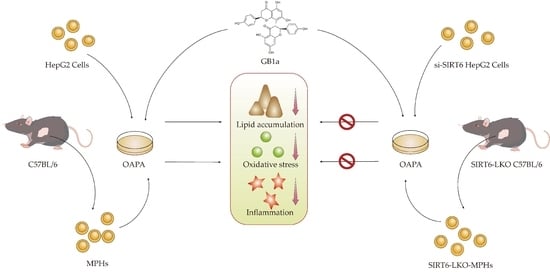
GB1a Activates SIRT6 to Regulate Lipid Metabolism in Mouse Primary Hepatocytes
Abstract
1. Introduction
2. Results
2.1. The Effect of GB1a on the HepG2 Adipophilin Model
2.1.1. GB1a Decreased OA&PA-Induced Lipid Accumulation in HepG2 Cells
2.1.2. GB1a Inhibited Lipogenic Genes and Promoted the Expression of Lipid Oxidation Genes in HepG2 Cells
2.1.3. GB1a Improved the Oxidative Stress Induced by OA&PA in HepG2 Cells
2.1.4. GB1a Alleviated the OA&PA-Induced Inflammatory Response in HepG2 Cells
2.1.5. GB1a Acted as an Agonist of SIRT6
2.1.6. GB1a Has Good Combination with SIRT6
2.2. Regulation Effect of GB1a on Lipid Metabolism in MPHs
2.2.1. GB1a Failed to Reduce OA&PA-Induced Lipid Accumulation in SIRT6-LKO MPHs
2.2.2. GB1a Lost Protection against Oxidative Stress and the Inflammatory Response Due to OA&PA in SIRT6-LKO MPHs
3. Discussion
4. Materials and Methods
4.1. The Source of GB1a and Cells
4.2. Cell Viability and GB1a Administration
4.3. Western Blot
4.4. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.5. Isolation and Culture of Mouse Primary Hepatocytes
4.6. Lipid Probe Staining
4.7. Cellular Immunofluorescence
4.8. Detection of Reactive Oxygen Species (ROS)
4.9. Detection of Mitochondrial Membrane Potential (MMP)
4.10. The morphology and Structure of Cell Mitochondria Were Observed by Transmission Electron Microscopy
4.11. Molecular Docking
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chalasani, N.; Younossi, Z.; LaVine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- de Alwis, N.M.W.; Day, C.P. Non-alcoholic fatty liver disease: The mist gradually clears. J. Hepatol. 2008, 48, S104–S112. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Oakley, F.; Anstee, Q.M.; Day, C.P. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 451–496. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; James, O.F. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fan, J.-G.; Qiao, L. Potential Epigenetic Mechanism in Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2015, 16, 5161–5179. [Google Scholar] [CrossRef] [PubMed]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Cobbina, E.; Akhlaghi, F. Non-alcoholic fatty liver disease (NAFLD)—Pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef]
- Tasselli, L.; Xi, Y.; Zheng, W.; Tennen, R.I.; Odrowaz, Z.; Simeoni, F.; Li, W.; Chua, K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016, 23, 434–440. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.A.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef]
- Yang, B.; Zwaans, B.M.M.; Eckersdorff, M.; Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 2009, 8, 2662–2663. [Google Scholar] [CrossRef]
- Kugel, S.; Mostoslavsky, R. Chromatin and beyond: The multitasking roles for SIRT6. Trends Biochem. Sci. 2014, 39, 72–81. [Google Scholar] [CrossRef]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.-X.; Dong, X.C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 2013, 54, 2745–2753. [Google Scholar] [CrossRef]
- Kuang, J.; Chen, L.; Tang, Q.; Zhang, J.; Li, Y.; He, J. The Role of Sirt6 in Obesity and Diabetes. Front. Physiol. 2018, 9, 135. [Google Scholar] [CrossRef]
- Dong, X.C. Sirtuin 6—A Key Regulator of Hepatic Lipid Metabolism and Liver Health. Cells 2023, 12, 663. [Google Scholar] [CrossRef]
- Bittar, M.; de Souza, M.M.; Yunes, R.A.; Lento, R.; Monache, F.D.; Filho, V.C. Antinociceptive Activity of I3,II8-Binaringenin, a Biflavonoid Present in Plants of the Guttiferae. Planta Med. 2000, 66, 84–86. [Google Scholar] [CrossRef]
- Oyagbemi, A.; Bester, D.; Esterhuyse, J.; Farombi, E. Kolaviron, a biflavonoid of Garcinia kola seed mitigates ischaemic/reperfusion injury by modulation of pro-survival and apoptotic signaling pathways. J. Intercult. Ethnopharmacol. 2017, 6, 42–49. [Google Scholar] [CrossRef]
- Tajuddeen, N.; Van Heerden, F.R. Antiplasmodial natural products: An update. Malar. J. 2019, 18, 404. [Google Scholar] [CrossRef]
- Abarikwu, S.O. Kolaviron, a natural flavonoid from the seeds of Garcinia kola, reduces LPS-induced inflammation in macrophages by combined inhibition of IL-6 secretion, and inflammatory transcription factors, ERK1/2, NF-κB, p38, Akt, p-c-JUN and JNK. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2373–2381. [Google Scholar] [CrossRef]
- Tsai, S.-Y.; Chung, P.-C.; Owaga, E.E.; Tsai, I.-J.; Wang, P.-Y.; Tsai, J.-I.; Yeh, T.-S.; Hsieh, R.-H. Alpha-mangostin from mangosteen (Garcinia mangostana Linn.) pericarp extract reduces high fat-diet induced hepatic steatosis in rats by regulating mitochondria function and apoptosis. Nutr. Metab. 2016, 13, 88. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, Y.; Sun, N.; Zhou, L.; Zhang, D.; Chen, H.; Miao, W.; Gao, W.; Zhang, C.; Liu, C.; et al. Ginsenosides Rc, as a novel SIRT6 activator, protects mice against high fat diet induced NAFLD. J. Ginseng Res. 2020, 47, 376–384. [Google Scholar] [CrossRef]
- Farombi, E.O.; Adedara, I.A.; Ajayi, B.O.; Ayepola, O.R.; Egbeme, E.E. Kolaviron, a Natural Antioxidant and Anti-Inflammatory Phytochemical Prevents Dextran Sulphate Sodium-Induced Colitis in Rats. Basic Clin. Pharmacol. Toxicol. 2013, 113, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Farombi, E.O.; Adepoju, B.F.; Ola-Davies, O.E.; Emerole, G.O. Chemoprevention of aflatoxin B1-induced genotoxicity and hepatic oxidative damage in rats by kolaviron, a natural biflavonoid of Garcinia kola seeds. Eur. J. Cancer Prev. 2005, 14, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Farombi, E.; Tahnteng, J.; Agboola, A.; Nwankwo, J.; Emerole, G. Chemoprevention of 2-acetylaminofluorene-induced hepatotoxicity and lipid peroxidation in rats by kolaviron—A Garcinia kola seed extract. Food Chem. Toxicol. 2000, 38, 535–541. [Google Scholar] [CrossRef]
- Yu, Y.Y.; Zheng, C.M.; Lu, X.; Deng, C.S.; Xu, Q.; Guo, W.F.; Wu, Q.Y.; Wang, Q.; Liu, C.H.; Huang, X.A.; et al. GB1a Ameliorates Ulcerative Colitis via Regulation of the NF-κB and Nrf2 Signaling Pathways in an Experimental Model. Front. Med. 2021, 8, 654867. [Google Scholar]
- Chen, H.-X.; Yang, F.; He, X.-Q.; Li, T.; Sun, Y.-Z.; Song, J.-P.; Huang, X.-A.; Guo, W.-F. Garcinia Biflavonoid 1 Improves Lipid Metabolism in HepG2 Cells via Regulating PPARα. Molecules 2022, 27, 1978. [Google Scholar] [CrossRef]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human Fatty Liver Disease: Old Questions and New Insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef]
- Mendez-Sanchez, N.; Cruz-Ramon, V.C.; Ramirez-Perez, O.L.; Hwang, J.P.; Barranco-Fragoso, B.; Cordova-Gallardo, J. New Aspects of Lipotoxicity in Nonalcoholic Steatohepatitis. Int. J. Mol. Sci. 2018, 19, 2034. [Google Scholar] [CrossRef]
- Chen, Z.; Tian, R.; She, Z.; Cai, J.; Li, H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2020, 152, 116–141. [Google Scholar] [CrossRef]
- Spahis, S.; Delvin, E.; Borys, J.-M.; Levy, E. Oxidative Stress as a Critical Factor in Nonalcoholic Fatty Liver Disease Pathogenesis. Antioxid. Redox Signal. 2017, 26, 519–541. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Afonso, M.B.; Simão, A.L.; Carvalho, C.C.; Trindade, A.; Duarte, A.; Borralho, P.M.; Machado, M.V.; Cortez-Pinto, H.; Rodrigues, C.M.; et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017, 8, e2748. [Google Scholar] [CrossRef]
- Hui, J.M.; Hodge, A.; Farrell, G.C.; Kench, J.G.; Kriketos, A.; George, J. Beyond insulin resistance in NASH: TNF-α or adiponectin? Hepatology 2004, 40, 46–54. [Google Scholar] [CrossRef]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef]
- Xiong, X.; Tao, R.; DePinho, R.; Dong, X.C. Deletion of Hepatic FoxO1/3/4 Genes in Mice Significantly Impacts on Glucose Metabolism through Downregulation of Gluconeogenesis and Upregulation of Glycolysis. PLoS ONE 2013, 8, e74340. [Google Scholar] [CrossRef]
- Dominy, J.E., Jr.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.-B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The Deacetylase Sirt6 Activates the Acetyltransferase GCN5 and Suppresses Hepatic Gluconeogenesis. Mol. Cell 2012, 48, 900–913. [Google Scholar] [CrossRef]
- Kim, H.-S.; Xiao, C.; Wang, R.-H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.-I.; Park, O.; Ki, S.H.; et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.-X.; Dong, X.C. FoxO3 Transcription Factor and Sirt6 Deacetylase Regulate Low Density Lipoprotein (LDL)-cholesterol Homeostasis via Control of the Proprotein Convertase Subtilisin/Kexin Type 9 (Pcsk9) Gene Expression. J. Biol. Chem. 2013, 288, 29252–29259. [Google Scholar] [CrossRef]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Gibor, G.; Cohen, H.Y. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef]
- Ka, S.-O.; Bang, I.H.; Bae, E.J.; Park, B.-H. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J. 2017, 31, 3999–4010. [Google Scholar] [CrossRef]











| Chemical | Resolution | PDB_ID | Total Score | Crash | Polar | C_SCORE |
|---|---|---|---|---|---|---|
| GB1a | 1.97Å | 5X16 | 12.1993 | −3.8572 | 4.2729 | 3 |
| Primer | Sequences |
|---|---|
| SIRT6 (h) | F1:5′-CCTCTCTCTAATCAGCCCTCTG-3′ |
| R1:5′-GAGGACCTGGGAGTAGATGAG-3′ | |
| SIRT6 (m) | F1:5′-ATGTCGGTGAATTATGCAGCA-3′ |
| SREBP-1c (h) | R1:5′-GCTGGAGGACTGCCACATTA-3′ F1:5′-ACAGTGACTTCCCTGGCCTAT-3′ R1:5′-GCATGGACGGGTACATCTTCAA-3′ |
| SREBP-1c (m) | F1:5′-TGACCCGGCTATTCCGTGA-3′ |
| R1:5′-CTGGGCTGAGCAATACAGTTC-3′ | |
| PPARα (m) | F1:5′-AGAGCCCCATCTGTCCTCTC-3′ |
| ACC (h) | R1:5′-ACTGGTAGTCTGCAAAACCAAA-3′ F1:5′-ATGTCTGGCTTGCACCTAGTA-3′ R1:5′-CCCCAAAGCGAGTAACAAATTCT-3′ |
| ACC (m) | F1:5′-CTTGGGTGCTGACTACAACC-3′ |
| Cpt1a (h) | R1:5′-GCCCTCCCGTACACTCACTC-3′ F1:5′-TCCAGTTGGCTTATCGTGGTG-3′ R1:5′-TCCAGAGTCCGATTGATTTTTGC-3′ |
| Cpt1a (m) | F1:5′-CTCCGCCTGAGCCATGAAG-3′ |
| Cpt1b (h) | R1:5′-CACCAGTGATGATGCCATTCT-3′ F1:5′-GCGCCCCTTGTTGGATGAT-3′ R1:5′-CCACCATGACTTGAGCACCAG-3′ |
| Cpt1b (m) | F1:5′-GCACACCAGGCAGTAGCTTT-3′ |
| TNF-α (h) | R1:5′-CAGGAGTTGATTCCAGACAGGTA-3′ F1:5′-CCTCTCTCTAATCAGCCCTCTG-3′ R1:5′-GAGGACCTGGGAGTAGATGAG-3′ |
| TNF-α (m) | F1:5′-CCCTCACACTCAGATCATCTTCT-3′ |
| IL-1β (h) | R1:5′-GCTACGACGTGGGCTACAG-3′ F1:5′-ATGATGGCTTATTACAGTGGCAA-3′ R1:5′-GTCGGAGATTCGTAGCTGGA-3′ |
| IL-1β (m) | F1:5′-GCAACTGTTCCTGAACTCAACT-3′ |
| IL-6 (h) | R1:5′-ATCTTTTGGGGTCCGTCAACT-3′ F1:5′-ACTCACCTCTTCAGAACGAATTG-3′ R1:5′-CCATCTTTGGAAGGTTCAGGTTG-3′ |
| IL-6 (m) | F1:5′-TAGTCCTTCCTACCCCAATTTCC-3′ |
| Nrf2 (h) | R1:5′-TTGGTCCTTAGCCACTCCTTC-3′ F1:5′-TCAGCGACGGAAAGAGTATGA-3′ R1:5′-CCACTGGTTTCTGACTGGATGT-3′ |
| Nrf2 (m) | F1:5′-TCTTGGAGTAAGTCGAGAAGTGT-3′ |
| Keap1 (h) | R1:5′-GTTGAAACTGAGCGAAAAAGGC-3′ F1:5′-CTGGAGGATCATACCAAGCAGG-3′ R1:5′-GGATACCCTCAATGGACACCAC-3′ |
| Keap1 (m) | F1:5′-TGCCCCTGTGGTCAAAGTG-3′ |
| HO-1 (h) | R1:5′-GGTTCGGTTACCGTCCTGC-3′ F1:5′-AAGACTGCGTTCCTGCTCAAC-3′ R1:5′-AAAGCCCTACAGCAACTGTCG-3′ |
| HO-1 (m) | F1:5′-AAGCCGAGAATGCTGAGTTCA-3′ |
| NQO1 (h) | R1:5′-GCCGTGTAGATATGGTACAAGGA-3′ F1:5′-GAAGAGCACTGATCGTACTGGC-3′ R1:5′-GGATACTGAAAGTTCGCAGGG-3′ |
| NQO1 (m) | F1:5′-AGGATGGGAGGTACTCGAATC-3′ |
| R1:5′-AGGCGTCCTTCCTTATATGCTA-3′ | |
| β-actin (m) | F1:5′-GGCTGTATTCCCCTCCATCG-3′ |
| R1:5′-CCAGTTGGTAACAATGCCATGT-3′ | |
| β-actin (h) | F1:5′-CATGTACGTTGCTATCCAGGC-3′ |
| R1:5′-CTCCTTAATGTCACGCACGAT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Zheng, C.; Li, T.; He, X.; Yang, F.; Guo, W.; Song, J.; Gao, Y.; Deng, C.; Huang, X. GB1a Activates SIRT6 to Regulate Lipid Metabolism in Mouse Primary Hepatocytes. Int. J. Mol. Sci. 2023, 24, 9540. https://doi.org/10.3390/ijms24119540
Sun Y, Zheng C, Li T, He X, Yang F, Guo W, Song J, Gao Y, Deng C, Huang X. GB1a Activates SIRT6 to Regulate Lipid Metabolism in Mouse Primary Hepatocytes. International Journal of Molecular Sciences. 2023; 24(11):9540. https://doi.org/10.3390/ijms24119540
Chicago/Turabian StyleSun, Yongzhi, Congmin Zheng, Ting Li, Xinqian He, Fan Yang, Wenfeng Guo, Jianping Song, Yong Gao, Changsheng Deng, and Xinan Huang. 2023. "GB1a Activates SIRT6 to Regulate Lipid Metabolism in Mouse Primary Hepatocytes" International Journal of Molecular Sciences 24, no. 11: 9540. https://doi.org/10.3390/ijms24119540
APA StyleSun, Y., Zheng, C., Li, T., He, X., Yang, F., Guo, W., Song, J., Gao, Y., Deng, C., & Huang, X. (2023). GB1a Activates SIRT6 to Regulate Lipid Metabolism in Mouse Primary Hepatocytes. International Journal of Molecular Sciences, 24(11), 9540. https://doi.org/10.3390/ijms24119540