3. Discussion
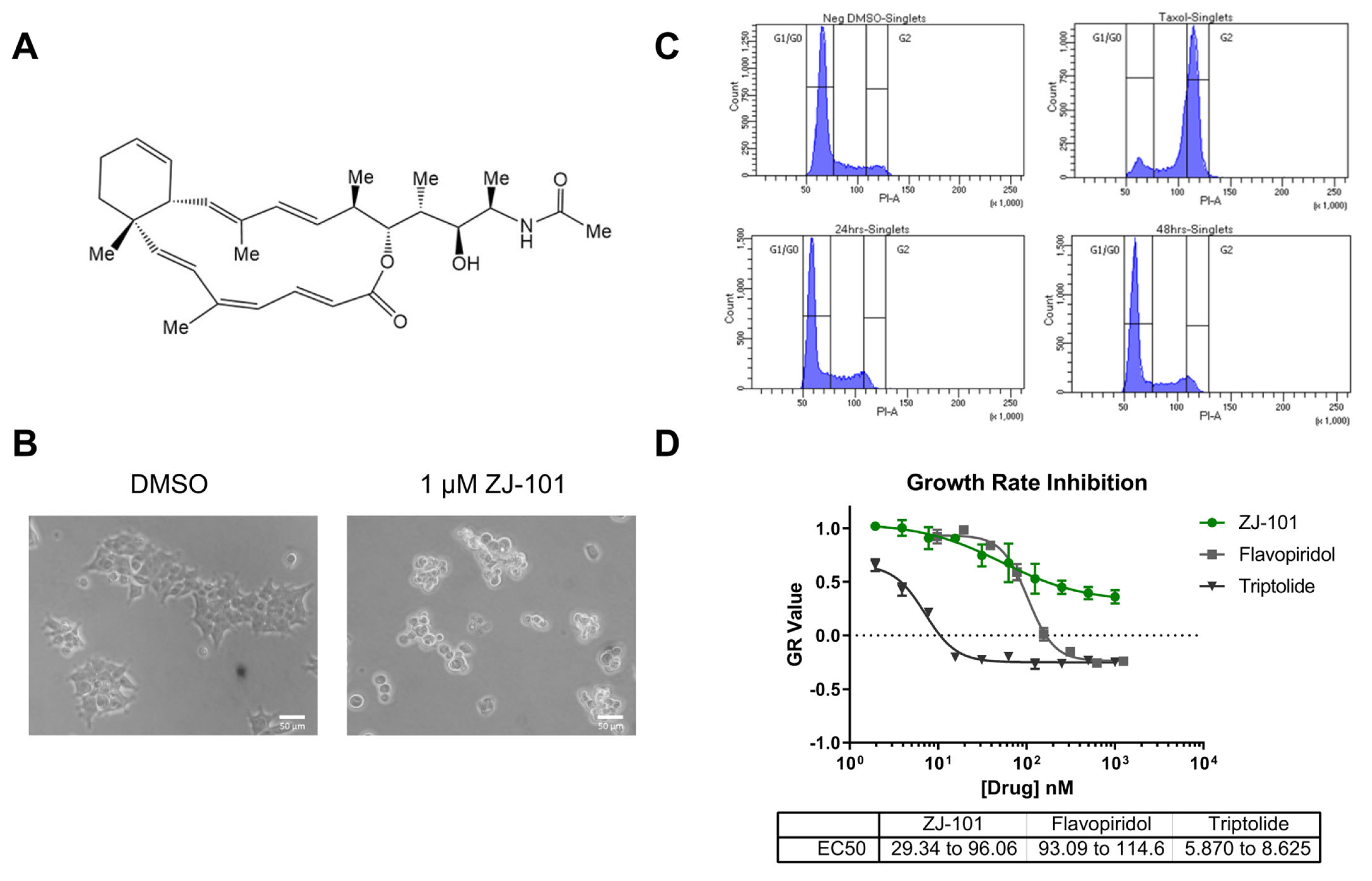
Our group has identified several key phenotypes that suggest a distinct mechanism of action for the compound ZJ-101 by acting through the endomembrane system. We first identified the clear ability of ZJ-101 to cause cell rounding and dislodging from a growth substrate within a few hours. Cellular adhesion occurs in primarily two modalities: cell–cell and cell–substrate adhesion. To determine if ZJ-101 had preferential activity for either of these, we assessed its activity in a 3D spheroid model. To our amazement, extremely low doses of ZJ-101 could halt the formation of spheroids. At approximately 1 nM of the compound, we observed a halt in the progression of standard spheroid formation in the triple-negative breast cancer cell line MDA-MB-231. The disparity in EC50s for the antiproliferative effect previously established versus anti-adhesion in the spheroid model was approximately 30-fold. This unexpected enhancement in potency against the spheroid model challenged our previous assumptions about the molecule.
We utilized the growth rate normalized metric assay (GR assay) to minimize variation between experiments and dependence on cell growth rates. A major benefit of this assay is its ability to discern cytostatic versus cytotoxic effects by comparing cell viability to cellular proliferation. Since all cell types have their own intrinsic growth rate kinetics, we can accurately compare responses to drugs between cell types by normalizing them to this rate. Using this live/dead cell image-based assay, we were able to determine that the effect of ZJ-101 on cancer cells is entirely cytostatic. In GR metrics, cytotoxic compounds are identified by their ability to deplete cell populations below initial seeding densities, denoted by a GR value less than 0. At a GR value greater than 0 but less than 1, cellular proliferation is suppressed in a cytostatic manner. ZJ-101 suppresses cellular growth with a GR value of 0.5 at the highest concentration of 1 μM using the calcein-AM/ethidium homodimer dye staining assay. We obtained the same EC
50 value (30–90 nM) for ZJ-101 using GR metrics as we previously determined using resazurin-based viability assays (
Figure S1A). These results confirmed that one of ZJ-101′s primary novel phenotypes is cytostatic suppression of cellular proliferation.
Cytostasis is often a temporary cellular state induced by exogenous signals or chemicals that suppress cell division. Senescence, a physiological state of non-dividing cells that can be induced by oxidative stress or DNA damage, has all the hallmarks of cellular cytostasis. A key characteristic of senescence is a sharp increase in G1-phase cell populations. Because of the lack of change in any population (G1, S, or G2) during prolonged treatment with ZJ-101, cellular senescence could potentially be ruled out as a mechanism for the cytostatic activity. At time points of up to 48 h, MDA-MB-231 cells remained dislodged from their substrate under the sustained treatment of ZJ-101, further underscoring the distinct lack of any sub-G1 populations. Typically, cells will undergo apoptosis following sustained senescence or cell adhesion loss, a unique form of apoptosis termed anoikis [
17]. To further explore senescence using GR metrics, we tested isogenic cell lines of HCT-116 containing knockouts of p21 and p53, key senescence potentiators [
18]. We found no reversal of anti-proliferative activity under treatment with ZJ-101 in these knockout lines. Instead, p21 knockout potentiates the effect of ZJ-101′s suppression of cell proliferation. This result runs counter to an activation of senescence by ZJ-101, given that p21 expression is protective against cytostasis. Although the outcome of ZJ-101 treatment results in the halting of cellular proliferation, more work is required to determine the specific pathway(s) leading to this long-term cytostatic suppression.
To identify cellular pathways regulated by ZJ-101, transcriptome sequencing was undertaken across four doses in two contexts: before 2D adhesion loss at 6 h, and after adhesion loss at 24 h in 3D spheroids (
Figure S4). Gene Ontology (GO) analysis identified increasing significance for the endoplasmic reticulum (GO:005783), cell adhesion (GO:007155), and the endomembrane system (GO:0012505) as the dose of ZJ-101 increases. Notably, endomembrane system genes represent the highest differentially expressed genes by significance between 2D and 3D culture formats. Genes related to the endoplasmic reticulum follow a paradoxical response to ZJ-101 treatment. Despite a general upregulation of heat-shock genes such as HSP90AA1, unfolded protein response (UPR) target genes were suppressed at 6 h. UPR-related genes remained suppressed at the 24 h time point as well, indicating a context-independent downregulation of ER-stress-responsive genes. Likewise, cell adhesion and extracellular matrix (ECM) genes were suppressed. Among these, several integrins, such as ITGA5, ITGAV, and ITGB5, were downregulated. These integrins are primarily responsible for cell–ECM adhesion and cannot solely explain the dual inhibition of cell–substrate and cell–cell adhesion caused by ZJ-101. Due to this unique transcriptional response, we reasoned that the stress caused by ZJ-101 may localize to a source within the wider endomembrane system, such as the Golgi and endosomes.
Genes involved in endosome homeostasis, such as ATP6V0A1, were significantly dysregulated by ZJ-101. V-type ATPases, such as ATP6V0A1, regulate endosome and vesicle pH. We compared ZJ-101 to the well-known inhibitor of V-type ATPases, Bafilomycin A1, through lysotracker staining, which fluoresces in the acidic compartments of endosomes and lysosomes. As observed in
Figure 5B, bafilomycin A1 potently decreases lysotracker staining intensity relative to control, while ZJ-101 does not. Additionally, ZJ-101 treatment decreases endolysosome size in a similar manner to brefeldin A and kifunensine. Brefeldin A’s unique mechanism of redistributing the Golgi membrane to the ER through uncompetitive inhibition of the Arf1 GDP exchange cycle poses the most curious parallel for ZJ-101′s potential mechanism of action [
19]. Although ZJ-101 does not redistribute GM130 as BFA does, they both likely enhance endolysosomal fission events, leading to the decreased puncta size observed after treatment. It is possible that ZJ-101 inhibits a target in opposition to the BFA mechanism, preserving the Golgi structurally but resulting in similar outcomes. Kifunensine also affects endolysosomal size, though it acts through inhibition of the ER mannosidase I enzyme. By inhibiting N-glycan trimming in the ER, protein sorting through the ER-associated degradation (ERAD) pathway is increased, leading to a loss of successful traversal of cargo through the secretory pathway [
20].
We initially identified a dysregulation of glycosylation localized to the Golgi apparatus through lectin staining assays. Lectin staining determined a selectivity for decreasing N-acetyl-galactosamine (GalNAc) residues, which are typically added to substrates by GalNAc transferases within the Golgi [
21]. Despite no clear structural alteration to the Golgi itself, GalNAc accumulation was suppressed at short time points by ZJ-101. Glycomics analysis confirmed a general downregulation of
O- and
N-glycan-bearing proteins (
Figure S5), with several exceptions. Poly-sialylated high-molecular-weight N-glycans, bearing N-acetyl-lactosamine (LacNAc), were upregulated to a relatively high degree. These glycans are produced by the enzyme βGALT4 in the
trans-Golgi, which indicates this region remains active during ZJ-101 treatment [
22]. This result is counter to that of the transcriptomic signature, where transcripts of B4GALT1 and other galactosyltransferases are found to be suppressed by ZJ-101. It is possible this downregulation is caused by a feedback loop from these LacNAc residues accumulating in the
trans-Golgi. Additional poly-sialylated O-glycan residues, such as the sialyl-Tn antigen, known to be processed by ST6GalNAc, were also found to be upregulated (
Table 1), indicating that sialyl-transferase activity in the Golgi also remained. Despite the global decrease in N-glycan abundance, lectin staining of both high-mannose and N-acetyl-glucosamine (GlcNAc) residues was unchanged by ZJ-101 treatment. This discrepancy may stem from the long half-lives of proteins bearing these residues, which may not yet have turned over. It’s likely that new proteins exiting the Golgi, or held within the trans-Golgi network, bear the more complex LacNAc residues identified in the glycomics analysis. A similar accrual of poly-sialylated N-glycans in the trans-Golgi was recently observed by Kitano et al. following Rab11 knockdown, which may provide a possible explanation for ZJ-101′s effect [
23]. Further inquiry is required to determine whether glycosyltransferase localization at the Golgi is affected by ZJ-101.
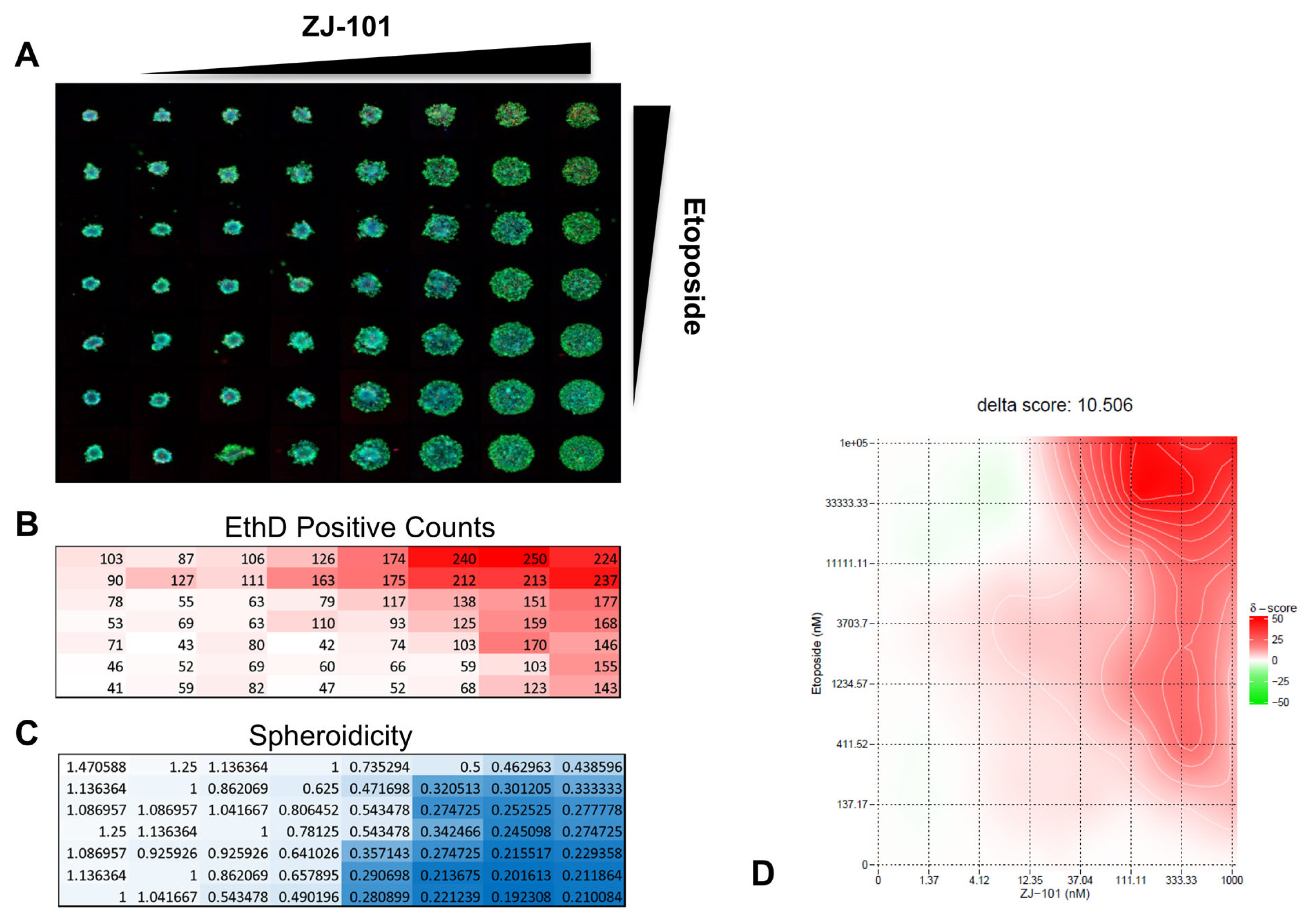
We performed a high-content 3D-spheroid drug combination synergy assay using a modified spheroid disassembly assay. Our objective was to identify whether the anti-adhesive effect of ZJ-101 could reverse a common mechanism of drug resistance mediated by tight cellular adhesion [
24,
25]. In a 2D context, etoposide treatment induces high BRCA1 expression in MDA-MB-231 cells, making them naturally resistant to etoposide’s mechanism of action [
26]. For MDA-MB-231 spheroids in particular, ECM interactions further limit drug accessibility, providing a key context to assess 3D-induced chemoresistance to topoisomerase II inhibitors [
27]. Broad suppression of extracellular matrix and cell adhesion gene transcripts by ZJ-101 lends further support to a possible reversal of chemoresistance mediated by such factors. Our combination synergy assay utilizes the zero interaction potency (ZIP) synergy score, which evaluates individual drug combination pairs relative to their separate dose–response curves [
28]. ZIP synergy normalizes drug response to assume a “zero interaction” or minimal shift in dose–response between combination pairs. Etoposide synergizes with ZJ-101 with an average δ score of 10.5 across all combination pairs, indicating positive synergy. δ-scores describe the excess percentage at which combination pairs are synergistic or antagonistic. For etoposide, the average δ-score for concentrations above 10 μM is between 40 and 50, indicating 40–50% excess synergy with ZJ-101 over etoposide alone.
In conclusion, we have identified several unique phenotypes produced by treatment with the marine natural product-derived compound, ZJ-101. We discovered a strongly cytostatic and antiadhesive phenotype at single-digit nanomolar concentrations, which is unique and unprecedented. Through transcriptomic analysis, we identified dysregulation of the endomembrane system, which was later confirmed by lectin staining and glycomics analysis. Our work has established a potential synergistic mechanism for the reversal of chemoresistance mediated by 3D cell adhesion. Other mechanisms of multidrug resistance mediated by endolysosomal trafficking provide a future direction for compounds such as ZJ-101, which dysregulate glycosylation [
29]. Recent screens for regulators of cellular glycosylation have indicated potential use against SARS-CoV-2 viral entry, offering another potential path for further ZJ-101 investigation [
30].
4. Materials and Methods
4.1. Cell Culture
Cells were cultured in DMEM media (Gibco Cat # 11885, Billings, MT, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
4.2. Cell Cycle Analysis
Cell cycle analysis was performed using propidium iodide staining of fixed MDA-MB-231 cells treated with ZJ-101. Briefly, a 70% confluent 10 cm dish of MDA-MB-231 was collected via trypsinization and washed with 1× PBS. Pelleted cells were fixed by the dropwise addition of 2 mL of ice-cold 75% EtOH. Fixed cells were washed again with PBS and stained with 1 mg/mL propidium iodide solution prior to FACS analysis.
4.3. Growth-Rate Inhibition Metric Analysis
Control populations of MDA-MB-231 cells were prepared for growth rate normalization at seeding densities of 50, 200, 500, and 1000 cells per well in a 384-well flat bottom plate (Corning Inc., Corning, NY, USA). Following compound treatment, cells were stained with 1 μM Calcein AM (for live cells), 20 μg/mL Hoechst 33342 (for nucleus), and 1 μM ethidium homodimer (a cell membrane impermeable dye for dead cells) for 15 min at 37 °C prior to imaging. Imaging was performed using the 4× objective on the ImageXpress Micro (Molecular Devices, San Jose, CA, USA) with image-based focusing. Collected images were assessed using the live/dead program in MetaXpress (version 6.1), and data were organized and entered into grcalculator.org, where growth rate normalized inhibition calculations were performed for each compound tested.
4.4. 3D Spheroid Assays
Three-dimensional spheroid formation was assessed using Corning ultra-low attachment (ULA) plates. MDA-MB-231 cells were seeded in ULA plates at a density of 2 × 103 cells/well (96-well) or 5 × 102 cells/well (384-well) in DMEM media supplemented with 10% FBS and centrifuged for 5 min at 400× g. Cells were then left undisturbed in a 37 °C, 5% CO2 incubator for 72 h to form spheroids. ZJ-101 was added at the indicated concentrations and incubated for a further 72 h. Pretreatment of ZJ-101 was also performed with basic light images taken every 24 h for up to 72 h. Spheroidicity was determined based on the overall diameter normalized to an untreated control using ImageJ (version 1.52r).
The 3D spheroid combination assay was performed as a spheroid disassembly assay requiring 72 h of pre-formed spheroids prior to a further 72 h of drug addition. ZJ-101 and etoposide were arranged with 1:3 dilutions in an 8 × 8 matrix format at the indicated concentrations. Fluorescent images were acquired after staining using the same protocol and instrumentation outlined in the GR assay. Laser-based focusing was utilized to obtain clear spheroid images from the bottom of each well. Ten z-stack images were taken and combined into a single maximum-intensity image. Images were processed with the live/dead program in MetaExpress.
4.5. Cell Staining and Imaging
Basic light microscopy was performed on HEK293T cells cultured in 6-well TC-treated plates (Greiner #657160, Kremsmünster, Austria) by capturing images through the 20X objective of a Zeiss Axiovert 25 (Carl Zeiss Microscopy, White Plains NY, USA) with a 12.2 megapixel camera. For MDA-MB-231 spheroid light microscopy, the same protocol was used with a 10X objective.
For all staining assays, HeLa cells were cultured on 96-well CellView TC-treated microplates (Greiner #655891, Kremsmünster, Austria). Cells were washed with cold PBS prior to fixation with 4% PFA for 10 min at RT. After fixation, cells were washed twice with PBS and permeabilized with 0.1% Triton-X for 10 min at RT. Cells were again washed twice with PBS and blocked with cell staining buffer for 30 min. Antibodies for GM130 (Cell Signaling #12480, Danvers, MA, USA) were added at 1:1000 in staining buffer for 1 h at RT. After three washes with PBS, secondary anti-rabbit AlexaFluor-488, or AlexaFluor-647 (ThermoFisher, Waltham, MA, USA) were added along with the indicated fluorescent lectin conjugates (HPA-647, WGA-488, and PNA-555 from ThermoFisher) at 1:1000 dilution for 1 h RT. Cells were again washed and stained with 1:104 Hoechst 33,342 for 3 min prior to imaging with an OperaPhenix (PerkinElmer, Waltham, MA, USA). Images were uploaded to the Columbus Analyzer (version 2.9.1.699) and processed for high-content analysis.
For the live cell lysotracker assay, HeLa cells were cultured as above during treatment with the specified compounds. Cells were loaded with DMEM media containing 100 nM Lysotracker Deep Red for 30 min at 37 °C. After lysotracker staining, media was exchanged for Live Cell Imaging Solution (Invitrogen #A14291DJ, Waltham, MA, USA) containing 1:104 Hoechst 33342 for nuclei staining. Images were uploaded to the Columbus Analyzer and processed for high-content analysis using default settings for spot detection and intensity calculation.
4.6. Transcriptome Analysis
MDA-MB-231 cells from 10 cm dishes were harvested through scraping (for 2D) or spheroids collected (for 3D) using a wide-gauge pipette and subjected to RNA extraction via the RNeasy mini kit using the manufacturer’s instructions. Biological replicates of N = 3 were used for both sets of analyses, with N = 96 spheroids representing a single biological replicate for the 3D populations. RNA-sequencing was performed by Genewiz as paired-end 150 bp reads following poly-A selection to enrich mRNA transcripts. Paired-end FASTQ files were uploaded to Galaxy using the public server at usegalaxy.org (accessed on 7 April 2021) and aligned to hg38 using HISAT2. Transcripts were assembled and counted using htseq-count, and differential gene expression was evaluated with DESeq2 with default settings. Fold changes were assessed against the DMSO vehicle controls. Transcripts were annotated using the most current GENCODE release. Gene Ontology analysis was performed using g:Profiler at
https://biit.cs.ut.ee/gprofiler/gost (accessed on 8 April 2021) by inputting the top 200 significant genes for each concentration of ZJ-101 and arranging them by descending order of significance [
9]. Heatmaps and PCA plots were generated using ClustVis v1.0 at
https://biit.cs.ut.ee/clustvis/ (accessed on 8 April 2021) [
10].
4.7. Glycan Analysis
4.7.1. Glycan Incorporation Assay
Briefly, 50 μM azido-modified sugars (Invitrogen) tetraacetylated N-azidoacetylglucosamine (GlcNAz), tetraacetylated N-azidoacetylgalactosamine (GalNAz), tetraacetylated N-azidoacetyl-D-mannosamine (ManNAz), and 100 μM alkynyl-fucose (Invitrogen) were added to 96-well plates in combination with the indicated doses of ZJ-101. Cells were then washed, fixed, and permeabilized prior to the click reaction. Copper-catalyzed click reactions were performed using the Click-iT Cell Reaction Buffer Kit (Invitrogen C10269) per the manufacturer’s instructions, containing 5 μM of either Fluor alkyne-647 (Invitrogen A10278) to label azido-incorporated sugars or Fluor Azide-488 to label incorporated alkynyl-fucose. Plates were washed five times before Hoechst counterstaining and imaging. Glycan incorporation was determined by the total intensity of each signal normalized to untreated control cells.
4.7.2. Glycome Profiling
Glycomics profiling was performed by Creative Proteomics. N-glycans were prepared from fresh cell pellets washed with PBS, resuspended in 1 mL of lysis buffer, and sonicated (5 pulses of 10 s). Samples were then dialyzed in 50 mM ammonium bicarbonate for 24 h at 4 °C, with the buffer changed three times. Following dialysis, the samples were lyophilized. To the lyophilized powder, 1 mL of 2 mg/mL DTT was added, and the solution was incubated at 50 °C for 2 h. Briefly, 1 mL of a 12 mg/mL IAA (iodoacetamide, Sigma, St. Louis, MO, USA) solution was then added and incubated at RT in the dark for 2 h. The DTT and IAA-treated proteins were then dialyzed against 50 mM ammonium bicarbonate (Sigma) for 24 h at 4 °C. Samples were next resuspended in 1 mL of 500 µg/mL TPCK-treated trypsin (Sigma) solution and incubated at 37 °C overnight. The trypsin reaction mixture was purified over C18 Sep-Pak columns (Waters, Milford, MA, USA) by 1-propanol elution. Fractions containing peptides were pooled and lyophilized. The lyophilized peptides were resuspended in 200 µL of 50 mM ammonium bicarbonate, to which 3 µL of PNGaseF (New England Biolabs, Ipswich, MA, USA) was added for a 4 h incubation at 37 °C. Following this initial incubation, another 5 µL of PNGaseF was added for overnight incubation at 37 °C. The enzymatic reaction was stopped by the addition of two drops of 5% acetic acid, and the released N-glycans were purified over C18 Sep-Pak columns. Flow-through and 5% acetic acid washing fractions containing the released N-glycans were pooled and lyophilized and were subject to permethylation. For O-glycan analysis, the PNGaseF-treated glycopeptides were eluted from the C18 column with 1-propanol. The lyophilized eluted peptides were subjected to O-glycan preparation.
O-glycan-containing powder was solubilized by 400 µL of a sodium borohydride (Sigma-Aldrich) solution in 0.1 M NaOH (55 mg NaBH4/1 mL 0.1 M NaOH) and incubated overnight at 45 °C. The reaction was stopped by adding drops of pure acetic acid until the fizzing stopped. The samples were passed through a Dowex 50W X8 resin (Sigma-Aldrich) column, and the pooled fractions were dried by lyophilization. The lyophilized samples were next resuspended in 1 mL of an acetic acid:methanol solution (1:9 v/v) and co-evaporated under nitrogen flow. This step was repeated 3 more times, and the dried samples were resuspended in 200 μL of 50% methanol prior to being loaded onto the C18 Sep-Pak column. Free O-glycans were collected in the flow-through and 5% acetic acid wash fractions. These fractions were pooled, lyophilized, and subjected to permethylation.
Permethylation was performed as follows. Seven pellets of NaOH in 3 mL of DMSO were ground with a mortar and pestle. One milliliter of this slurry solution was added to the dry sample in a glass tube with a screw cap. Five hundred microliters of iodomethane was then added to the slurry and shaken at RT for ~30 min. After the reaction reaches completion, noting the formation of white solids, the cap is released slowly to relieve the gas pressure that has built up. One milliliter of MilliQ water was added to stop the reaction, and the sample was vortexed until all solids were dissolved. To the sample, 1 mL of chloroform and an additional 3 mL of MilliQ water were added with continuous vortexing to mix both phases. The samples were then centrifuged briefly to separate the chloroform and the water phases (~5000 rpm, <20 s). The aqueous top layer was discarded, and washing was repeated twice with the addition of 3 mL of MilliQ water. The chloroform fraction was then dried with a SpeedVac (~20–30 min). A C18 Spe-Pak (200 mg) column was prepared with methanol, MiliQ water, acetonitrile, and MilliQ water. The dry sample was resuspended with 200 µL of 50% methanol and loaded onto the column. The column is then washed with 2 mL of 15% acetonitrile and eluted into a clean glass tube with 3 mL of 50% acetonitrile. Finally, the eluted fraction was subjected to MS analysis.
MS data were acquired on a Bruker UltraFlex II MALDI-TOF mass spectrometer instrument (Bruker Scientific LLC, Billerica, MA, USA). Reflective positive mode was used, and data were usually recorded between 500
m/
z and 6000
m/
z for
N-linked glycans and between 500
m/
z and 4000
m/
z for
O-glycans. For each MS N- and
O-glycan profile, the aggregation of 20,000 laser shots or more was considered for data extraction. Mass signals with a signal/noise ratio of at least 2 were considered, and only MS signals matching an
N- and
O-glycan composition were considered for further analysis and annotation. Subsequent MS post-data acquisition analysis was made using mMass [
32].
4.8. Statistical Analysis
Statistical tests were performed on GraphPad Prism version 9.5. The normalized data from n = 3 independent experiments were analyzed for significance using the Brown–Forsythe and Welch one-way ANOVA. Statistical significance is set for * p < 0.05, ** p ≤ 0.01, *** p ≤ 0.001.