Biofilm Formation and Genetic Diversity of Microbial Communities in Anaerobic Batch Reactor with Polylactide (PLA) Addition
Abstract
:1. Introduction
2. Results and Discussion
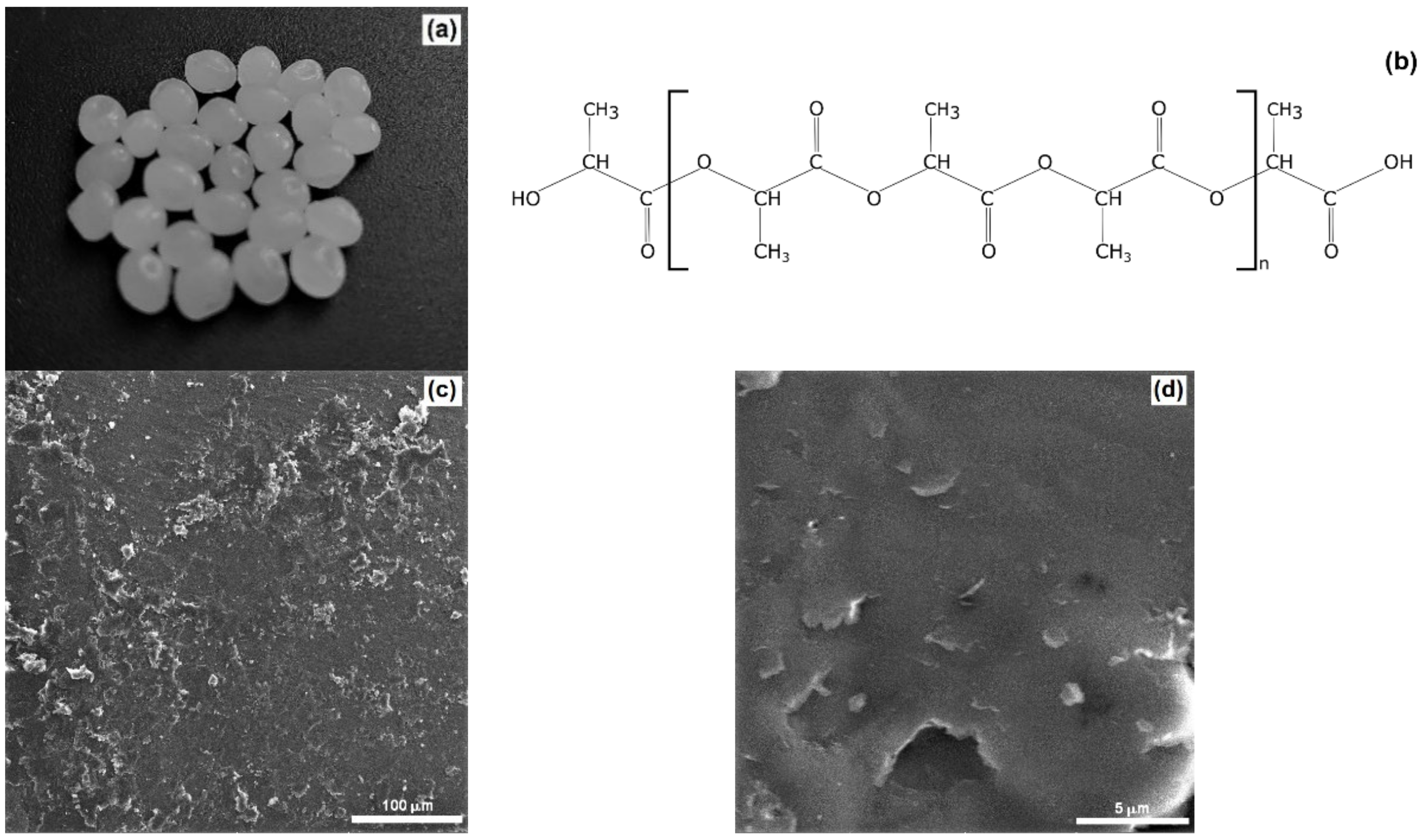
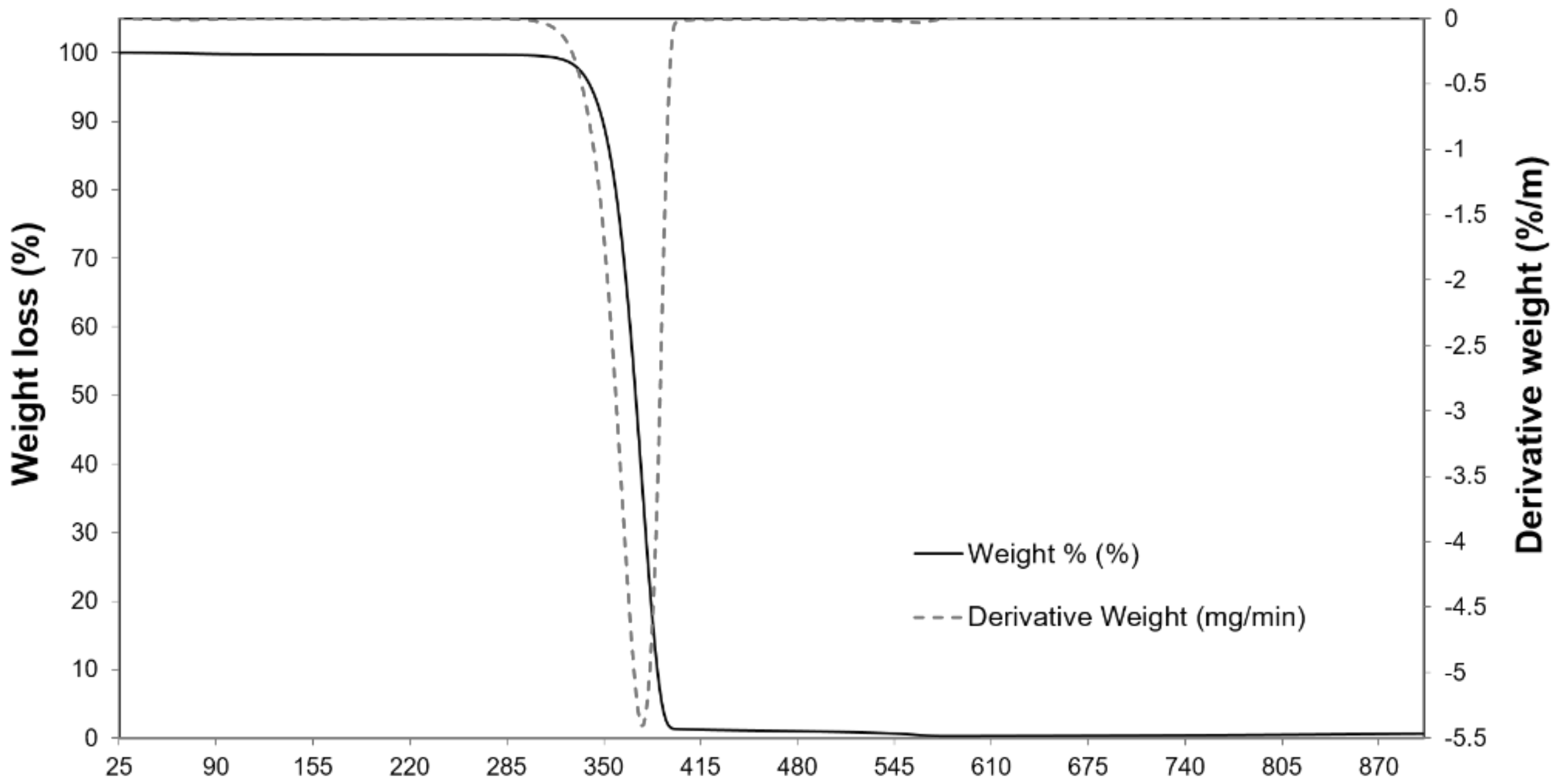
2.1. Physicochemical Properties of the Carrier
2.2. Physicochemical Parameters and Total Bacterial Count of Digested Samples
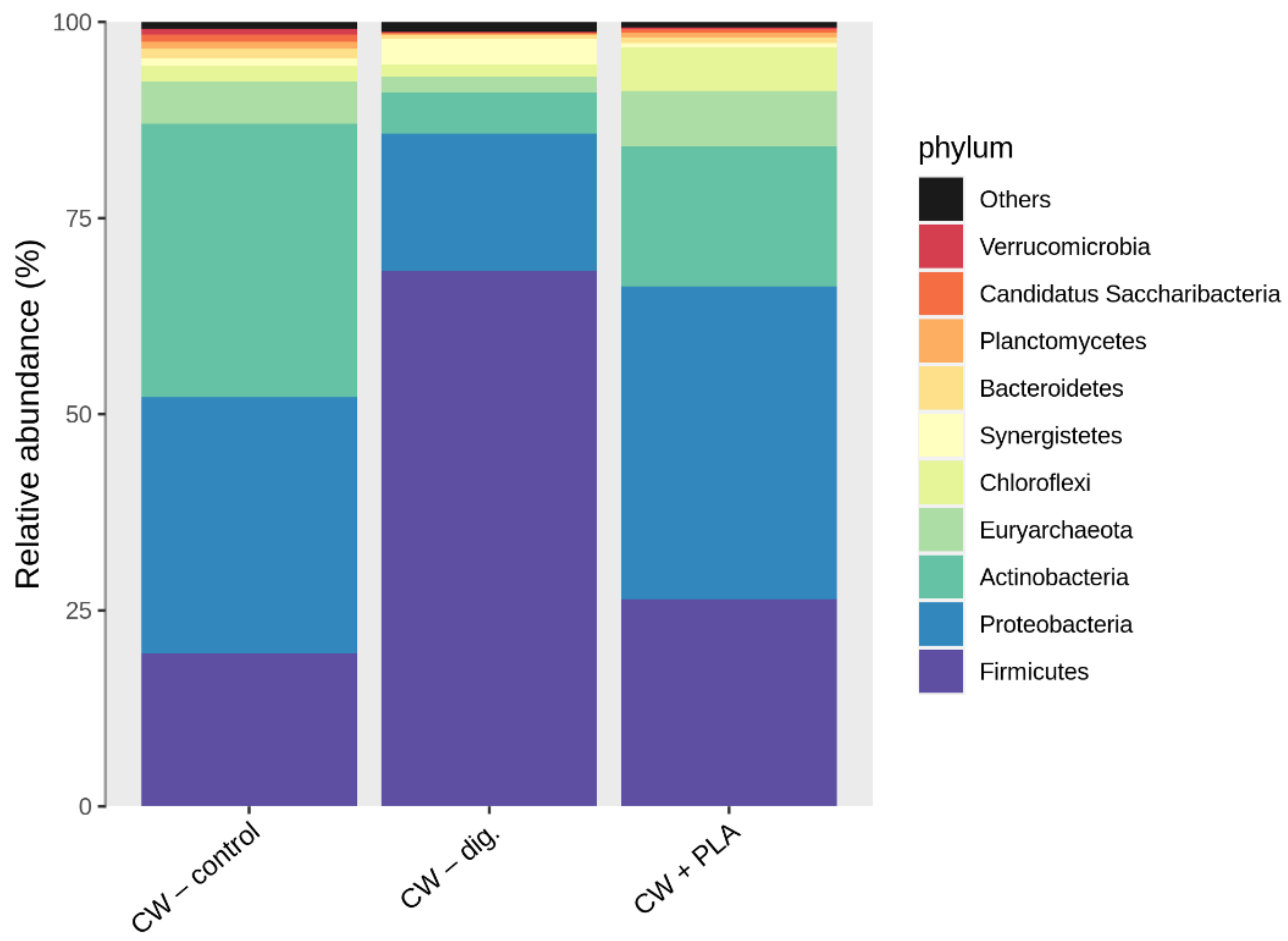
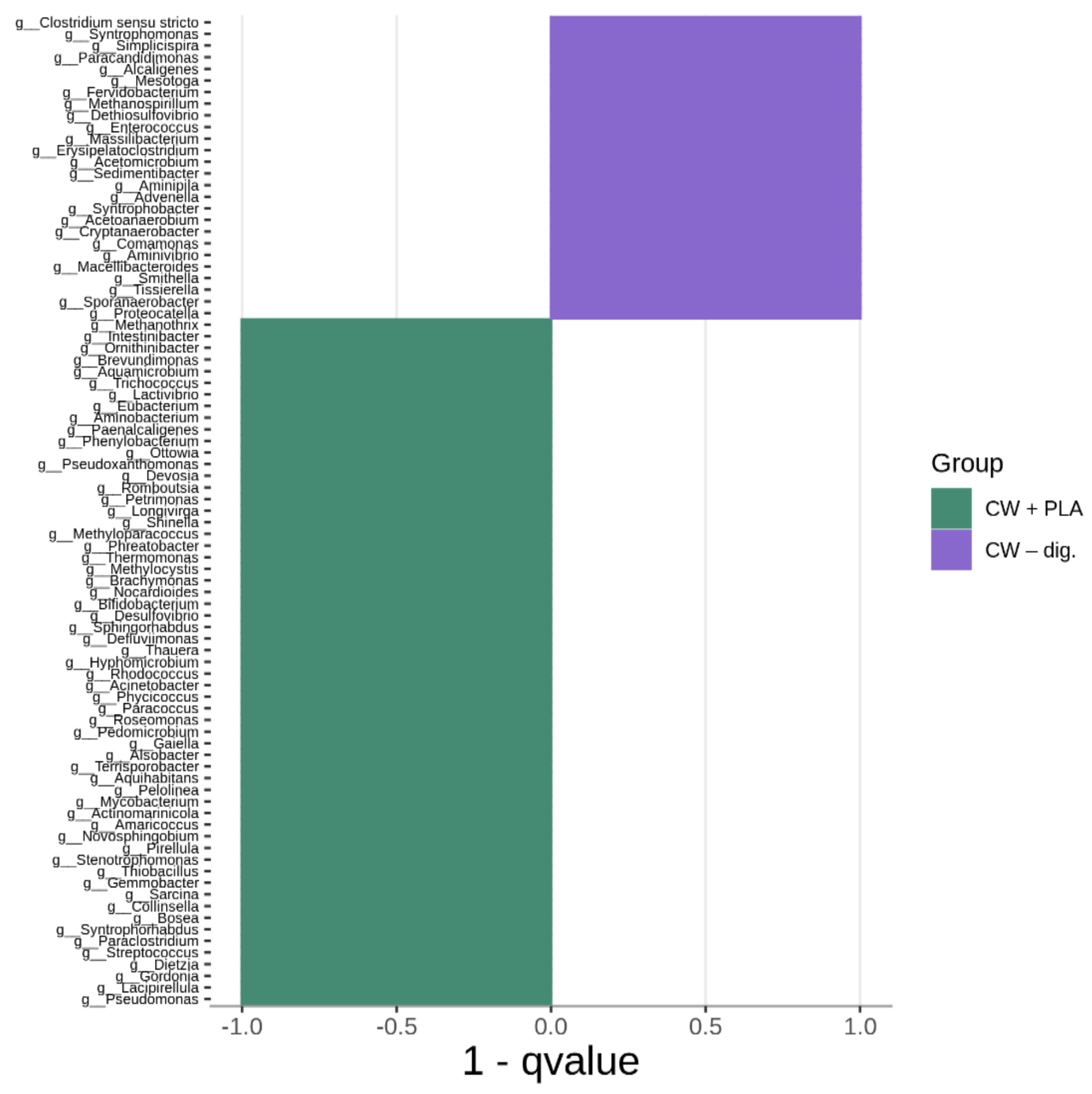
2.3. Bacterial Community Abundance and Composition
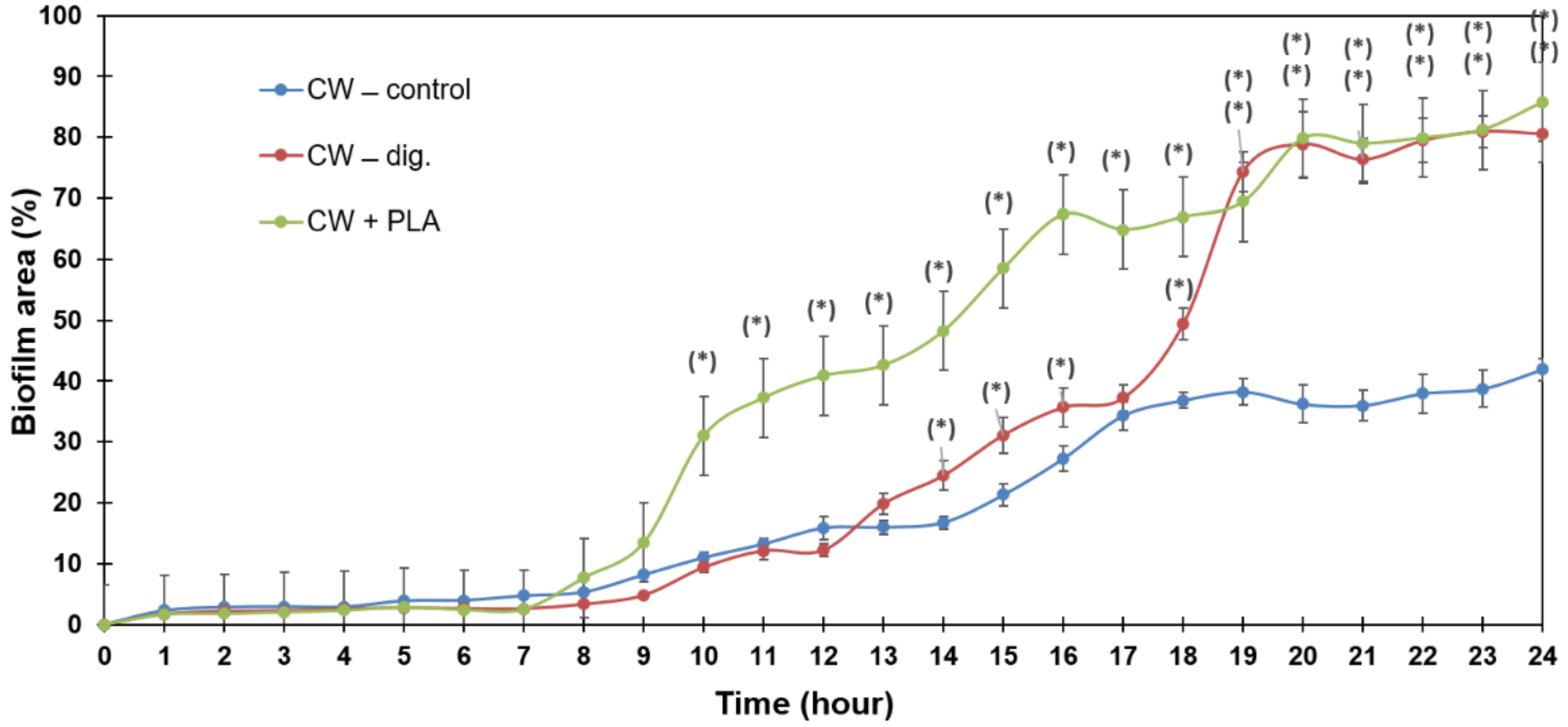
2.4. Biofilm Formation Dynamics
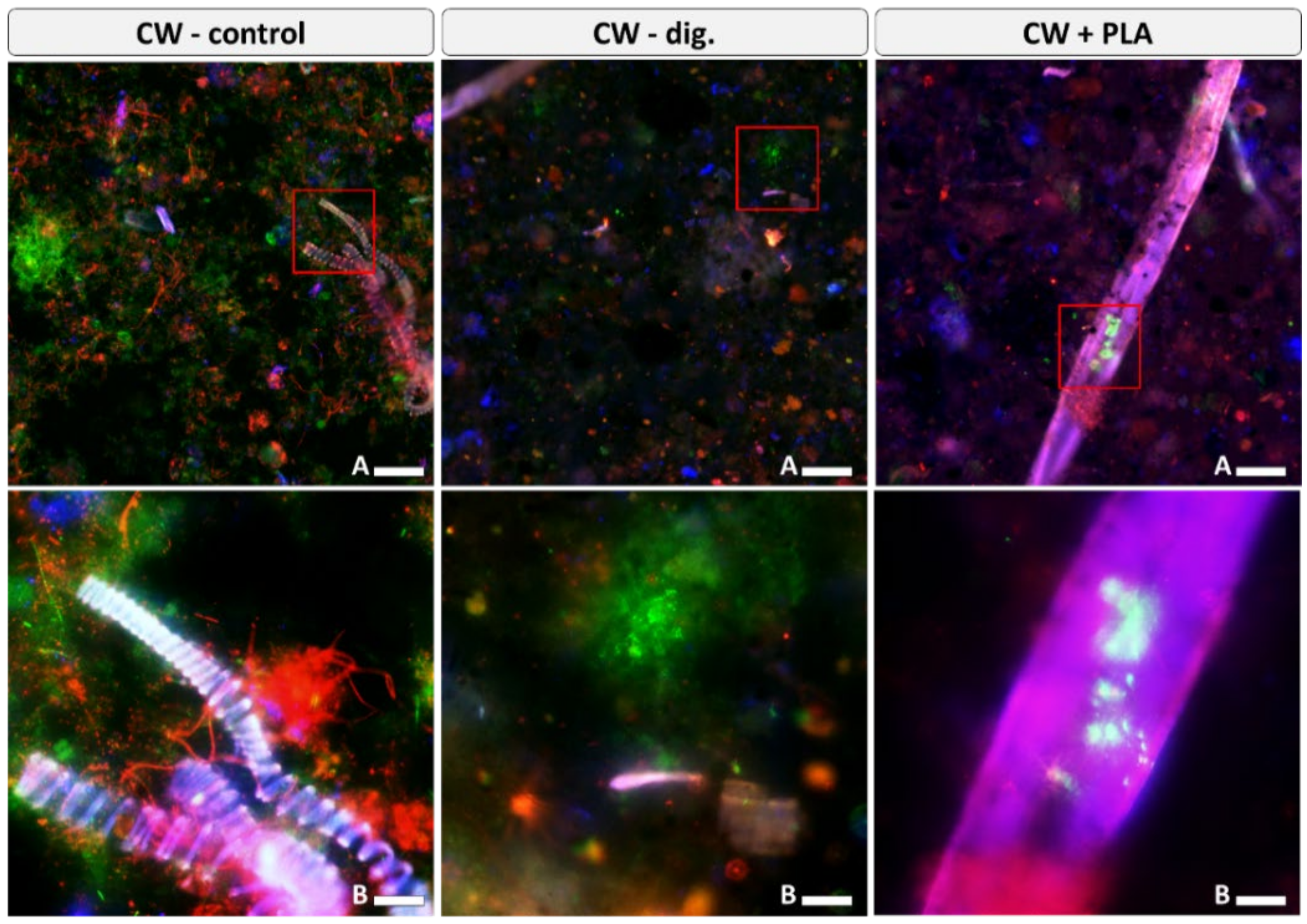
2.5. Visualisation of Microbiome
3. Materials and Methods
3.1. Materials
3.2. Bioreactor Configuration and Operation
3.3. Physicochemical Analysis
3.4. Microbial Analysis
3.4.1. Analysis of the Total Bacterial Count
3.4.2. DNA Extraction and Next Generation Sequencing (NGS)
3.5. Bioinformatics and Statistical Analysis
3.6. Microfluidic Flow System BioFlux 1000z
3.7. Microscopic Visualisation of Microbiome
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kulkarni, V.S.; Butte, K.D.; Rathod, S.S. Natural polymers–A comprehensive review. Int. J. Res. Pharm. Biomed. Sci. 2012, 3, 1597–1613. [Google Scholar]
- Rogovina, S.Z. Biodegradable polymer composites based on synthetic and natural polymers of various classes. Polym. Sci. Ser. C 2016, 58, 62–73. [Google Scholar] [CrossRef]
- Murariu, M.; Bonnaud, L.; Yoann, P.; Fontaine, G.; Bourbigot, S.; Dubois, P. New trends in polylactide (PLA)-based materials: “Green” PLA: Calcium sulfate (nano)composites tailored with flame retardant properties. Polym. Degrad. Stab. 2010, 95, 374–381. [Google Scholar] [CrossRef]
- Rasselet, D.; Ruellan, A.; Guinault, A.; Miquelard-Garnier, G.; Sollogoub, C.; Fayolle, B. Oxidative degradation of polylactide (PLA) and its effects on physical and mechanical properties. Eur. Polym. J. 2014, 50, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.M.; Afrin, S.; Haque, P.; Islam, M.M.; Islam, M.S.; Gafur, M.A. Preparation and characterization of jute cellulose crystals-reinforced poly(L-lactic acid) biocomposite for biomedical applications. Int. J. Chem. Eng. 2014, 2014, 842147. [Google Scholar] [CrossRef] [Green Version]
- Tyler, B.; Gullotti, D.; Mangraviti, A.; Utsuki, T.; Brem, H. Polylactic acid (PLA) controlled delivery carriers for biomedical applications. Adv. Drug Deliv. Rev. 2016, 107, 163–175. [Google Scholar] [CrossRef]
- Nampoothiri, K.M.; Nair, N.R.; John, R.P. An overview of the recent developments in polylactide (PLA) research. Bioresour. Technol. 2010, 101, 8493–8501. [Google Scholar] [CrossRef]
- Najafi, N.; Heuzey, M.C.; Carreau, P.J.; Therriault, D.; Park, C.B. Mechanical and morphological properties of injection molded linear and branched-polylactide (PLA) nanocomposite foams. Eur. Polym. J. 2015, 73, 455–465. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.Q.; Wei, Q.Y.; Chen, Y.; Jia, D.Z.; Lin, H.; Zhong, G.J.; Li, Z.M. Light weight, low dielectric constant, super-robust polylactide film based on stress-induced cavitation aided by crystallization. Polymer 2022, 256, 125234. [Google Scholar] [CrossRef]
- Cazaudehore, G.; Guyoneaud, R.; Evon, P.; Martin-Closas, L.; Pelacho, A.M.; Raynaud, C.; Monlau, F. Can anaerobic digestion be a suitable end-of-life scenario for biodegradable plastics? A critical review of the current situation, hurdles, and challenges. Biotechnol. Adv. 2022, 56, 107916. [Google Scholar] [CrossRef] [PubMed]
- Kolstad, J.J.; Vink, E.T.H.; Wilde, B.D.; Debeer, L. Assessment of anaerobic degradation of IngeoTM polylactides under accelerated landfill conditions. Polym. Degrad. Stab. 2012, 97, 1131–1141. [Google Scholar] [CrossRef]
- Wang, F.; Hidaka, T.; Tsuno, H.; Tsubota, J. Co-digestion of polylactide and kitchen garbage in hyperthermophilic and thermophilic continuous anaerobic process. Bioresour. Technol. 2012, 112, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Tsuno, H.; Hidaka, T.; Tsubota, J. Promotion of polylactide degradation by ammonia under hyperthermophilic anaerobic conditions. Bioresour. Technol. 2011, 102, 9933–9941. [Google Scholar] [CrossRef]
- Benn, N.; Zitome, D. Pretreatment and anaerobic co-digestion of selected PHB and PLA bioplastics. Front. Environ. Sci. 2018, 5, 93. [Google Scholar] [CrossRef] [Green Version]
- Alqaralleh, R.M.; Kennedy, K.; Delatolla, R. Improving biogas production from anaerobic co-digestion of Thickened Waste Activated Sludge (TWAS) and fat, oil and grease (FOG) using a dual-stage hyper-thermophilic/thermophilic semi-continuous reactor. J. Environ. Manag. 2018, 217, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.R.; Sekhar, V.C.; Nampoothiri, K.M. Augmentation of a microbial consortium for enhanced polylactide (PLA) degradation. Indian J. Microbiol. 2016, 56, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Pilarska, A.A.; Bula, K.; Pilarski, K.; Adamski, M.; Wolna-Maruwka, A.; Tomasz Kałuża, T.; Magda, P.; Boniecki, P. Polylactide (PLA) as a cell carrier in mesophilic anaerobic digestion–A new strategy in the management of PLA. Materials 2022, 15, 8113. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, M.; Szara, E.; Sosulski, T.; Stępień, W.; Pilarski, K.; Pilarska, A. Chemical properties and fertilizer value of ten different anaerobic digestates. Fresen. Environ. Bull. 2018, 27, 3425–3432. [Google Scholar]
- Walczak, N.; Walczak, Z.; Kałuża, T.; Hämmerling, M.; Stachowski, P. The impact of shrubby floodplain vegetation growth on the discharge capacity of river valleys. Water 2018, 10, 556. [Google Scholar] [CrossRef] [Green Version]
- Yadav, M.; Joshi, C.; Paritosh, K.; Thakur, J.; Pareek, N.; Masakapalli, S.K.; Vivekanand, V. Reprint of organic waste conversion through anaerobic digestion: A critical insight into the metabolic pathways and microbial interactions. Metab. Eng. 2022, 71, 62–76. [Google Scholar] [CrossRef]
- Cayetano, R.D.A.; Kim, G.B.; Park, J.; Yang, Y.H.; Jeon, B.H.; Jang, M.; Kim, S.H. Biofilm formation as a method of improved treatment during anaerobic digestion of organic matter for biogas recovery. Bioresour. Technol. 2022, 344, 126309. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Waliszewska, B.; Zborowska, M.; Witaszek, K.; Waliszewska, H.; Kolasiński, M.; Szwarc-Rzepka, K. Evaluation of bio-methane yields for high-energy organic waste and sewage sludge: A pilot-scale study for a wastewater treatment plant. Environ. Eng. Manag. J. 2019, 18, 2019–2030. [Google Scholar] [CrossRef]
- Pilarski, K.; Pilarska, A.A.; Boniecki, P.; Niedbała, G.; Durczak, K.; Witaszek, K.; Mioduszewska, N.; Kowalik, I. The efficiency of industrial and laboratory anaerobic digesters of organic substrates: The use of the Biochemical Methane Potential Correction Coefficient. Energies 2020, 13, 1280. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Ren, L.; Yan, B.; Luo, L.; Zhang, J.; Awasthi, M.K. Electron transfer and mechanism of energy production among syntrophic bacteria during acidogenic fermentation: A review. Bioresour. Technol. 2021, 323, 124637. [Google Scholar] [CrossRef]
- Shen, L.; Zhao, Q.; Wu, X.; Li, X.; Li, Q.; Wang, Y. Interspecies electron transfer in syntrophic methanogenic consortia: From cultures to bioreactors. Renew. Sust. Energ. Rev. 2016, 54, 1358–1367. [Google Scholar] [CrossRef]
- Sieber, J.R.; Crable, B.R.; Sheik, C.S.; Hurst, G.B.; Rohlin, L.; Gunsalus, R.P.; McInerney, M.J. Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front. Microbiol. 2015, 6, 115. [Google Scholar] [CrossRef]
- Boonmee, C.; Kositanont, C.; Leejarkpai, T. Degradation of poly (lactic acid) under simulated landfill conditions. Environ. Nat. Res. J. 2016, 14, 1–9. [Google Scholar]
- Grząbka-Zasadzińska, A.; Klapiszewski, Ł.; Bula, K.; Jesionowski, T.; Borysiak, S. Supermolecular structure and nucleation ability of polylactide based composites with silica/lignin hybrid fillers. J. Therm. Anal. Calorim. 2016, 126, 263–275. [Google Scholar] [CrossRef] [Green Version]
- Signori, F.; Coltelli, M.B.; Bronco, S. Thermal degradation of poly(lactic acid) (PLA) and poly(butylene adipate-co-terephthalate) (PBAT) and their blends upon melt processing. Polym. Degrad. Stab. 2009, 94, 74–82. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Wolna-Maruwka, A.; Boniecki, P.; Zaborowicz, M. Use of confectionery waste in biogas production by the anaerobic digestion process. Molecules 2019, 24, 37. [Google Scholar] [CrossRef] [Green Version]
- Feng, K.; Li, H.; Deng, Z.; Wang, Q.; Zhang, Y.; Zheng, C. Effect of pre-fermentation types on the potential of methane production and energy recovery from food waste. Renew. Energy 2020, 146, 1588–1595. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Olesienkiewicz, A. A Comparison of the influence of kraft lignin and the kraft lignin/silica system as cell carriers on the stability and efficiency of the anaerobic digestion process. Energies 2020, 13, 5803. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, Y.; Jia, H.; Yong, X.; Zhang, L.; Zhou, J.; Cao, Z.; Kruse, A.; Wei, P. Effects of different biofilm carriers on biogas production during anaerobic digestion of corn straw. Bioresour. Technol. 2017, 244, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Adamski, M.; Grzyb, A.; Grządziel, J.; Gałązka, A. Silica/lignin carrier as a factor increasing the process performance and genetic diversity of microbial communities in laboratory scale anaerobic digesters. Energies 2021, 14, 4429. [Google Scholar] [CrossRef]
- Choromański, P.; Karwowska, E.; Łebkowska, M. The influence of petroleum products on the methane fermentation process. J. Hazard. Mater. 2016, 301, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.F.; Ebie, Y.; Xu, K.Q.; Li, Y.Y.; Inamori, Y. Characterization of microbial community in the two-stage process for hydrogen and methane production from food waste. Int. J. Hydr. Energy 2010, 35, 8253–8261. [Google Scholar] [CrossRef]
- Vu, H.T.; Min, B. Integration of submersible microbial fuel cell in anaerobic digestion for enhanced production of methane and current at varying glucose levels. Int. J. Hydr. Energy 2019, 44, 7574–7582. [Google Scholar] [CrossRef]
- Zhang, L.; Loh, K.C.; Lim, J.W.; Zhang, J. Bioinformatics analysis of metagenomics data of biogas-producing microbial communities in anaerobic digesters: A review. Renew. Sust. Energ. Rev. 2019, 100, 110–126. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Jarosław Grządziel, J.; Gałązka, A.; Paluch, E.; Borowiak, K.; Pilarski, K. Quantitative and qualitative changes in the genetic diversity of bacterial communities in anaerobic bioreactors with the diatomaceous earth/peat cell carrier. Cells 2022, 11, 2571. [Google Scholar] [CrossRef]
- Fukuyama, Y.; Inoue, M.; Omae, K.; Yoshida, T.; Sako, Y. Chapter Three–Anaerobic and hydrogenogenic carbon monoxide-oxidizing prokaryotes: Versatile microbial conversion of a toxic gas into an available energy. Adv. Appl. Microbiol. 2020, 110, 99–148. [Google Scholar]
- Nakasaki, K.; Koyama, M.; Maekawa, T.; Fujita, J. Jo Fujita Changes in the microbial community during the acclimation process of anaerobic digestion for treatment of synthetic lipid-rich wastewater. J. Biotechnol. 2019, 306, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Poirier, S.; Madigou, C.; Bouchez, T.; Chapleur, O. Improving anaerobic digestion with support media: Mitigation of ammonia inhibition and effect on microbial communities. Bioresour. Technol. 2017, 235, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.L.; Hanada, S.; Ohashi, A.; Harada, H.; Kamagata, Y.; Sekiguchi, Y. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Microb. Ecol. 2008, 74, 2051–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Dong, F.; Zhang, D.; Zhang, J.; Wang, X. Effect of microfluidic channel geometry on Bacillus subtilis biofilm formation. Biomed. Microdevices 2022, 24, 11. [Google Scholar] [CrossRef]
- Lawson, C.E.; Harcombe, W.R.; Hatzenpichler, R.; Lindemann, S.R.; Loffler, F.E.; O’Malley, M.A.; Martín, H.G.; Pfleger, B.F.; Raskin, L.; Venturelli, O.S. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 2019, 17, 725–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.K.; Lee, S.H.; Kim, Y.; Park, H.D. Current understanding and perspectives in anaerobic digestion based on genome-resolved metagenomic approaches. Bioresour. Technol. 2022, 344, 126350. [Google Scholar] [CrossRef]
- Campanaro, S.; Treu, L.; Kougias, P.G.; Luo, G.; Angelidaki, I. Metagenomic binning reveals the functional roles of core abundant microorganisms in twelve full scale biogas plants. Water Res. 2018, 140, 123–134. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Witaszek, K.; Waliszewska, H.; Zborowska, M.; Waliszewska, B.; Kolasiński, M.; Szwarc-Rzepka, K. Treatment of dairy waste by anaerobic digestion with sewage sludge. Ecol. Chem. Eng. 2016, 23, 99–115. [Google Scholar] [CrossRef] [Green Version]
- Norm VDI 4630; Fermentation of Organic Materials Characterization of the Substrate, Sampling, Collection of Material Data, Fermentation Tests. German Engineers Club: Düsseldorf, Germany, 2006.
- Kałuża, T.; Sojka, M.; Wróżyński, R.; Jaskuła, J.; Zaborowski, S.; Hämmerling, M. Modeling of River Channel Shading as a Factor for Changes in Hydromorphological Conditions of Small Lowland Rivers. Water 2020, 12, 527. [Google Scholar] [CrossRef] [Green Version]
- Pilarska, A.A.; Pilarski, K.; Ryniecki, A.; Tomaszyk, K.; Dach, J.; Wolna-Maruwka, A. Utilization of vegetable dumplings waste from industrial production by anaerobic digestion. Int. Agrophys. 2017, 31, 93–102. [Google Scholar] [CrossRef]
- DIN Guideline 38 414-S8; Characterisation of the Substrate, Sampling, Collection of Material Data, Fermentation Tests. German Institute for Standardization: Berlin, Germany, 1985.
- Pilarska, A.; Linda, I.; Wysokowski, M.; Paukszta, D.; Jesionowski, T. Synthesis of Mg(OH)2 from manesium salts and NH4OH by direct functionalisation with poly(ethylene glycols). Physicochem. Probl. Miner. Process. 2012, 48, 631–643. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Wright, E.S. RDP v16 Modified Training Set for 16S rRNA Classification. 2019. Available online: http://www2.decipher.codes/Classification/TrainingSets/RDP_v16_March2018.RData (accessed on 2 January 2019).
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. cPhyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Paluch, E.; Okińczyc, P.; Zwyrzykowska-Wodzińska, A.; Szperlik, J.; Żarowska, B.; Duda-Madej, A.; Bąbelewski, P.; Włodarczyk, M.; Wojtasik, W.; Kupczyński, R.; et al. Composition and antimicrobial activity of Ilex leaves water extracts. Molecules 2021, 26, 7442. [Google Scholar] [CrossRef] [PubMed]
- Paluch, E.; Sobierajska, P.; Okińczyc, P.; Widelski, J.; Duda-Madej, A.; Krzyżanowska, B.; Krzyżek, P.; Ogórek, R.; Szperlik, J.; Chmielowiec, J.; et al. Nanoapatites doped and co-doped with noble metal ions as modern antibiofilm materials for biomedical applications against drug-resistant clinical strains of Enterococcus faecalis VRE and Staphylococcus aureus MRSA. Int. J. Mol. Sci. 2022, 23, 1533. [Google Scholar] [CrossRef]



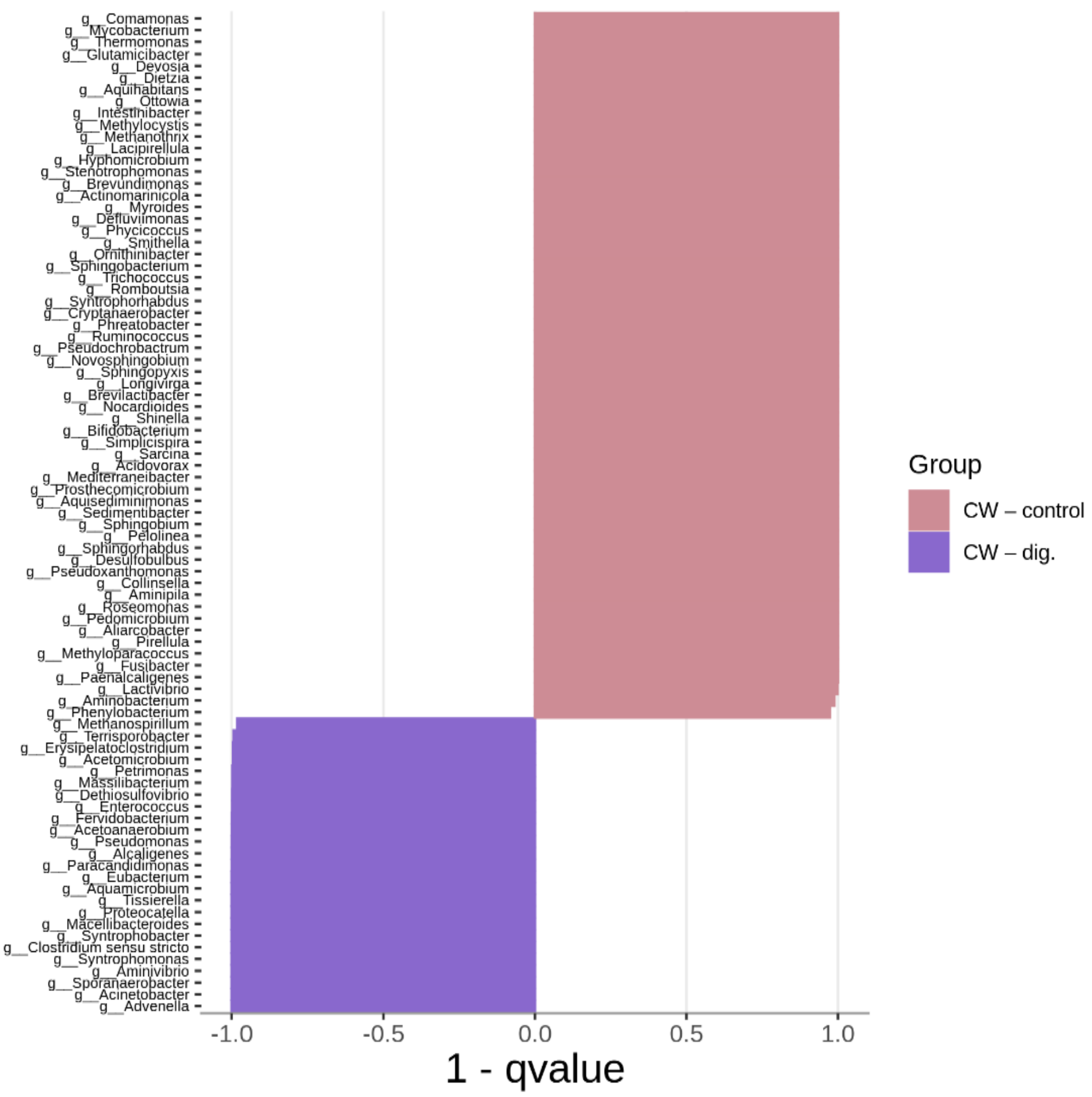


| CW + PLA | CW–dig. | CW + PLA | CW–dig. | ||
|---|---|---|---|---|---|
| Unidentified_003 | ↑ 0.4 | ↓ 2.7 | Unidentified_040 | ↑ – | ≈ – |
| Unidentified_006 | ↑ 0.2 | ↓ 3.6 | Unidentified_042 | ↑ 0.4 | ↓ – |
| Unidentified_007 | ↑ 0.7 | ↓ 1.1 | Unidentified_043 | ↑ – | ≈ – |
| Unidentified_010 | ↑ 0.9 | ↓ 5.8 | Unidentified_046 | ↑ – | ≈ – |
| Unidentified_011 | ↑ 0.6 | ↓ 2.5 | Unidentified_047 | ↑ – | ≈ – |
| Unidentified_014 | ↓ 5.9 | ↑ 0.2 | Unidentified_048 | ↓ 2.1 | ↑ 0.6 |
| Unidentified_016 | ↑ 0.9 | ↓ 3.4 | Unidentified_049 | ↑ – | ≈ – |
| Unidentified_019 | ↑ 0.9 | ↓ 3.4 | Unidentified_050 | ↑ – | ≈ – |
| Unidentified_022 | ↓ 2.3 | ↑ 0.5 | Unidentified_056 | ↑ – | ≈ – |
| Unidentified_024 | ↑ 0.6 | ↓ – | Unidentified_057 | ↑ – | ≈ – |
| Unidentified_025 | ↓ 1.1 | ↑ 0.5 | Unidentified_061 | ↑ 0.6 | ↓ – |
| Unidentified_028 | ↑ 0.7 | ↓ 4.0 | Unidentified_064 | ↑ – | ≈ – |
| Unidentified_029 | ↑ 0.6 | ↓ – | Unidentified_069 | ↑ 0.6 | ↓ – |
| Unidentified_038 | ↑ 0.5 | ↓ – | Unidentified_070 | ↓ 2.2 | ↑ 0.4 |
| Unidentified_039 | ↑ 0.9 | ↓ – |
| Chao1 | Shannon | Simpson | |
|---|---|---|---|
| CW–control | 127 | 3.787 | 0.955 |
| CW–dig. | 101 | 2.857 | 0.846 |
| CW + PLA | 125 | 3.799 | 0.965 |
| CW–Control | CW–dig. | |
|---|---|---|
| CW–dig. | 0.649 | – |
| CW + PLA | 0.367 | 0.606 |
| Materials | pH | Cond. | TS | VS | C/N Ratio | C | N | N-NH4 |
|---|---|---|---|---|---|---|---|---|
| − | (mS cm−1) | (wt %) | (wt %TS) | − | (wt %TS) | (wt %TS) | (wt %TS) | |
| CW | 7.01 | 3.29 | 94.85 | 98.31 | 49.90 | 45.91 | 0.92 | 0.28 |
| Inoculum | 6.94 | 28.62 | 2.96 | 78.65 | 3.20 | 26.72 | 8.34 | 3.96 |
| Batches | WF (g) | Carrier (g) | Inoculum (g) | pH | TS (%) | VS (%) |
|---|---|---|---|---|---|---|
| CW–control | 10.6 | − | 900.0 | 6.95 | 4.23 | 70.15 |
| CW + PLA | 10.6 | 20.0 | 900.0 | 7.08 | 4.05 | 67.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilarska, A.A.; Marzec-Grządziel, A.; Paluch, E.; Pilarski, K.; Wolna-Maruwka, A.; Kubiak, A.; Kałuża, T.; Kulupa, T. Biofilm Formation and Genetic Diversity of Microbial Communities in Anaerobic Batch Reactor with Polylactide (PLA) Addition. Int. J. Mol. Sci. 2023, 24, 10042. https://doi.org/10.3390/ijms241210042
Pilarska AA, Marzec-Grządziel A, Paluch E, Pilarski K, Wolna-Maruwka A, Kubiak A, Kałuża T, Kulupa T. Biofilm Formation and Genetic Diversity of Microbial Communities in Anaerobic Batch Reactor with Polylactide (PLA) Addition. International Journal of Molecular Sciences. 2023; 24(12):10042. https://doi.org/10.3390/ijms241210042
Chicago/Turabian StylePilarska, Agnieszka A., Anna Marzec-Grządziel, Emil Paluch, Krzysztof Pilarski, Agnieszka Wolna-Maruwka, Adrianna Kubiak, Tomasz Kałuża, and Tomasz Kulupa. 2023. "Biofilm Formation and Genetic Diversity of Microbial Communities in Anaerobic Batch Reactor with Polylactide (PLA) Addition" International Journal of Molecular Sciences 24, no. 12: 10042. https://doi.org/10.3390/ijms241210042
APA StylePilarska, A. A., Marzec-Grządziel, A., Paluch, E., Pilarski, K., Wolna-Maruwka, A., Kubiak, A., Kałuża, T., & Kulupa, T. (2023). Biofilm Formation and Genetic Diversity of Microbial Communities in Anaerobic Batch Reactor with Polylactide (PLA) Addition. International Journal of Molecular Sciences, 24(12), 10042. https://doi.org/10.3390/ijms241210042








