophoresis were purified using the EasyPure Quick Gel Extraction Kit (Transgen, Beijing, China) for sequencing (RNAsyn Biottech Co., Ltd., Jiangsu, Suzhou, China) to verify whether the cloned fragment base sequences corresponded to dsRNA. The sequenced data were analyzed by Chromas (Technelysium Pty Ltd., South Brisbane, Queensland, Australia). DsGFP-designed primers were used for independent stability detection; the stability detection method for dsGFP was identical to that of dsVg4.
4.5. Interference Efficiency of dsVg4 and dsVgR Treatment and Ovarian Morphological Changes in D. citri
Six rearing containers were allocated to each of the dsVg4, dsVgR, and dsGFP treatments. Fifteen one-day-old females and males were randomly selected, paired, and reared in each container as previously described. Females from one container per treatment were selected every 5 days and dissected for ovary sampling, while M. odorifera tender shoots and dsVg4 solution were replaced in other containers. The dissected ovaries were first photographed with a microscopic measurement system coupled to a stereoscope (SMZ800N, Nikon, Japan) for further egg measurements (Figure 7) and then sampled for RNA extraction. In this way, ovaries were collected from 5-, 10-, 15-, 20-, 25-, and 30-day-old females that were treated with dsVg4, dsVgR, and dsGFP. The cDNA from these samples was prepared as previously described and used for qRT–PCR assays to check RNAi efficiency.
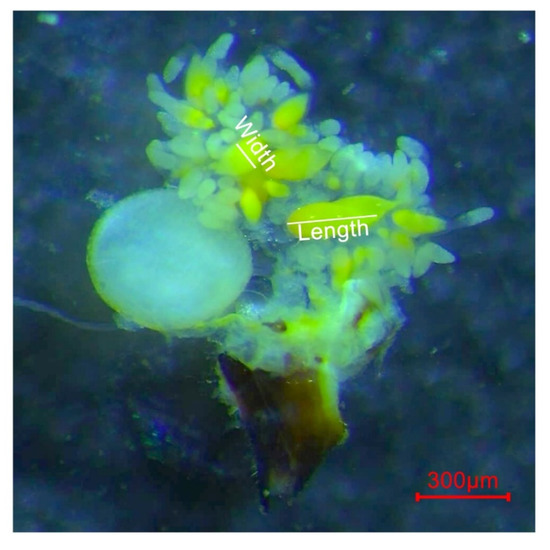
Figure 7.
Measurement method of D. citri eggs. The length and width of the eggs are marked with white lines.
qRT-PCR was carried out using 2 × Q3 SYBR qPCR Master Mix (High Rox) (TOLOBIO, Shanghai, China) following the manufacturer’s instructions: (1) prepare the following mixture in an RNase-free Eppendorff tube, 2 × Q3 SYBR qPCR Master Mix 10 μL, primer 1 (10 µM) 0.4 μL, primer 2 (10 µM) 0.4 μL, template cDNA 1 μL, ddH2O to 20 μL; (2) place the prepared solution into 96-well plates and perform qRT-PCR on the qRT-PCR machine. The qRT-PCR reaction conditions were as follows: pre-denaturation, cycle 1 at 95 °C for 30 s; circular reaction, cycle 40 at 95 °C for 10 s; melting curves, cycle 1 at 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. All reactions were performed with the QuantStudioTM Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) using the primers listed in Table 1. The expression analyses of dsVgR and dsGFP were the same as that of dsVg4. Relative gene expression data from qRT-PCR were analyzed using the 2−ΔΔCT method.
4.6. Interference of Vg and VgR on Oviposition in D. citri
Newly emerged, unmated females (n = 30) were randomly selected, fed dsVg4 (300 ng), and paired with 7- to 12-day-old unmated males (n = 30). A pair of male and female was placed in each rearing container. The dsVgR, dsGFP, and blank control (water) treatments were carried out as described for the dsVg4 treatment. Each day, M. odorifera shoots were replaced when eggs were detected on the shoots. The replaced M. odorifera shoots were stored separately after counting the number of eggs. The number of nymphs was also counted after egg hatching. The fecundity, egg hatchability, and deformity rate of eggs and nymphs were assessed after all females had died (the lifetime of D. citri females is 30–90 days).
4.7. Statistical Analysis
Data analysis was performed using SPSS 25.0 (IBM Corp., Armonk, New York, NY, USA). Data on the length and width of eggs, number of mature eggs, mature egg percentage, number of eggs per female, and egg hatchability were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) multiple tests. The effects of dsGFP and dsVg4 on the relative expression of Vg4 and VgR were analyzed using independent samples t-tests. The difference was considered statistically significant at the 5% level (p < 0.05).
5. Conclusions
In summary, interference with Vg4 and VgR gene expression can prevent the accumulation of Vitellogenin in the ovary of D. citri, leading to an inability to form mature eggs normally. Consequently, most eggs remain immature and egg formation is suppressed, resulting in the production of abnormal eggs and nymphs. The expression of the Vg4 and VgR genes was significantly affected by dsVg4 and dsVgR. DsVgR was more effective than dsVg4 at suppressing egg formation in D. citri, suggesting that dsVgR is more suitable than dsVg4 for use in the integrated management of insect populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24119497/s1.
Author Contributions
H.L., X.W., X.Z. and W.L. designed the research. B.P., S.X., J.M. and S.L. conducted the experiments. H.L., X.W. and X.Z. wrote the first draft of the manuscript. X.W., X.Z. and W.L. revised the final draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by a grant from the Foundation of Guangdong Provincial Key Laboratory of High Technology for Plant Protection (grant no. Zhizhong 2022-04), the Training Programs of Innovation and Entrepreneurship for Undergraduates of Guangxi University (grant no. 202210593902) in China and Guangxi Natural Science Foundation, China (grant no. 2023GXNSFBA026131).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge experimental assistance from Huili Ouyang and Ranran Su (Guangxi University), and professional language editing from Fanzhou Liu (Cornell University).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stockton, D.G.; Martini, X.; Stelinski, L.L. Male psyllids differentially learn in the context of copulation. Insects 2017, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.D.; Killiny, N. Effect of silencing a boule homologue on the survival and reproduction of Asian citrus psyllid Diaphorina citri. Physiol. Entomol. 2018, 43, 268–275. [Google Scholar] [CrossRef]
- Li, H.L.; Zheng, X.L.; Wang, X.Y.; Lu, W. Prophase of reproductive behavior and activity rhythm in adults of Diaphorina citri (Kuwayama). J. South. Agric. 2019, 50, 2009–2014. [Google Scholar]
- Li, H.L.; Wang, X.L.; Zheng, X.L.; Huang, Z.Y.; Lu, W. Review of reproductive behavior in Diaphorina citri (Kuwayama) (Homoptera: Liviidae). J. Plant Dis. Protect. 2020, 127, 601–606. [Google Scholar] [CrossRef]
- Tsai, J.H.; Liu, Y.H. Biology of Diaphorina citri (Homoptera: Psyllidae) on four host plants. J. Econ. Entomol. 2000, 93, 1721–1725. [Google Scholar] [CrossRef] [PubMed]
- Nava, D.E.; Torres, M.L.G.; Rodrigues, M.D.L.; Bento, J.M.S.; Parra, J.R.P. Biology of Diaphorina citri (Hem. Psyllidae) on different hosts and at different temperatures. J. Appl. Entomol. 2007, 131, 709–715. [Google Scholar] [CrossRef]
- De-León, J.H.; Sétamou, M.; Gastaminza, G.A.; Buenahora, J.; Ceceres, S.; Yamamoto, P.T.; Bouvet, J.P.; Logarzo, G.A. Two separate introductions of Asian citrus psyllid populations found in the American continents. Ann. Entomol. Soc. Am. 2011, 104, 1392–1398. [Google Scholar] [CrossRef]
- Abdullaha, A.B.M.; Leeb, D.W.; Jungc, J.; Kima, Y. Deletion mutant of sPLA2 using CRISPR/Cas9 exhibits immunosuppression, developmental retardation, and failure of oocyte development in legume pod borer, Maruca vitrata. Dev. Comp. Immunol. 2019, 103, 103500. [Google Scholar] [CrossRef]
- Fandino, R.A.; Haverkamp, A.; Bisch-Knaden, S.; Zhang, J.; Bucks, S.; Nguyen, T.A.T.; Schröder, K.; Werckenthin, A.; Rybak, J.; Stengl, M.; et al. Mutagenesis of odorant coreceptor Orco fully disrupts foraging but not oviposition behaviors in the hawkmoth Manduca sexta. Proc. Natl. Acad. Sci. USA 2019, 116, 15677–15685. [Google Scholar] [CrossRef]
- Hao, K.; Tu, X.B.; Ullah, H.; McNeill, M.R.; Zhang, Z.H. Novel Lom-dh genes play potential role in promoting egg diapause of Locusta migratoria L. Front. Physiol. 2019, 10, 767. [Google Scholar] [CrossRef]
- Lu, J.B.; Zhang, M.Q.; Li, L.C.; Zhang, C.X. DDC plays vital roles in the wing spot formation, egg production, and chorion tanning in the brown planthopper. Arch. Insect Biochem. Physiol. 2019, 101, e21552. [Google Scholar] [CrossRef]
- Ortega, Y.S.; Killiny, N. Silencing of sucrose hydrolase causes nymph mortality and disturbs adult osmotic homeostasis in Diaphorina citri (Hemiptera: Liviidae). Insect Biochem. Mol. Biol. 2019, 101, 131–143. [Google Scholar] [CrossRef]
- Hu, Y.Y.; Sun, Y.; Zou, Z.W.; Huang, S.J. Resistance of Diaphorina citri to five conventional insecticides in Jiangxi province. Chin. J. Appl. Entomol. 2022, 6, 1556. [Google Scholar]
- Tufail, M.; Takeda, M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008, 54, 1447–1458. [Google Scholar] [CrossRef]
- Ibanez, F.; Levy, J.; Tamborindeguy., C. Identification and expression analyses of vitellogenin in Bactericera cockerelli (Šulc). J. Insect Physiol. 2017, 98, 205–213. [Google Scholar] [CrossRef]
- Xie, Y.F.; Shang, F.; Ding, B.Y.; Wu, Y.B.; Niu, J.Z.; Wei, D.; Dou, W.; Christiaens, O.; Smagghe, G.; Wang, J.J. Tudor knockdown disrupts ovary development in Bactrocera dorsalis. Insect Mol. Biol. 2019, 28, 136–144. [Google Scholar] [CrossRef]
- Peng, L.; Wang, Q.; Zou, M.M.; Qin, Y.D.; Vasseur, L.; Chu, L.N.; Zhai, Y.L.; Dong, S.J.; Liu, L.L.; He, W.Y.; et al. CRISPR/Cas9-mediated vitellogenin receptor knockout leads to functional deficiency in the reproductive development of Plutella xylostella. Front. Physiol. 2020, 10, 1585. [Google Scholar] [CrossRef]
- Lu, K.; Wang, Y.; Chen, X.; Zhang, X.Y.; Li, W.R.; Cheng, Y.B.; Li, Y.; Zhou, J.M.; You, K.K.; Song, Y.Y.; et al. Adipokinetic hormone receptor mediates trehalose homeostasis to promote vitellogenin uptake by oocytes in Nilaparvata lugens. Front. Physiol. 2019, 9, 1904. [Google Scholar] [CrossRef]
- Li, H.L.; Wang, X.Y.; Zheng, X.L.; Dong, Z.S.; Yi, X.L.; Lu, W. Transcriptome and proteome analysis of oviposition- and spermatogenesis-related genes of Diaphorina citri. Anim. Gene 2022, 23, 200120. [Google Scholar] [CrossRef]
- Killiny, N.; Kishk, A. Delivery of dsRNA through topical feeding for RNA interference in the citrus sap piercing-sucking hemipteran, Diaphorina citri. Arch. Insect Biochem. 2017, 95, e21394. [Google Scholar] [CrossRef]
- Marques, V.V.; Angelotti-Mendonça, J.; Roberto, S.R. Advances and challenges in RNA interference technology for citrus Huanglongbing vector control. Horticulturae 2021, 7, 277. [Google Scholar] [CrossRef]
- Andrade, E.C.; Hunter, W.B. RNAi feeding bioassay: Development of a non-transgenic approach to control Asian citrus psyllid and other hemipterans. Entomol. Exp. Appl. 2016, 162, 389–396. [Google Scholar] [CrossRef]
- Moriyama, M.; Hosokawa, T.; Tanahashi, M.; Nikoh, N.; Fukatsu, T. Suppression of bedbug’s reproduction by RNA interference of vitellogenin. PLoS ONE 2016, 11, e0153984. [Google Scholar] [CrossRef]
- Rika, U.S.; Ryo, M.; Kozo, F.; Hiroshi, S. Intracellular localization of vitellogenin receptor mRNA and protein during oogenesis of a parthenogenetic tick, Haemaphysalis longicornis. Parasite Vector 2019, 12, 205. [Google Scholar]
- Zhang, J.J.; Xi, G.S.; Zhao, J. Vitellogenin regulates estrogen-related receptor expression by crosstalk with the JH and IIS-TOR signaling pathway in Polyrhachis vicina Roger (Hymenoptera, Formicidae). Gen. Comp. Endocr. 2021, 310, 113836. [Google Scholar] [CrossRef]
- Bartlett, D.W.; Davis, M.E. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol. Bioeng. 2007, 97, 909–921. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Zhang, X.C.; Chen, L.F.; Li, J.H.; Yin, J.G.; Liu, Q.; Gong, P.T. Cloning and sequence analysis of a partial gene of Trichomonas vaginalis dsRNA virus. Chin. J. Parasitol. Parasit. Dis. 2006, 24, 389–390. [Google Scholar]
- Sun, G.X. Construction of Brassica Oleracea Expressing ds RNA and Its Resistance against Plutella xylostella. Doctor’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2020. [Google Scholar]
- Ghosh, S.K.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double-stranded RNA oral delivery methods to induce RNA interference in phloem and plant-sap-feeding hemipteran insects. J. Vis. Exp. 2018, 135, e57390. [Google Scholar]
- Garbutt, J.S.; Bellés, X.; Richards, E.H.; Reynolds, S.E. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 2013, 59, 171–178. [Google Scholar] [CrossRef]
- Chen, J.Z.; Jiang, Y.X.; Li, M.W.; Li, J.W.; Zha, B.H.; Yang, G. Double-stranded RNA-degrading enzymes reduce the efficiency of RNA interference in Plutella xylostella. Insects 2021, 12, 712. [Google Scholar] [CrossRef]
- Swevers, L.; Broeck, J.V.; Smagghe, G. The possible impact of persistent virus infection on the function of the RNAi machinery in insects: A hypothesis. Front. Physiol. 2013, 4, 319. [Google Scholar] [CrossRef]
- Rasool, K.G.; Mehmood, K.; Tufail, M.; Husain, M.; Alwaneen, W.S.; Aldawood, A.S. Silencing of vitellogenin gene contributes to the promise of controlling red palm weevil, Rhynchophorus ferrugineus (Olivier). Sci. Rep. 2021, 11, 21695. [Google Scholar] [CrossRef]
- Yao, Q.; Xu, S.; Dong, Y.Z.; Que, Y.L.; Quan, L.F.; Chen, B.X. Characterization of vitellogenin and vitellogenin receptor of Conopomorpha sinensis bradley and their responses to sublethal concentrations of insecticide. Front. Physiol. 2018, 9, 1250. [Google Scholar] [CrossRef]
- Zhu, Z.D.; Tong, C.M.; Qiu, B.B.; Yang, H.G.; Xu, J.H.; Zheng, S.C.; Song, Q.S.; Feng, Q.L.; Deng, H.M. 20E-mediated regulation of BmKr-h1 by BmKRP promotes oocyte maturation. BMC Biol. 2021, 19, 39. [Google Scholar] [CrossRef]
- Chen, X.; Cao, Y.; Zhan, S.; Tan, A.; Palli, S.R.; Huang, Y. Disruption of sex-specific doublesex exons results in male- and female specific defects in the black cutworm, Agrotis ipsilon. Pest. Manag. Sci. 2019, 75, 1697–1706. [Google Scholar] [CrossRef]
- Lou, Y.H.; Shen, Y.; Li, D.T.; Huang, H.J.; Lu, J.B.; Zhang, C.X. A Mucin-like protein is essential for oviposition in Nilaparvata lugens. Front. Physiol. 2019, 10, 551. [Google Scholar] [CrossRef]
- Ellango, R.; Asokan, R.; Sharath Chandra, G.; Kumar, N.K.K.; Mahmood, R.; Ramamurthy, V.V. Tyrosine hydroxylase, a potential target for the RNAi-mediated management of diamondback moth (Lepidoptera: Plutellidae). BioOne 2018, 101, 1–5. [Google Scholar] [CrossRef]
- Lanno, S.M.; Lam, I.; Drum, Z.; Linde, S.C.; Gregory, S.M.; Shimshak, S.J.; Becker, M.V.; Brew, K.E.; Budhiraja, A.; Carter, E.A. Genomics analysis of l-DOPA exposure in Drosophila sechellia. G3 (Bethesda) 2019, 9, 3973–3980. [Google Scholar] [CrossRef]
- Li, H.L.; Wang, X.Y.; Zheng, X.L.; Lu, W. Research progress on oviposition-related genes in insects. J. Insect Sci. 2020, 20, 36. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Zhu, Y.K. Chitosan/doublestranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef]
- Cong, L.; Yang, W.J.; Jiang, X.Z.; Niu, J.Z.; Shen, G.M.; Ran, C.; Wang, J.J. The essential role of vitellogenin receptor in ovary development and vitellogenin uptake in Bactrocera dorsalis (Hendel). Int. J. Mol. Sci. 2015, 16, 18368–18383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mysore, K.; Flannery, E.; Michel, K.; David, W.; Severson, D.W.; Zhu, K.Y.; Molly, D.S. Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J. Vis. Exp. 2015, 97, e52523. [Google Scholar]
- Li, H.H.; Kong, L.F.; Yu, R.; Yu, H.; Li, Q. Characterization, expression, and functional analysis of testis-specific serine/threonine kinase 1 (Tssk1) in the pen shell Atrina pectinata. Invertebr. Reprod. Dev. 2016, 60, 118–125. [Google Scholar] [CrossRef]
- Zhen, C.A.; Miao, L.; Gao, X.W. Sublethal effects of sulfoxaflor on biological characteristics and vitellogenin gene (AlVg) expression in the mirid bug, Apolygus lucorum (Meyer-Dür). Pestic. Biochem. Phys. 2018, 144, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, S.J.; Park, C.J.; Nam, Y.K. Characterization of testis-specific serine/threonine kinase 1-like (TSSK1-like) gene and expression patterns in diploid and triploid Pacific abalone (Haliotis discus hannai; Gastropoda; Mollusca) males. PLoS ONE 2019, 14, e0226022. [Google Scholar] [CrossRef]
- Zhao, J.H.; Guo, H.S. RNA silencing: From discovery and elucidation to application and perspectives. J. Integr. Plant Biol. 2021, 64, 476–498. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).