DNA Methylation in Alcohol Use Disorder
Abstract
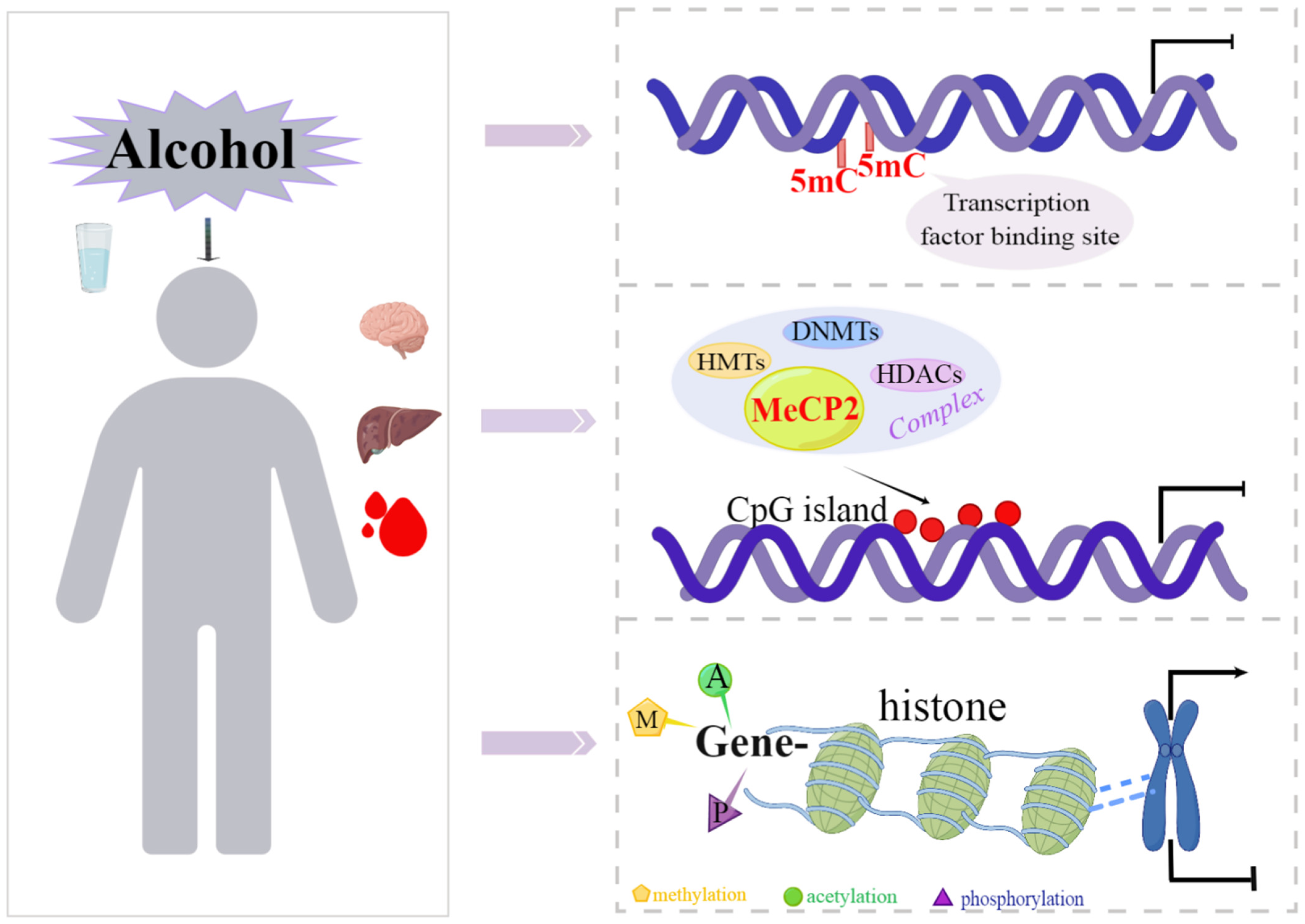
1. Introduction
2. DNA Methylation and Its Regulatory Mechanisms
2.1. Methylation and Demethylation
2.2. Mechanism of DNA Methylation Regulating Gene Transcription
2.3. DNA Methylation Inhibitors
3. DNA Methylation and Alcohol Abuse
3.1. DNA Methylation Changes Ethanol Oxidation System
3.2. DNA Methylation Profiles in Prenatal Alcohol Exposure (PAE)
3.3. DNA Methylation Changes during Adolescent Alcohol Exposure
3.4. DNA Methylation Changes Induced by Alcohol Abuse in Adulthood
4. DNA Methylation as a Therapeutic Target for AUD
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Status Report on Alcohol and Health; WHO: Geneva, Switzerland, 2018.
- Oliveira de Araújo Melo, C.; Cidália Vieira, T.; Duarte Gigonzac, M.A.; Soares Fortes, J.; Moreira Duarte, S.S.; da Cruz, A.D.; Silva, D.M.E. Evaluation of polymorphisms in repair and detoxification genes in alcohol drinkers and non-drinkers using capillary electrophoresis. Electrophoresis 2020, 41, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Heath, A.C.; Bucholz, K.K.; Madden, P.A.; Dinwiddie, S.H.; Slutske, W.S.; Bierut, L.J.; Statham, D.J.; Dunne, M.P.; Whitfield, J.B.; Martin, N.G. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol. Med. 1997, 27, 1381–1396. [Google Scholar] [CrossRef]
- Morozova, T.V.; Mackay, T.F.; Anholt, R.R. Genetics and genomics of alcohol sensitivity. Mol. Genet. Genom. 2014, 289, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Reilly, M.T.; Noronha, A.; Goldman, D.; Koob, G.F. Genetic studies of alcohol dependence in the context of the addiction cycle. Neuropharmacology 2017, 122, 3–21. [Google Scholar] [CrossRef]
- Farris, S.P.; Mayfield, R.D. Epigenetic and non-coding regulation of alcohol abuse and addiction. Int. Rev. Neurobiol. 2021, 156, 63–86. [Google Scholar] [PubMed]
- Boschen, K.E.; Keller, S.M.; Roth, T.L.; Klintsova, A.Y. Epigenetic mechanisms in alcohol- and adversity-induced developmental origins of neurobehavioral functioning. Neurotoxicol. Teratol. 2018, 66, 63–79. [Google Scholar] [CrossRef]
- Mahna, D.; Puri, S.; Sharma, S. DNA methylation signatures: Biomarkers of drug and alcohol abuse. Mutat. Res. Rev. Mutat. Res. 2018, 777, 19–28. [Google Scholar] [CrossRef]
- Stevenson, T.J.; Prendergast, B.J. Reversible DNA methylation regulates seasonal photoperiodic time measurement. Proc. Natl. Acad. Sci. USA 2013, 110, 16651–16656. [Google Scholar] [CrossRef]
- Kim, M.; Costello, J. DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell. Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Planques, A.; Kerner, P.; Ferry, L.; Grunau, C.; Gazave, E.; Vervoort, M. DNA methylation atlas and machinery in the developing and regenerating annelid Platynereis dumerilii. BMC Biol. 2021, 19, 148. [Google Scholar] [CrossRef] [PubMed]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar]
- Morris, M.J.; Monteggia, L.M. Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues Clin. Neurosci. 2014, 16, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Takai, D.; Jones, P.A. The CpG island searcher: A new WWW resource. Silico Biol. 2003, 3, 235–240. [Google Scholar]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef]
- Heberle, E.; Bardet, A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019, 63, 727–741. [Google Scholar]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Turek-Plewa, J.; Jagodziński, P.P. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell. Mol. Biol. Lett. 2005, 10, 631–647. [Google Scholar]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Aapola, U.; Kawasaki, K.; Scott, H.S.; Ollila, J.; Vihinen, M.; Heino, M.; Shintani, A.; Kawasaki, K.; Minoshima, S.; Krohn, K.; et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 2000, 65, 293–298. [Google Scholar] [CrossRef]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Sadri-Vakili, G. Cocaine triggers epigenetic alterations in the corticostriatal circuit. Brain Res. 2015, 1628, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 2004, 15, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Shiota, K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003, 278, 4806–4812. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Oslin, D.W.; Cary, M.S. Alcohol-related dementia: Validation of diagnostic criteria. Am. J. Geriatr. Psychiatry 2003, 11, 441–447. [Google Scholar] [CrossRef]
- Penas, C.; Navarro, X. Epigenetic Modifications Associated to Neuroinflammation and Neuropathic Pain After Neural Trauma. Front. Cell. Neurosci. 2018, 12, 158. [Google Scholar] [CrossRef]
- Gray, S.G.; Dangond, F. Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics 2006, 1, 67–75. [Google Scholar] [CrossRef]
- Roberto, M.; Nelson, T.E.; Ur, C.L.; Gruol, D.L. Long-term potentiation in the rat hippocampus is reversibly depressed by chronic intermittent ethanol exposure. J. Neurophysiol. 2002, 87, 2385–2397. [Google Scholar] [CrossRef]
- Zhang, Y.; Rohde, C.; Tierling, S.; Jurkowski, T.P.; Bock, C.; Santacruz, D.; Ragozin, S.; Reinhardt, R.; Groth, M.; Walter, J.; et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009, 5, e1000438. [Google Scholar] [CrossRef]
- Chhabra, R. miRNA and methylation: A multifaceted liaison. Chembiochem 2015, 16, 195–203. [Google Scholar] [CrossRef]
- Fuso, A.; Lucarelli, M. CpG and Non-CpG Methylation in the Diet-Epigenetics-Neurodegeneration Connection. Curr. Nutr. Rep. 2019, 8, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kaminskas, E.; Farrell, A.T.; Wang, Y.C.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 2005, 10, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Sarkisjan, D.; Julsing, J.R.; El Hassouni, B.; Honeywell, R.J.; Kathmann, I.; Matherly, L.H.; Lee, Y.B.; Kim, D.J.; Peters, G.J. RX-3117 (Fluorocyclopentenyl-Cytosine)-Mediated Down-Regulation of DNA Methyltransferase 1 Leads to Protein Expression of Tumor-Suppressor Genes and Increased Functionality of the Proton-Coupled Folate Carrier. Int. J. Mol. Sci. 2020, 21, 2717. [Google Scholar] [CrossRef] [PubMed]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Zhang, X.L. DNA Methyltransferase Inhibitor 5-AZA-DC Regulates TGFβ1-Mediated Alteration of Neuroglial Cell Functions after Oxidative Stress. Oxidative Med. Cell. Longev. 2022, 2022, 9259465. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Fu, Y.; Tian, S.; Huang, S.; Luo, X.; Lin, L.; Zhang, X.; Wang, H.; Lin, Z.; Zhao, H.; et al. Zebularine elevates STING expression and enhances cGAMP cancer immunotherapy in mice. Mol. Ther. 2021, 29, 1758–1771. [Google Scholar] [CrossRef]
- Brueckner, B.; Garcia Boy, R.; Siedlecki, P.; Musch, T.; Kliem, H.C.; Zielenkiewicz, P.; Suhai, S.; Wiessler, M.; Lyko, F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005, 65, 6305–6311. [Google Scholar] [CrossRef]
- Rilova, E.; Erdmann, A.; Gros, C.; Masson, V.; Aussagues, Y.; Poughon-Cassabois, V.; Rajavelu, A.; Jeltsch, A.; Menon, Y.; Novosad, N.; et al. Design, synthesis and biological evaluation of 4-amino-N-(4-aminophenyl)benzamide analogues of quinoline-based SGI-1027 as inhibitors of DNA methylation. ChemMedChem 2014, 9, 590–601. [Google Scholar] [CrossRef]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M.; et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef]
- Halby, L.; Champion, C.; Sénamaud-Beaufort, C.; Ajjan, S.; Drujon, T.; Rajavelu, A.; Ceccaldi, A.; Jurkowska, R.; Lequin, O.; Nelson, W.G.; et al. Rapid synthesis of new DNMT inhibitors derivatives of procainamide. Chembiochem 2012, 13, 157–165. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, Y.; Li, D.D.; Zhang, Y.; Zhao, T.C.; Li, C.F. Procaine is a specific DNA methylation inhibitor with anti-tumor effect for human gastric cancer. J. Cell. Biochem. 2018, 119, 2440–2449. [Google Scholar] [CrossRef]
- Plummer, R.; Vidal, L.; Griffin, M.; Lesley, M.; de Bono, J.; Coulthard, S.; Sludden, J.; Siu, L.L.; Chen, E.X.; Oza, A.M.; et al. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin. Cancer Res. 2009, 15, 3177–3183. [Google Scholar] [CrossRef] [PubMed]
- Starlard-Davenport, A.; Kutanzi, K.; Tryndyak, V.; Word, B.; Lyn-Cook, B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: A novel epigenetic therapeutic approach. J. Carcinog. 2013, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Hu, G.; Luo, C.; Liang, Z. DNA methyltransferase inhibitors: An updated patent review (2012-2015). Expert Opin. Ther. Patents 2016, 26, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Aguilera, O.; Depreux, P.; Halby, L.; Arimondo, P.B.; Goossens, L. DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge. Biomolecules 2017, 7, 3. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Jones, W.; Pavlovicz, R.E.; Liu, S.; Yu, J.; Li, P.K.; Lin, J.; Fuchs, J.R.; Marcucci, G.; et al. Curcumin is a potent DNA hypomethylation agent. Bioorg. Med. Chem. Lett. 2009, 19, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Lee, K.W.; Choi, H.D. Rice bran constituents: Immunomodulatory and therapeutic activities. Food Funct. 2017, 8, 935–943. [Google Scholar] [CrossRef]
- Enoch, M.A.; Goldman, D. The genetics of alcoholism and alcohol abuse. Curr. Psychiatry Rep. 2001, 3, 144–151. [Google Scholar] [CrossRef]
- Mead, E.A.; Sarkar, D.K. Fetal alcohol spectrum disorders and their transmission through genetic and epigenetic mechanisms. Front. Genet. 2014, 5, 154. [Google Scholar] [CrossRef]
- Oroszi, G.; Goldman, D. Alcoholism: Genes and mechanisms. Pharmacogenomics 2004, 5, 1037–1048. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, F.; Kranzler, H.R.; Zhao, H.; Gelernter, J. Profiling of childhood adversity-associated DNA methylation changes in alcoholic patients and healthy controls. PLoS ONE 2013, 8, e65648. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, T.; Kusumanchi, P.; Han, S.; Yang, Z.; Liangpunsakul, S. Alcohol Metabolizing Enzymes, Microsomal Ethanol Oxidizing System, Cytochrome P450 2E1, Catalase, and Aldehyde Dehydrogenase in Alcohol-Associated Liver Disease. Biomedicines 2020, 8, 50. [Google Scholar] [CrossRef]
- Lu, Y.; Cederbaum, A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2008, 44, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.M.; Boobis, A.R.; Moore, G.E.; Stanier, P.M. Expression of CYP2E1 during human fetal development: Methylation of the CYP2E1 gene in human fetal and adult liver samples. Biochem. Pharm. 1992, 43, 1876–1879. [Google Scholar] [CrossRef]
- Kronfol, M.M.; Jahr, F.M.; Dozmorov, M.G.; Phansalkar, P.S.; Xie, L.Y.; Aberg, K.A.; McRae, M.; Price, E.T.; Slattum, P.W.; Gerk, P.M.; et al. DNA methylation and histone acetylation changes to cytochrome P450 2E1 regulation in normal aging and impact on rates of drug metabolism in the liver. Geroscience 2020, 42, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Mueller, S. Alcohol and cancer: An overview with special emphasis on the role of acetaldehyde and cytochrome P450 2E1. Adv. Exp. Med. Biol. 2015, 815, 59–70. [Google Scholar]
- Penaloza, C.G.; Cruz, M.; Germain, G.; Jabeen, S.; Javdan, M.; Lockshin, R.A.; Zakeri, Z. Higher sensitivity of female cells to ethanol: Methylation of DNA lowers Cyp2e1, generating more ROS. Cell. Commun. Signal. 2020, 18, 111. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.; Peng, H.; Barbier-Torres, L.; Robinson, A.E.; Li, T.W.H.; Fan, W.; Tomasi, M.L.; Gottlieb, R.A.; Van Eyk, J.; Lu, Z.; et al. Methionine Adenosyltransferase α1 Is Targeted to the Mitochondrial Matrix and Interacts with Cytochrome P450 2E1 to Lower Its Expression. Hepatology 2019, 70, 2018–2034. [Google Scholar] [CrossRef]
- Hwang, P.H.; Lian, L.; Zavras, A.I. Alcohol intake and folate antagonism via CYP2E1 and ALDH1: Effects on oral carcinogenesis. Med. Hypotheses 2012, 78, 197–202. [Google Scholar] [CrossRef]
- Zhang, P.; Li, Y.; Wang, K.; Huang, J.; Su, B.B.; Xu, C.; Wang, Z.; Tan, S.; Yang, F.; Tan, Y. Altered DNA methylation of CYP2E1 gene in schizophrenia patients with tardive dyskinesia. BMC Med. Genom. 2022, 15, 253. [Google Scholar] [CrossRef]
- Kaut, O.; Schmitt, I.; Stahl, F.; Fröhlich, H.; Hoffmann, P.; Gonzalez, F.J.; Wüllner, U. Epigenome-Wide Analysis of DNA Methylation in Parkinson’s Disease Cortex. Life 2022, 12, 502. [Google Scholar] [CrossRef]
- Liu, Y.; Balaraman, Y.; Wang, G.; Nephew, K.P.; Zhou, F.C. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics 2009, 4, 500–511. [Google Scholar] [CrossRef]
- Schuckit, M.A. Alcohol-use disorders. Lancet 2009, 373, 492–501. [Google Scholar] [CrossRef]
- Hemberger, M.; Dean, W.; Reik, W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat. Rev. Mol. Cell. Biol. 2009, 10, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511, 611–615. [Google Scholar] [CrossRef]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef]
- Amiri, S.; Davie, J.R.; Rastegar, M. Chronic Ethanol Exposure Alters DNA Methylation in Neural Stem Cells: Role of Mouse Strain and Sex. Mol. Neurobiol. 2020, 57, 650–667. [Google Scholar] [CrossRef] [PubMed]
- Lussier, A.A.; Bodnar, T.S.; Mingay, M.; Morin, A.M.; Hirst, M.; Kobor, M.S.; Weinberg, J. Prenatal Alcohol Exposure: Profiling Developmental DNA Methylation Patterns in Central and Peripheral Tissues. Front. Genet. 2018, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Jarmasz, J.S.; Stirton, H.; Basalah, D.; Davie, J.R.; Clarren, S.K.; Astley, S.J.; Del Bigio, M.R. Global DNA Methylation and Histone Posttranslational Modifications in Human and Nonhuman Primate Brain in Association with Prenatal Alcohol Exposure. Alcohol. Clin. Exp. Res. 2019, 43, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.R.; Ho, M.F.; Vega, M.C.; Burne, T.H.; Chong, S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenetics Chromatin 2015, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Otero, N.K.; Thomas, J.D.; Saski, C.A.; Xia, X.; Kelly, S.J. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol. Clin. Exp. Res. 2012, 36, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ozturk, N.C.; Zhou, F.C. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE 2013, 8, e60503. [Google Scholar] [CrossRef]
- Cravo, M.L.; Camilo, M.E. Hyperhomocysteinemia in chronic alcoholism: Relations to folic acid and vitamins B(6) and B(12) status. Nutrition 2000, 16, 296–302. [Google Scholar] [CrossRef]
- Bayerlein, K.; Hillemacher, T.; Reulbach, U.; Mugele, B.; Sperling, W.; Kornhuber, J.; Bleich, S. Alcoholism-associated hyperhomocysteinemia and previous withdrawal seizures. Biol. Psychiatry 2005, 57, 1590–1593. [Google Scholar] [CrossRef]
- Wallén, E.; Auvinen, P.; Kaminen-Ahola, N. The Effects of Early Prenatal Alcohol Exposure on Epigenome and Embryonic Development. Genes 2021, 12, 1095. [Google Scholar] [CrossRef]
- Hicks, S.D.; Middleton, F.A.; Miller, M.W. Ethanol-induced methylation of cell cycle genes in neural stem cells. J. Neurochem. 2010, 114, 1767–1780. [Google Scholar] [CrossRef]
- Mattson, S.N.; Crocker, N.; Nguyen, T.T. Fetal alcohol spectrum disorders: Neuropsychological and behavioral features. Neuropsychol. Rev. 2011, 21, 81–101. [Google Scholar] [CrossRef]
- LaSalle, J.M.; Powell, W.T.; Yasui, D.H. Epigenetic layers and players underlying neurodevelopment. Trends Neurosci. 2013, 36, 460–470. [Google Scholar] [CrossRef]
- Frey, S.; Eichler, A.; Stonawski, V.; Kriebel, J.; Wahl, S.; Gallati, S.; Goecke, T.W.; Fasching, P.A.; Beckmann, M.W.; Kratz, O.; et al. Prenatal Alcohol Exposure Is Associated With Adverse Cognitive Effects and Distinct Whole-Genome DNA Methylation Patterns in Primary School Children. Front. Behav. Neurosci. 2018, 12, 125. [Google Scholar] [CrossRef] [PubMed]
- Kernohan, K.D.; Bérubé, N.G. Genetic and epigenetic dysregulation of imprinted genes in the brain. Epigenomics 2010, 2, 743–763. [Google Scholar] [CrossRef] [PubMed]
- Marjonen, H.; Toivonen, M.; Lahti, L.; Kaminen-Ahola, N. Early prenatal alcohol exposure alters imprinted gene expression in placenta and embryo in a mouse model. PLoS ONE 2018, 13, e0197461. [Google Scholar] [CrossRef]
- Yurgelun-Todd, D. Emotional and cognitive changes during adolescence. Curr. Opin. Neurobiol. 2007, 17, 251–257. [Google Scholar] [CrossRef]
- Thorpe, H.H.A.; Hamidullah, S.; Jenkins, B.W.; Khokhar, J.Y. Adolescent neurodevelopment and substance use: Receptor expression and behavioral consequences. Pharmacol. Ther. 2020, 206, 107431. [Google Scholar] [CrossRef]
- Spear, L.P.; Varlinskaya, E.I. Adolescence. Alcohol sensitivity, tolerance, and intake. Recent. Dev. Alcohol. 2005, 17, 143–159. [Google Scholar]
- Teague, C.D.; Nestler, E.J. Teenage drinking and adult neuropsychiatric disorders: An epigenetic connection. Sci. Adv. 2022, 8, eabq5934. [Google Scholar] [CrossRef]
- Boschen, K.E.; McKeown, S.E.; Roth, T.L.; Klintsova, A.Y. Impact of exercise and a complex environment on hippocampal dendritic morphology, Bdnf gene expression, and DNA methylation in male rat pups neonatally exposed to alcohol. Dev. Neurobiol. 2017, 77, 708–725. [Google Scholar] [CrossRef] [PubMed]
- Sakharkar, A.J.; Kyzar, E.J.; Gavin, D.P.; Zhang, H.; Chen, Y.; Krishnan, H.R.; Grayson, D.R.; Pandey, S.C. Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology 2019, 157, 107679. [Google Scholar] [CrossRef]
- Timothy, A.; Benegal, V.; Shankarappa, B.; Saxena, S.; Jain, S.; Purushottam, M. Influence of early adversity on cortisol reactivity, SLC6A4 methylation and externalizing behavior in children of alcoholics. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 94, 109649. [Google Scholar] [CrossRef]
- Kyzar, E.J.; Bohnsack, J.P.; Zhang, H.; Pandey, S.C. MicroRNA-137 Drives Epigenetic Reprogramming in the Adult Amygdala and Behavioral Changes after Adolescent Alcohol Exposure. eNeuro 2019, 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Vetreno, R.P.; Bohnsack, J.P.; Kusumo, H.; Liu, W.; Pandey, S.C.; Crews, F.T. Neuroimmune and epigenetic involvement in adolescent binge ethanol-induced loss of basal forebrain cholinergic neurons: Restoration with voluntary exercise. Addict. Biol. 2020, 25, e12731. [Google Scholar] [CrossRef]
- Asimes, A.; Torcaso, A.; Pinceti, E.; Kim, C.K.; Zeleznik-Le, N.J.; Pak, T.R. Adolescent binge-pattern alcohol exposure alters genome-wide DNA methylation patterns in the hypothalamus of alcohol-naïve male offspring. Alcohol 2017, 60, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, Y.; Wu, H.; Song, Y.; Shivalila, C.S.; Markoulaki, S.; Jaenisch, R. Parent-of-Origin DNA Methylation Dynamics during Mouse Development. Cell. Rep. 2016, 16, 3167–3180. [Google Scholar] [CrossRef] [PubMed]
- Brocato, E.; Wolstenholme, J.T. Neuroepigenetic consequences of adolescent ethanol exposure. Int. Rev. Neurobiol. 2021, 160, 45–84. [Google Scholar]
- Boecker-Schlier, R.; Holz, N.E.; Hohm, E.; Zohsel, K.; Blomeyer, D.; Buchmann, A.F.; Baumeister, S.; Wolf, I.; Esser, G.; Schmidt, M.H.; et al. Association between pubertal stage at first drink and neural reward processing in early adulthood. Addict. Biol. 2017, 22, 1402–1415. [Google Scholar] [CrossRef]
- Islam, M.M. Exploring the relationship between age at first drink, low-risk drinking knowledge and drinks counting: Six rounds of a country-wide survey in Australia. Public Health 2020, 179, 160–168. [Google Scholar] [CrossRef]
- Veerbeek, M.A.; Ten Have, M.; van Dorsselaer, S.A.; Oude Voshaar, R.C.; Rhebergen, D.; Willemse, B.M. Differences in alcohol use between younger and older people: Results from a general population study. Drug. Alcohol. Depend. 2019, 202, 18–23. [Google Scholar] [CrossRef]
- Bray, B.C.; Dziak, J.J.; Lanza, S.T. Age trends in alcohol use behavior patterns among U.S. adults ages 18–65. Drug. Alcohol. Depend. 2019, 205, 107689. [Google Scholar] [CrossRef]
- Vore, A.S.; Doremus-Fitzwater, T.; Gano, A.; Deak, T. Adolescent Ethanol Exposure Leads to Stimulus-Specific Changes in Cytokine Reactivity and Hypothalamic-Pituitary-Adrenal Axis Sensitivity in Adulthood. Front. Behav. Neurosci. 2017, 11, 78. [Google Scholar] [CrossRef]
- Su, J.; Kuo, S.I.; Aliev, F.; Chan, G.; Edenberg, H.J.; Kamarajan, C.; McCutcheon, V.V.; Meyers, J.L.; Schuckit, M.; Tischfield, J.; et al. The associations between polygenic risk, sensation seeking, social support, and alcohol use in adulthood. J. Abnorm. Psychol. 2021, 130, 525–536. [Google Scholar] [CrossRef]
- Jimenez Chavez, C.L.; Van Doren, E.; Matalon, J.; Ogele, N.; Kharwa, A.; Madory, L.; Kazerani, I.; Herbert, J.; Torres-Gonzalez, J.; Rivera, E.; et al. Alcohol-Drinking Under Limited-Access Procedures During Mature Adulthood Accelerates the Onset of Cognitive Impairment in Mice. Front. Behav. Neurosci. 2022, 16, 732375. [Google Scholar] [CrossRef]
- Dugué, P.A.; Wilson, R.; Lehne, B.; Jayasekara, H.; Wang, X.; Jung, C.H.; Joo, J.E.; Makalic, E.; Schmidt, D.F.; Baglietto, L.; et al. Alcohol consumption is associated with widespread changes in blood DNA methylation: Analysis of cross-sectional and longitudinal data. Addict. Biol. 2021, 26, e12855. [Google Scholar] [CrossRef]
- Gatta, E.; Grayson, D.R.; Auta, J.; Saudagar, V.; Dong, E.; Chen, Y.; Krishnan, H.R.; Drnevich, J.; Pandey, S.C.; Guidotti, A. Genome-wide methylation in alcohol use disorder subjects: Implications for an epigenetic regulation of the cortico-limbic glucocorticoid receptors (NR3C1). Mol. Psychiatry 2021, 26, 1029–1041. [Google Scholar] [CrossRef]
- Wang, F.; Xu, H.; Zhao, H.; Gelernter, J.; Zhang, H. DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci. Rep. 2016, 6, 19430. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Montalvo-Ortiz, J.L.; Zhang, X.; Southwick, S.M.; Krystal, J.H.; Pietrzak, R.H.; Gelernter, J. Epigenome-Wide DNA Methylation Association Analysis Identified Novel Loci in Peripheral Cells for Alcohol Consumption Among European American Male Veterans. Alcohol. Clin. Exp. Res. 2019, 43, 2111–2121. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, B.; Nymberg, C.; Vuoksimaa, E.; Lourdusamy, A.; Wong, C.P.; Carvalho, F.M.; Jia, T.; Cattrell, A.; Macare, C.; Banaschewski, T.; et al. Association of Protein Phosphatase PPM1G With Alcohol Use Disorder and Brain Activity During Behavioral Control in a Genome-Wide Methylation Analysis. Am. J. Psychiatry 2015, 172, 543–552. [Google Scholar] [CrossRef]
- Grace, A.A. The tonic/phasic model of dopamine system regulation and its implications for understanding alcohol and psychostimulant craving. Addiction 2000, 95 (Suppl. S2), S119–S128. [Google Scholar] [CrossRef]
- Nieratschker, V.; Grosshans, M.; Frank, J.; Strohmaier, J.; von der Goltz, C.; El-Maarri, O.; Witt, S.H.; Cichon, S.; Nothen, M.M.; Kiefer, F.; et al. Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addict. Biol. 2014, 19, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Barbier, E.; Tapocik, J.D.; Juergens, N.; Pitcairn, C.; Borich, A.; Schank, J.R.; Sun, H.; Schuebel, K.; Zhou, Z.; Yuan, Q.; et al. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 2015, 35, 6153–6164. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.Z.; Sun, M.Z.; Wang, R.Z.; Li, C.Y.; Huang, Y.X.; Huang, Q.J.; Qiao, X.M. DNA methylation in the medial prefrontal cortex regulates alcohol-related behavior in rats. Yi Chuan 2020, 42, 112–125. [Google Scholar] [PubMed]
- Niinep, K.; Anier, K.; Eteläinen, T.; Piepponen, P.; Kalda, A. Repeated Ethanol Exposure Alters DNA Methylation Status and Dynorphin/Kappa-Opioid Receptor Expression in Nucleus Accumbens of Alcohol-Preferring AA Rats. Front. Genet. 2021, 12, 750142. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, K.K. Alcoholic liver disease and methionine metabolism. Semin. Liver Dis. 2009, 29, 155–165. [Google Scholar] [CrossRef]
- Auta, J.; Zhang, H.; Pandey, S.C.; Guidotti, A. Chronic Alcohol Exposure Differentially Alters One-Carbon Metabolism in Rat Liver and Brain. Alcohol. Clin. Exp. Res. 2017, 41, 1105–1111. [Google Scholar] [CrossRef]
- Stragier, E.; Martin, V.; Davenas, E.; Poilbout, C.; Mongeau, R.; Corradetti, R.; Lanfumey, L. Brain plasticity and cognitive functions after ethanol consumption in C57BL/6J mice. Transl. Psychiatry 2015, 5, e696. [Google Scholar] [CrossRef]
- Yang, M.; Barrios, J.; Yan, J.; Zhao, W.; Yuan, S.; Dong, E.; Ai, X. Causal roles of stress kinase JNK2 in DNA methylation and binge alcohol withdrawal-evoked behavioral deficits. Pharm. Res. 2021, 164, 105375. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, S.; Le Roy, T.; Furgiuele, S.; Coste, V.; Bindels, L.B.; Leyrolle, Q.; Neyrinck, A.M.; Quoilin, C.; Amadieu, C.; Petit, G.; et al. Gut Microbiota-Induced Changes in β-Hydroxybutyrate Metabolism Are Linked to Altered Sociability and Depression in Alcohol Use Disorder. Cell. Rep. 2020, 33, 108238. [Google Scholar] [CrossRef]
- de Assis Pinheiro, J.; Freitas, F.V.; Borçoi, A.R.; Mendes, S.O.; Conti, C.L.; Arpini, J.K.; Dos Santos Vieira, T.; de Souza, R.A.; Dos Santos, D.P.; Barbosa, W.M.; et al. Alcohol consumption, depression, overweight and cortisol levels as determining factors for NR3C1 gene methylation. Sci. Rep. 2021, 11, 6768. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.; Halby, L.; Arimondo, P.B. DNA Methyltransferase Inhibitors: Development and Applications. Adv. Exp. Med. Biol. 2016, 945, 431–473. [Google Scholar]
- Hernández, J.A.; López-Sánchez, R.C.; Rendón-Ramírez, A. Lipids and Oxidative Stress Associated with Ethanol-Induced Neurological Damage. Oxidative Med. Cell. Longev. 2016, 2016, 1543809. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Garcia-Milian, R.; Golla, J.P.; Charkoftaki, G.; Lam, T.T.; Thompson, D.C.; Vasiliou, V. Proteomic profiling reveals an association between ALDH and oxidative phosphorylation and DNA damage repair pathways in human colon adenocarcinoma stem cells. Chem. Biol. Interact. 2022, 368, 110175. [Google Scholar] [CrossRef] [PubMed]
- Rulten, S.L.; Hodder, E.; Ripley, T.L.; Stephens, D.N.; Mayne, L.V. Alcohol induces DNA damage and the Fanconi anemia D2 protein implicating FANCD2 in the DNA damage response pathways in brain. Alcohol. Clin. Exp. Res. 2008, 32, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Nepal, M.; Che, R.; Ma, C.; Zhang, J.; Fei, P. FANCD2 and DNA Damage. Int. J. Mol. Sci. 2017, 18, 1804. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Han, M.; Matsumoto, A.; Wang, Y.; Thompson, D.C.; Vasiliou, V. Glutathione and Transsulfuration in Alcohol-Associated Tissue Injury and Carcinogenesis. Adv. Exp. Med. Biol. 2018, 1032, 37–53. [Google Scholar]
- Mc Auley, M.T.; Mooney, K.M.; Salcedo-Sora, J.E. Computational modelling folate metabolism and DNA methylation: Implications for understanding health and ageing. Brief. Bioinform. 2018, 19, 303–317. [Google Scholar] [CrossRef]
- Zhao, H.; Guo, P.; Zuo, Y.; Wang, Y.; Zhao, H.; Lan, T.; Xue, M.; Zhang, H.; Liang, H. Folic acid intervention changes liver Foxp3 methylation and ameliorates the damage caused by Th17/Treg imbalance after long-term alcohol exposure. Food Funct. 2022, 13, 5262–5274. [Google Scholar] [CrossRef]
- Yiu, T.T.; Li, W. Pediatric cancer epigenome and the influence of folate. Epigenomics 2015, 7, 961–973. [Google Scholar] [CrossRef]
- Kirkbride, J.B.; Susser, E.; Kundakovic, M.; Kresovich, J.K.; Davey Smith, G.; Relton, C.L. Prenatal nutrition, epigenetics and schizophrenia risk: Can we test causal effects? Epigenomics 2012, 4, 303–315. [Google Scholar] [CrossRef]
- Sogut, I.; Uysal, O.; Oglakci, A.; Yucel, F.; Kartkaya, K.; Kanbak, G. Prenatal alcohol-induced neuroapoptosis in rat brain cerebral cortex: Protective effect of folic acid and betaine. Childs Nerv. Syst. 2017, 33, 407–417. [Google Scholar] [CrossRef]
- An, Y.; Feng, L.; Zhang, X.; Wang, Y.; Wang, Y.; Tao, L.; Qin, Z.; Xiao, R. Dietary intakes and biomarker patterns of folate, vitamin B(6), and vitamin B(12) can be associated with cognitive impairment by hypermethylation of redox-related genes NUDT15 and TXNRD1. Clin. Epigenet. 2019, 11, 139. [Google Scholar] [CrossRef]
- Rasmussen, E.M.K.; Seier, K.L.; Pedersen, I.K.; Kreibich, C.; Amdam, G.V.; Münch, D.; Dahl, J.A. Screening bioactive food compounds in honey bees suggests curcumin blocks alcohol-induced damage to longevity and DNA methylation. Sci. Rep. 2021, 11, 19156. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.K.; Chava, S.; Perumal, S.K.; Paal, M.C.; Rasineni, K.; Ganesan, M.; Donohue, T.M., Jr.; Osna, N.A.; Kharbanda, K.K. Acute ethanol-induced liver injury is prevented by betaine administration. Front. Physiol. 2022, 13, 940148. [Google Scholar] [CrossRef]
- Lai, G.; Guo, Y.; Chen, D.; Tang, X.; Shuai, O.; Yong, T.; Wang, D.; Xiao, C.; Zhou, G.; Xie, Y.; et al. Alcohol Extracts From Ganoderma lucidum Delay the Progress of Alzheimer’s Disease by Regulating DNA Methylation in Rodents. Front. Pharm. 2019, 10, 272. [Google Scholar] [CrossRef]
- Zhao, C.; Fan, J.; Liu, Y.; Guo, W.; Cao, H.; Xiao, J.; Wang, Y.; Liu, B. Hepatoprotective activity of Ganoderma lucidum triterpenoids in alcohol-induced liver injury in mice, an iTRAQ-based proteomic analysis. Food Chem. 2019, 271, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Buenz, E.J.; Weaver, J.G.; Bauer, B.A.; Chalpin, S.D.; Badley, A.D. Cordyceps sinensis extracts do not prevent Fas-receptor and hydrogen peroxide-induced T-cell apoptosis. J. Ethnopharmacol. 2004, 90, 57–62. [Google Scholar] [CrossRef]
- Lyko, F.; Brown, R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J. Natl. Cancer Inst. 2005, 97, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Yin, F.; Ji, Y.; Li, Y.; Yan, P.; Lai, J. 5-Aza-2’-deoxycytidine in the medial prefrontal cortex regulates alcohol-related behavior and Ntf3-TrkC expression in rats. PLoS ONE 2017, 12, e0179469. [Google Scholar] [CrossRef]
- Ji, C.; Nagaoka, K.; Zou, J.; Casulli, S.; Lu, S.; Cao, K.Y.; Zhang, H.; Iwagami, Y.; Carlson, R.I.; Brooks, K.; et al. Chronic ethanol-mediated hepatocyte apoptosis links to decreased TET1 and 5-hydroxymethylcytosine formation. FASEB J. 2019, 33, 1824–1835. [Google Scholar] [CrossRef]
- Tammen, S.A.; Park, J.E.; Shin, P.K.; Friso, S.; Chung, J.; Choi, S.W. Iron Supplementation Reverses the Reduction of Hydroxymethylcytosine in Hepatic DNA Associated With Chronic Alcohol Consumption in Rats. J. Cancer Prev. 2016, 21, 264–270. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Butter, R.; Spijker, R.; Medema, J.P.; van Laarhoven, H.W.M. Clinical and biological effects of demethylating agents on solid tumours—A systematic review. Cancer Treat. Rev. 2017, 54, 10–23. [Google Scholar] [CrossRef]
- Warnault, V.; Darcq, E.; Levine, A.; Barak, S.; Ron, D. Chromatin remodeling--a novel strategy to control excessive alcohol drinking. Transl. Psychiatry 2013, 3, e231. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozcan, E.; Wert, S.L.; Lim, P.H.; Ferreira, A.; Redei, E.E. Hippocampus-dependent memory and allele-specific gene expression in adult offspring of alcohol-consuming dams after neonatal treatment with thyroxin or metformin. Mol. Psychiatry 2018, 23, 1643–1651. [Google Scholar] [CrossRef]
- Gangisetty, O.; Wynne, O.; Jabbar, S.; Nasello, C.; Sarkar, D.K. Fetal Alcohol Exposure Reduces Dopamine Receptor D2 and Increases Pituitary Weight and Prolactin Production via Epigenetic Mechanisms. PLoS ONE 2015, 10, e0140699. [Google Scholar] [CrossRef] [PubMed]
- Witkiewitz, K.; Litten, R.Z.; Leggio, L. Advances in the science and treatment of alcohol use disorder. Sci. Adv. 2019, 5, eaax4043. [Google Scholar] [CrossRef] [PubMed]
| Classification | Drugs | Mechanisms | Status of Clinical Use | Refs. |
|---|---|---|---|---|
| Cytosine nucleoside derivatives | 5-aza RX-3117 | Incorporate DNA and participate in DNA replication | clinical application preclinical study | [34,35] |
| Deoxyribose analogs | 5-aza-dc | incorporate DNA and participate in DNA replication | clinical application | [37] |
| Benzoamide | Zebularine RG108 SGI-1027 | bind non-covalently to the active sites of DNMTs | preclinical study preclinical study preclinical study | [38,39,40] |
| Aminobenzoic acid derivatives | Procainamide Procaine | bind to the CpG sites | Clinical phase II Clinical phase II | [42,43] |
| Antisense oligonucleotide Polyphenols | MG-98 miR29a EGCG curcumin γ-oryzanol | act on DNMT1 mRNA Bind the active site of DNMT enzyme, bind to DNMT sulfhydryl | Clinical phase II preclinical study preclinical study preclinical study preclinical study | [44,45,47,48,49] |
| Drugs | Mechanism | Function | Status of Clinical Use | Refs |
|---|---|---|---|---|
| ALDH FANCD2 Glutathione Folic acid Vitamin B6/B12 | Reduce DNA damage and maintain DNA activity Reduce DNA damage and maintain DNA activity Neutralize free radicals, transport cysteine, and REDOX cells Methyl donor Methyl donor | Improve hematopoietic function, protect the liver Improve hematopoietic function, protect the liver Promote cell regeneration Improve oxidative damage and cognitive impairment Improve oxidative damage and cognitive impairment | preclinical study preclinical study clinical application clinical application clinical application | [122,123,124,125,127,131] |
| Curcumin betaine Lucidum Cordyceps sinensis | Increase methylation levels provide methyl groups to make S-adenosine Increase the expression of histone H3, DNMT3A and DNMT3B. Promote DNA methylation reprogramming | Resist cell oxidative damage and apoptosis Repairing damage to embryonic development Improve oxidative damage and cognitive impairment Improve brain atrophy and learning and memory function | clinical application clinical application clinical application clinical application | [132,133,134,135,136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Wang, H.; Yan, A.; Yin, F.; Qiao, X. DNA Methylation in Alcohol Use Disorder. Int. J. Mol. Sci. 2023, 24, 10130. https://doi.org/10.3390/ijms241210130
Zheng Q, Wang H, Yan A, Yin F, Qiao X. DNA Methylation in Alcohol Use Disorder. International Journal of Molecular Sciences. 2023; 24(12):10130. https://doi.org/10.3390/ijms241210130
Chicago/Turabian StyleZheng, Qingmeng, Heng Wang, An Yan, Fangyuan Yin, and Xiaomeng Qiao. 2023. "DNA Methylation in Alcohol Use Disorder" International Journal of Molecular Sciences 24, no. 12: 10130. https://doi.org/10.3390/ijms241210130
APA StyleZheng, Q., Wang, H., Yan, A., Yin, F., & Qiao, X. (2023). DNA Methylation in Alcohol Use Disorder. International Journal of Molecular Sciences, 24(12), 10130. https://doi.org/10.3390/ijms241210130





