Low-Abundance Protein Enrichment for Medical Applications: The Involvement of Combinatorial Peptide Library Technique
Abstract
:1. Introduction
2. Current Methods for Low-Abundance Protein Enrichment
3. Combinatorial Peptide Ligand Library Technology
4. Identification of Early-Stage Biomarkers of Human Diseases
5. Detection of Protein Impurity Traces from Recombinant Biopharmaceuticals
6. Discovery of Low-Concentration Allergens
7. Other Medical Involvements
8. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Alessandro, A.; Dzieciatkowska, M.; Nemkov, T.; Hansen, K.C. Red blood cell proteomics update: Is there more to discover? Blood Transfus. 2017, 15, 182–187. [Google Scholar] [PubMed]
- Bracher, A.; Whitney, S.M.; Hartl, F.U.; Hayer-Hartl, M. Biogenesis and metabolic maintenance of RuBisCO. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Wang, X.; Li, S.; Zhou, X.; Yue, L.; Fan, J.; Hao, D. Polyethylene glycol fractionation improved detection of low-abundant proteins by two-dimensional electrophoresis analysis of plant proteome. Phytochemistry 2006, 67, 2341–2348. [Google Scholar] [CrossRef]
- Pieper, R.; Su, Q.; Gatlin, C.L.; Huang, S.-T.; Anderson, N.L.; Steiner, S. Multi-component immunoaffinity subtraction chromatography: An innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 2003, 3, 422–432. [Google Scholar] [CrossRef]
- Levin, Y.; Schwarz, E.; Wang, L.; Leweke, F.M.; Bahn, S. Label-free LC-MS/MS quantitative proteomics for large-scale biomarker discovery in complex samples. J. Sep. Sci. 2007, 30, 2198–2203. [Google Scholar] [CrossRef]
- Gao, M.; Deng, C.; Yu, W.; Zhang, Y.; Yang, P.; Zhang, X. Large scale depletion of the high-abundance proteins and analysis of middle- and low-abundance proteins in human liver proteome by multidimensional liquid chromatography. Proteomics 2008, 8, 939–947. [Google Scholar] [CrossRef]
- Shen, Y.; Kim, J.; Strittmatter, E.F.; Jacobs, J.M.; Camp, D.G., II; Fang, R.; Tolié, N.; Moore, R.J.; Smith, R.D. Characterization of the human blood plasma proteome. Proteomics 2005, 5, 4034–4045. [Google Scholar] [CrossRef]
- Huang, B.Y.; Yang, C.K.; Liu, C.P.; Liu, C.Y. Stationary phases for the enrichment of glycoproteins and glycopeptides. Electrophoresis 2014, 35, 2091–2107. [Google Scholar]
- Low, T.Y.; Mohtar, M.A.; Lee, P.Y.; Omar, N.; Zhou, N.; Ye, M. Widening the bottleneck of phosphoproteomics: Evolving strategies for phosphopeptide enrichment. Mass Spectrom. Rev. 2021, 40, 309–333. [Google Scholar] [CrossRef]
- Llop, E.; Peracaula, R. Lectin affinity chromatography for the discovery of novel cancer glycobiomarkers: A case study with PSA glycoforms and prostate cancer. Methods Mol. Biol. 2022, 2370, 301–313. [Google Scholar]
- Nauom, S.; da Silva Neto, B.R.; Ribeiro, M.S.; Pedersoli, W.R.; Ulhoa, C.J.; Silva, R.N.; Monteiro, V.N. Biochemical and molecular study of Trichoderma harzianum enriched secretome protein profiles using lectin affinity chromatography. Appl. Biochem. Biotechnol. 2019, 187, 1–13. [Google Scholar]
- Chen, J.; Li, X.; Feng, M.; Luo, K.; Yang, J.; Zhang, B. Novel boronate material affords efficient enrichment of glycopeptides by synergized hydrophilic and affinity interactions. Anal. Bioanal. Chem. 2017, 409, 519–528. [Google Scholar]
- Pinto, G.; Caira, S.; Cuollo, M.; Lilla, S.; Fierro, O.; Addeo, F. Hydroxyapatite as a concentrating probe for phosphoproteomic analyses. J. Chromatogr. B 2010, 878, 2669–2678. [Google Scholar]
- Lin, H.; Deng, C. Development of immobilized Sn4+ affinity chromatography material for highly selective enrichment of phosphopeptides. Proteomics 2016, 16, 2733–2741. [Google Scholar]
- Wang, M.C.; Lee, Y.H.; Liao, P.C. Optimization of titanium dioxide and immunoaffinity-based enrichment procedures for tyrosine phosphopeptide using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 1343–1356. [Google Scholar] [PubMed]
- Di Girolamo, F.; D’Amato, A.; Lante, I.; Signore, F.; Muraca, M.; Putignani, L. Farm animal serum proteomics and impact on human health. Int. J. Mol. Sci. 2014, 15, 15396–15411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subba, P.; Narayana Kotimoole, C.; Prasad, T.S.K. Plant Proteome Databases and Bioinformatic Tools: An Expert Review and Comparative Insights. OMICS 2019, 23, 190–206. [Google Scholar] [PubMed]
- Righetti, P.G.; Fasoli, E.; D’Amato, A.; Boschetti, E. Making progress in plant proteomics for improved food safety. In Comprehensive Analytical Chemistry; Elsevier: Waltham, MA, USA, 2014; Volume 64, pp. 131–155. [Google Scholar]
- Johnson, R.O.; Greer, T.; Cejkov, M.; Zheng, X.; Li, N. Combination of FAIMS, Protein A depletion, and native digest conditions enables deep proteomic profiling of host cell proteins in monoclonal antibodies. Anal. Chem. 2020, 92, 10478–10484. [Google Scholar]
- Zhang, S.; Xiao, H.; Li, N. Degradation of polysorbate 20 by sialate O-acetylesterase in monoclonal antibody formulations. J. Pharm. Sci. 2021, 110, 3866–3873. [Google Scholar]
- Chen, I.H.; Xiao, H.; Daly, T.; Li, N. Improved host cell protein analysis in monoclonal antibody products through molecular weight cutoff enrichment. Anal. Chem. 2020, 92, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Abdubek, P.; Zhang, S.; Xiao, H.; Li, N. Analysis of host cell proteins in monoclonal antibody therapeutics through size exclusion chromatography. Pharm. Res. 2022, 39, 3029–3037. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, E.; Righetti, P.G. Host cell proteins enrichment for an in-depth analytical assessment of biopharmaceuticals quality. 2023; submitted for publication. [Google Scholar]
- Boschetti, E.; Righetti, P.G. Low-Abundance Protein Discovery: State of the Art and Protocols; Elsevier: Waltham, MA, USA, 2013. [Google Scholar]
- Boschetti, E.; Hernandez-Castellano, L.E.; Righetti, P.G. Progress in farm animal proteomics: The contribution of combinatorial peptide ligand libraries. J. Proteom. 2019, 197, 1–13. [Google Scholar]
- Righetti, P.G.; Boschetti, E. Low-abundance plant protein enrichment with peptide libraries to enlarge proteome coverage and related applications. Plant Sci. 2020, 290, 110302. [Google Scholar]
- Meng, R.; Gormley, M.; Bhat, V.B.; Rosenberg, A.; Quong, A.A. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J. Proteom. 2011, 75, 366–374. [Google Scholar] [CrossRef]
- Fonslow, B.R.; Carvalho, P.C.; Academia, K.; Freeby, S.; Xu, T.; Nakorchevsky, A.; Paulus, A.; Yates, J.R. Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J. Proteome Res. 2011, 10, 3690–3700. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.R.; Phelan, M.M.; Rubio-Martinez, L.M.; Fitzgerald, M.M.; Jones, S.W.; Clegg, P.D.; Peffers, M.J. Optimization of synovial fluid collection and processing for nmr metabolomics and LC-MS/MS proteomics. J. Proteome Res. 2020, 19, 2585–2597. [Google Scholar] [CrossRef]
- Beseme, O.; Fertin, M.; Drobecq, H.; Amouyel, P.; Pinet, F. Combinatorial peptide ligand library plasma treatment: Advantages for accessing low-abundance proteins. Electrophoresis 2010, 31, 2697–2704. [Google Scholar] [CrossRef]
- Fahiminiya, S.; Labas, V.; Roche, S.; Dacheux, J.-L.; Gérard, N. Proteomic analysis of mare follicular fluid during late follicle development. Proteome Sci. (Short Commun.) 2011, 9, 54–56. [Google Scholar]
- Pisanu, S.; Biosa, G.; Carcangiu, L.; Uzzau, S.; Pagnozzi, D. Comparative evaluation of seven commercial products for human serum enrichment/depletion by shotgun proteomics. Talanta 2018, 185, 213–220. [Google Scholar]
- Palstrøm, N.B.; Rasmussen, L.M.; Beck, H.C. Affinity capture enrichment versus affinity depletion: A comparison of strategies for increasing coverage of low-abundant human plasma proteins. Int. J. Mol. Sci. 2020, 21, 5903. [Google Scholar]
- Sennels, L.; Salek, M.; Lomas, L.; Boschetti, E.; Righetti, P.G.; Rappsilber, J. Proteomic analysis of human blood serum using peptide library beads. J. Proteome Res. 2007, 6, 4055–4062. [Google Scholar] [CrossRef]
- Roux-Dalvai, F.; Gonzalez de Peredo, A.; Simó, C.; Guerrier, L.; Bouyssie, D.; Zanella, A.; Citterio, A.; Burlet-Schiltz, O.; Boschetti, E.; Righetti, P.G.; et al. Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced spectrometry. Mol. Cell. Proteom. 2008, 7, 2254–2269. [Google Scholar] [CrossRef] [Green Version]
- D’Amato, A.; Cereda, A.; Bachi, A.; Pierce, J.C.; Righetti, P.G. In-depth exploration of the hemolymph of Limulus polyphemus via combinatorial peptide ligand libraries. J. Proteome Res. 2010, 9, 3260–3269. [Google Scholar] [CrossRef]
- Qu, Z.; Leung, T.C.N.; Nong, W.; Yip, H.Y.; Lee, I.H.T.; Cheung, S.G.; Ming, N.S.; So, W.L.; Bendena, W.G.; Tobe, S.S.; et al. Hemolymph proteomics and gut microbiota of horseshoe crabs Tachypleus tridentatus and Carcinoscorpius rotundicauda. Front. Mar. Sci. 2020, 7, 579706. [Google Scholar]
- Rodríguez, M.; Ajona, D.; Seijo, L.M.; Sanz, J.; Valencia, K.; Corral, J.; Mesa-Guzmán, M.; Pío, R.; Calvo, A.; Lozano, M.D.; et al. Molecular biomarkers in early stage lung cancer. Transl. Lung Cancer Res. 2021, 10, 1165–1185. [Google Scholar]
- Veenstra, T.D. Global and targeted quantitative proteomics for biomarker discovery. J. Chromatogr. B. 2007, 847, 3–11. [Google Scholar]
- Liu, N.Q.; Braakman, R.B.; Stingl, C.; Luider, T.M.; Martens, J.W.; Foekens JAUmar, A. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue, J. Mammary Gland Biol. Neoplasia 2012, 17, 155–164. [Google Scholar]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar]
- Sequeiros, T.; Rigau, M.; Chiva, C.; Montes, M.; Garcia-Grau, I.; Garcia, M.; Diaz, S.; Celma, A.; Bijnsdorp, I.; Campos, A.; et al. Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer. Oncotarget 2017, 8, 4960–4976. [Google Scholar] [PubMed] [Green Version]
- Salciccia, S.; Capriotti, A.L.; Laganà, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From current knowledge to the role of metabolomics and exosomes. Int. J. Mol. Sci. 2021, 22, 4367. [Google Scholar] [PubMed]
- Zhao, Z.; Liu, W. Pancreatic cancer: A review of risk factors, diagnosis, and treatment. Technol. Cancer Res. Treat. 2020, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Weng, Y.; Sui, Z.; Wu, Y.; Meng, X.; Wu, M.; Jin, H.; Tan, X.; Zhang, L.; Zhang, Y. Quantitative secretomic analysis of pancreatic cancer cells in serum-containing conditioned medium. Sci. Rep. 2016, 6, 37606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chidambaranathan-Reghupaty, S.; Fisher, P.B.; Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021, 149, 1–61. [Google Scholar]
- Mustafa, G.M.; Larry, D.; Petersen, J.R.; Elferink, C.J. Targeted proteomics for biomarker discovery and validation of hepatocellular carcinoma in hepatitis C infected patients. World J. Hepatol. 2015, 7, 1312–1324. [Google Scholar]
- Mustafa, M.G.; Petersen, J.R.; Ju, H.; Cicalese, L.; Snyder, N.; Haidacher, S.J.; Denner, L.; Elferink, C. Biomarker discovery for early detection of hepatocellular carcinoma in hepatitis C-infected patients. Mol. Cell. Proteom. 2013, 12, 3640–3652. [Google Scholar]
- Kimura, Y.; Nakai, Y.; Shin, J.; Hara, M.; Takeda, Y.; Kubo, S.; Jeremiah, S.S.; Ino, Y.; Akiyama, T.; Moriyama, K.; et al. Identification of serum prognostic biomarkers of severe COVID-19 using a quantitative proteomic approach. Sci. Rep. 2021, 11, 20638. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, Y.; Sun, X.; Li, Y.; Huang, C.; Deng, H.; Li, Z. Identification of potential serum biomarkers for rheumatoid arthritis by high-resolution quantitative proteomic analysis. Inflammation 2014, 37, 1459–1467. [Google Scholar]
- Zhao, X.; Chen, Y.; Wen, W.; Cheng, Y.; Li, R.; Liu, X.; Li, Y.; Jia, R.; Deng, H.; Li, Z.; et al. Identification of lipopolysaccharide-binding protein as a novel citrullinated autoantigen in rheumatoid arthritis. Rheumatol. Autoimmun. 2022, 2, 5–14. [Google Scholar]
- Malaud, E.; Piquer, D.; Merle, D.; Molina, F.; Guerrier, L.; Boschetti, E.; Saussine, M.; Marty-Ané, C.; Albat, B.; Fareh, J. Carotid atherosclerotic plaques: Proteomics study after a low-abundance protein enrichment step. Electrophoresis 2012, 33, 470–482. [Google Scholar]
- Eslava-Alcon, S.; Extremera-García, M.J.; González-Rovira, A.; Rosal-Vela, A.; Rojas-Torres, M.; Beltran-Camacho, L.; Sanchez-Gomar, I. Molecular signatures of atherosclerotic plaques: An up-dated panel of protein related markers. J Proteom. 2020, 221, 103757. [Google Scholar]
- Ulme, C.H.; Peffers, M.J.; Harrington, G.M.B.; Wilson, E.; Perry, J.; Roberts, S.; Gallacher, P.; Jermin, P.; Wright, K.T. Identification of candidate synovial fluid biomarkers for the prediction of patient outcome after microfracture or osteotomy. Am. J. Sports Med. 2021, 49, 1512–1523. [Google Scholar]
- Sivadasan, P.; Gupta, M.K.; Sathe, G.J.; Balakrishnan, L.; Palit, P.; Gowda, H.; Suresh, A.; Kuriakose, M.A.; Sirdeshmukh, R. Data from human salivary proteome—A resource of potential biomarkers for oral cancer. Data Brief. 2015, 4, 374–378. [Google Scholar]
- Celsi, F.; Monasta, L.; Arrigoni, G.; Battisti, I.; Licastro, D.; Aloisio, M.; Di Lorenzo, G.; Romano, F.; Ricci, G.; Ura, B. Gel-based proteomic identification of suprabasin as a potential new candidate biomarker in endometrial cancer. Int. J. Mol. Sci. 2022, 23, 2076. [Google Scholar]
- Jankovska, E.; Svitek, M.; Holada, K.; Petrak, J. Affinity depletion versus relative protein enrichment: A side-by-side comparison of two major strategies for increasing human cerebrospinal fluid proteome coverage. Clin. Proteom. 2019, 16, 9. [Google Scholar]
- Champion, K.; Madden, H.; Dougherty, J.; Shacter, E. Defining your product profile and maintaining control over it, part 2. Bioprocess Int. 2005, 9, 52–57. [Google Scholar]
- Zhu-Shimoni, J.; Yu, C.; Nishihara, J.; Wong, R.M.; Gunawan, F.; Lin, M.; Krawitz, D.; Liu, P.; Sandoval, W.; Vanderlaan, M. Host cell protein testing by ELISAs and the use of orthogonal methods. Biotechnol. Bioeng. 2014, 111, 2367–2379. [Google Scholar]
- Soderquist, R.G.; Trumbo, M.; Hart, R.A.; Zhang, Q.; Flynn, G.C. Development of advanced host cell protein enrichment and detection strategies to enable process relevant spike challenge studies. Biotechnol. Prog. 2015, 31, 983–989. [Google Scholar] [CrossRef]
- Madsen, J.A.; Farutin, V.; Carbeau, T.; Wudyka, S.; Yin, Y.; Smith, S.; Anderson, J.; Capila, I. Toward the complete characterization of host cell proteins in biotherapeutics via affinity depletions, LC-MS/MS, and multivariate analysis. MAbs 2015, 7, 1128–1137. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.-H.; Xiao, H.; Li, N. Improved host cell protein analysis in monoclonal antibody products through ProteoMiner. Anal. Biochem. 2020, 610, 113972. [Google Scholar] [CrossRef]
- Fortis, F.; Guerrier, G.; Areces, L.; Antonioli, P.; Hayes, T.; Carrick, K.; Hammond, D.; Boschetti, E.; Righetti, P.G. A new approach for the detection and identification of protein impurities using combinatorial solid phase ligand libraries. J. Proteome Res. 2006, 5, 2577–2585. [Google Scholar] [CrossRef] [PubMed]
- Antonioli PFortis, F.; Guerrier, L.; Rinalducci, S.; Zolla, L.; Righetti, P.G.; Boschetti, E. Capturing and amplifying impurities from purified recombinant monoclonal antibodies via peptide library beads: A proteomic study. Proteomics 2007, 7, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Mörtstedt, H.; Makower, A.; Edlund, P.O.; Sjöberg, K.; Tjernberg, A. Improved identification of host cell proteins in a protein biopharmaceutical by LC-MS/MS using the ProteoMiner enrichment kit. J. Pharm. Biomed. Anal. 2020, 185, 113256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xiao, H.; Li, N. Ultrasensitive method for profiling host cell proteins by coupling limited digestion to ProteoMiner technology. Anal. Biochem. 2022, 657, 114901. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.; Adaniya, S.; Xiao, H.; Li, N. Ultrasensitive quantification method for understanding biologically relevant concentrations of host cell proteins in therapeutics. Anal. Chem. 2023, 95, 6002–6008. [Google Scholar] [CrossRef]
- Bredehorst, R.; David, K. What establishes a protein as an allergen. J. Chromatogr. B. 2001, 756, 33–40. [Google Scholar] [CrossRef]
- Bousquet, J.; Anto, J.M.; Bachert, C.; Bousquet, P.J.; Colombo, P.; Crameri, R.; Daëron, M.; Fokkens, W.; Leynaert, B.; Lahoz, C.; et al. Factors responsible for differences between asymptomatic subjects and patients presenting an IgE sensitization to allergens. Allergy 2006, 61, 671–680. [Google Scholar] [CrossRef]
- Yagami, T. Allergies to cross-reactive plant proteins. Latex-fruit syndrome is comparable with pollen-food allergy syndrome. Int. Arch. Allergy Immunol. 2002, 128, 271–279. [Google Scholar] [CrossRef]
- Ramachandran, B.; Yang, C.T.; Downs, M.L. Parallel reaction monitoring mass spectrometry method for detection of both casein and whey milk allergens from a baked food matrix. J. Proteome Res. 2020, 19, 2964–2976. [Google Scholar] [CrossRef] [PubMed]
- Torres-Arroyo, A.; Martínez-Aguilar, J.; Castillo-Villanueva, A.; Zárate-Mondragón, F.; Cervantes-Bustamante, R.; Patiño-López, G.; Medina-Contreras, O.; Espinosa-Padilla, S.E.; Valencia-Rojas, S.; Romero-Guzmán, L.; et al. Immunoproteomics of cow’s milk allergy in Mexican pediatric patients. J. Proteom. 2023, 273, 104809. [Google Scholar] [CrossRef] [PubMed]
- Alisi, C.; Afferni, C.; Iacovacci, P.; Barletta, B.; Tinghino, R.; Butteroni, C.; Puggioni, E.M.; Wilson, I.B.; Federico, R.; Schininà, M.E.; et al. Rapid isolation, characterization, and glycan analysis of Cup a 1, the major allergen of Arizona cypress (Cupressus arizonica) pollen. Allergy 2001, 56, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Stott, D.I. Immunoblotting, dot-blotting, and ELISPOT assays: Methods and applications. J. Immunoass. 2000, 21, 273–296. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, A.; Bachi, A.; Fasoli, E.; Boschetti, E.; Peltre GSénéchal, H.; Righetti, P.G. In-depth exploration of cow’s whey proteome via combinatorial peptide ligand libraries. J. Proteome Res. 2009, 8, 3925–3936. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, R.A.; Mazzeo, M.F.; Arena, S.; Renzone, G.; Scaloni, A. Mass spectrometry for the analysis of protein lactosylation in milk products. Food Res. Int. 2013, 54, 988–1000. [Google Scholar] [CrossRef]
- Martos, G.; López-Fandiño, R.; Molina, E. Immunoreactivity of hen egg allergens: Influence on in-vitro gastrointestinal digestion of the presence of other egg white proteins and of egg yolk. Food Chem. 2013, 136, 775–781. [Google Scholar] [CrossRef]
- Coscia, A.; Orrù, S.; Di Nicola, P.; Giuliani, F.; Varalda, A.; Peila, C.; Fabris, C.; Conti, A.; Bertino, E. Detection of cow’s milk proteins and minor components in human milk using proteomics techniques. J. Matern. Fetal. Neonatal. Med. 2012, 25 (Suppl. S4), 54–56. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Brochetto Braga, M.R.; Magalhães de Abreu, R.M.; Blank, S. Standard methods for Apis mellifera venom research. J. Apic. Res. 2020, 60, 1–31. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Lorenzo, J.M.; Gagaoua, M.; Franco, D. Current Trends in Proteomic Adv. Food Allerg. Anal. Biol. 2020, 9, 247. [Google Scholar]
- Zimmermann, J.; Hubel, P.; Pfannstiel, J.; Afzal, M.; Longin, C.F.H.; Hitzmann, H.; Götz, H.; Bischoff, S.C. Comprehensive proteome analysis of bread deciphering the allergenic potential of bread wheat, spelt and rye. J. Proteom. 2021, 247, 104318. [Google Scholar] [CrossRef]
- Nikolić, J.; Nešić, A.; Kull, S.; Schocker, F.; Jappe, U.; Gavrović-Jankulović, M. Employment of proteomic and immunological based methods for the identification of catalase as novel allergen from banana. J. Proteom. 2018, 175, 87–94. [Google Scholar] [CrossRef]
- Gomez-Cardona, E.E.; Heathcote, K.; Teran, L.M.; Righetti, P.G.; Boschetti, E.; D’Amato, A. Novel low-abundance allergens from mango via combinatorial peptide libraries: A proteomics study. Food Chem. 2018, 269, 652–660. [Google Scholar] [CrossRef]
- Charpin, D.; Pichot, C.; Belmonte, J.; Sutra, J.P.; Zidkova, J.; Chanez, P.; Shahali, Y.; Sénéchal, H.; Poncet, P. Cypress pollinosis: From tree to clinic. Clinic Rev. Allerg. Immunol. 2019, 56, 174–195. [Google Scholar] [CrossRef] [PubMed]
- Shahali, Y.; Sénéchal, H.; Poncet, P. The use of combinatorial hexapeptide ligand library (CPLL) in allergomics. Funct. Proteom. 2019, 1871, 393–403. [Google Scholar]
- Poncet, P.; Sénéchal, H.; Charpin, D. Update on pollen-food allergy syndrome. Expert Rev. Clin. Immunol. 2020, 16, 561–578. [Google Scholar] [CrossRef] [PubMed]
- Ortea, I.; O’Connor, G.; Maquet, A. Review on proteomics for food authentication. J. Proteom. 2016, 147, 212–225. [Google Scholar] [CrossRef]
- Pedreschi, R.; Nørgaard, J.; Maquet, A. Current challenges in detecting food allergens by shotgun and targeted proteomic approaches: A case study on traces of peanut allergens in baked cookies. Nutrients 2012, 4, 132–150. [Google Scholar] [CrossRef] [Green Version]
- Binder, H.; Wirth, H.; Arakelyan, A.; Lembcke, K.; Tiys, E.S.; Ivanisenko, V.A.; Kolchanov, N.A.; Kononikhin, A.; Popov, I.; Nikolaev, E.N.; et al. Time-course human urine proteomics in space-flight simulation experiments. BMC Genom. 2014, 15, S2. [Google Scholar] [CrossRef] [Green Version]
- Brzhozovskiy, A.G.; Kononikhin, A.S.; Pastushkova, L.C.; Kashirina, D.N.; Indeykina, M.I.; Popov, I.A.; Custaud, M.A.; Larina, I.M.; Nikolaev, E.N. The effects of spaceflight factors on the human plasma proteome, including both real space missions and ground-based experiments. Int. J. Mol. Sci. 2019, 20, 3194–3210. [Google Scholar] [CrossRef] [Green Version]
- Kashirina, D.N.; Kononikhin, A.S.; Larina, I.M.; Buravkova, L.B. Secretome of cultured human endothelial cells in simulated microgravity. Exp. Bull. Biol. Med. 2019, 167, 35–38. [Google Scholar] [CrossRef]
- Kashirina, D.N.; Kononikhin, A.S.; Ratushnyy, A.Y.; Nikolaev, E.N.; Larina, I.M.; Buravkova, L.B. Proteomic profile of cultured human endothelial cells after exposition to simulated microgravity. Acta Astronaut. 2021, 179, 11–19. [Google Scholar] [CrossRef]
- Larina, I.M.; Brzhzovsky, A.G.; Nosovsky, A.M.; Kononikhin, A.S.; Orlov, O.I. Post-translational oxidation modifications of blood plasma proteins of cosmonauts after a long-term flight: Part I. Hum. Physiol. 2020, 46, 531–539. [Google Scholar] [CrossRef]
- Xia, Y.; Gao, L.; Guo, L.; Li, H.; Shao, M.; Yang, Q.; Liu, N.; Fang, M.; Xu, X.; Li, J.; et al. Identification of RPSA as a potential biomarker in bronchoalveolar lavage fluid for acute respiratory distress syndrome. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Boschetti, E.; Zilberstein, G.; Righetti, P.G. Combinatorial peptides: A library that continuously probes low-abundance proteins. Electrophoresis 2022, 43, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, E.; D’Amato, A.; Candiano, G.; Righetti, P.G. Protein biomarkers for early detection of diseases: The decisive contribution of combinatorial peptide ligand libraries. J. Proteom. 2018, 188, 1–14. [Google Scholar] [CrossRef]
- Bangy-Letheule, A.; Souab, F.; Bourgoin, S.; Michelland, S.; Cunin, V.; Seve, M.; Aillerie, V.; Dhot, J.; Montnach, J.; Persello, A.; et al. A non-targeted quantitative mass spectrometry approach for the identification of new blood biomarkers of septic shock in the secretory of a rat model of endotoxemic shock. Arch. Cardiovasc. Dis. Suppl. 2020, 12, 229–238. [Google Scholar] [CrossRef]
- Zhou, Y.; Qin, S.; Sun, M.; Tang, L.; Yan, X.; Kim, T.-K.; Caballero, J.; Glusman, G.; Brunkow, M.E.; Soloski, M.J.; et al. Measurement of organ-specific and acute-phase blood protein levels in early Lyme disease. J. Proteome Res. 2020, 19, 346–359. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, Z.; Tan, Y.; Bu, F.; Hao, P.; Zhang, K.; Yang, H.; Liu, S.; Ren, Y. Exploration of missing proteins by a combination approach to enrich the low-abundance hydrophobic proteins from four cancer cell lines. J. Proteome Res. 2020, 19, 401–408. [Google Scholar] [CrossRef]
- Gjoka, X.; Schofield, M.; Cvetkovic, A.; Gantier, R. Combined Protein A and size exclusion high performance liquid chromatography for the single-step measurement of mAb, aggregates and host cell proteins. J. Chromatogr B 2014, 972, 48–52. [Google Scholar] [CrossRef] [PubMed]


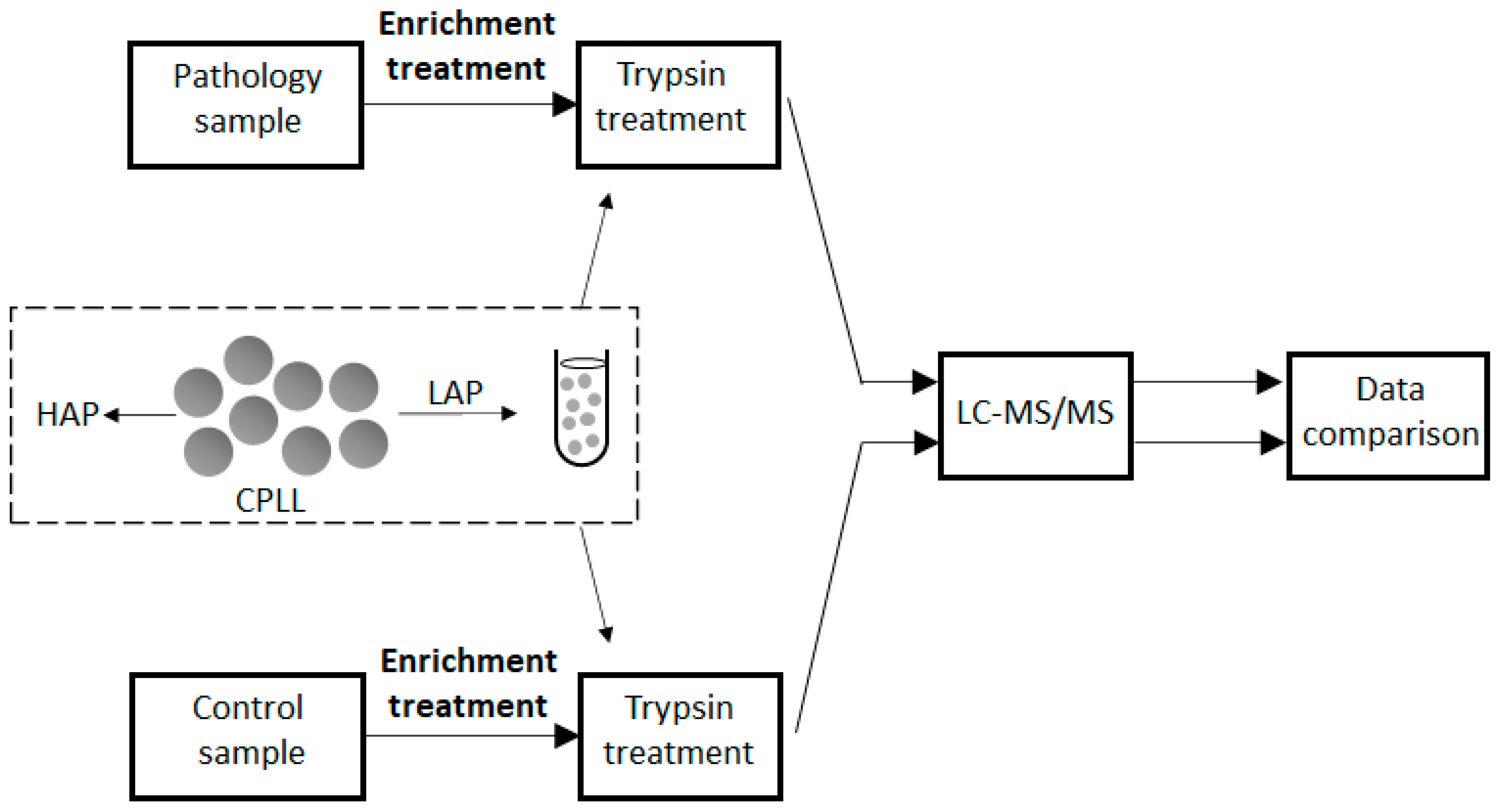
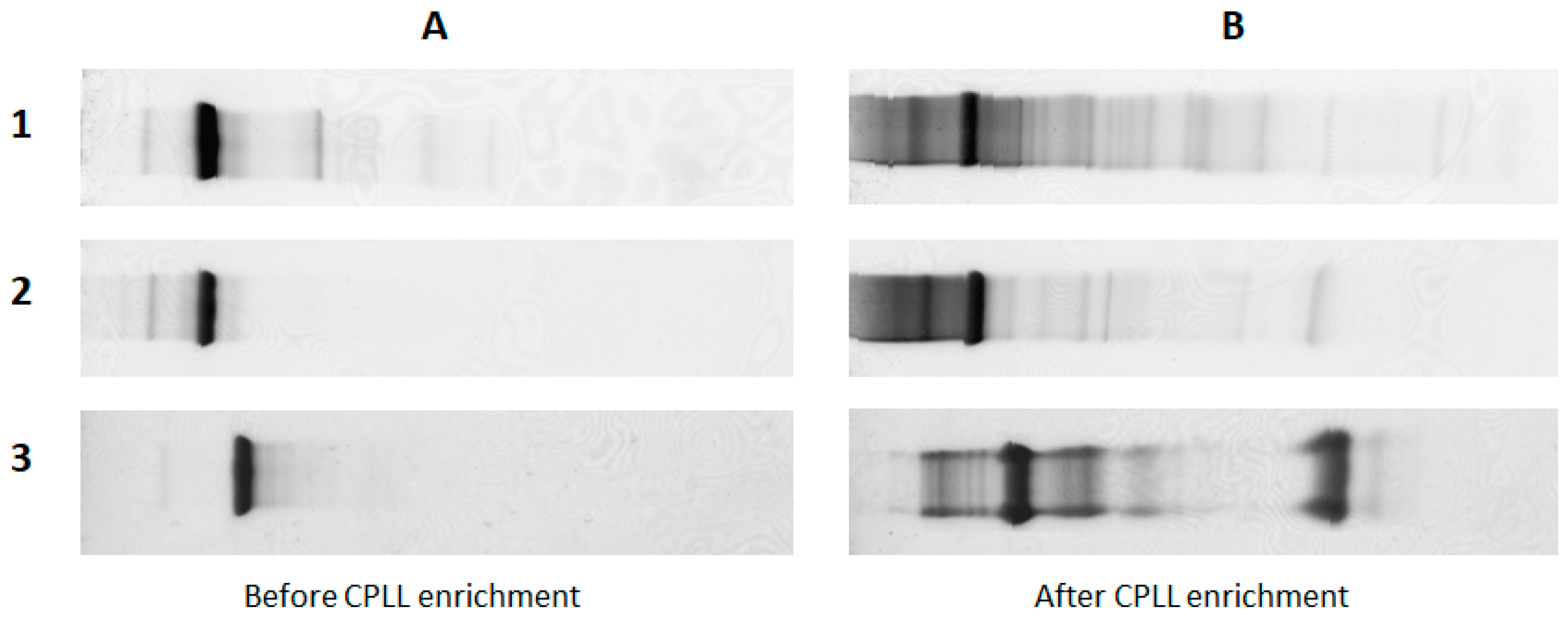
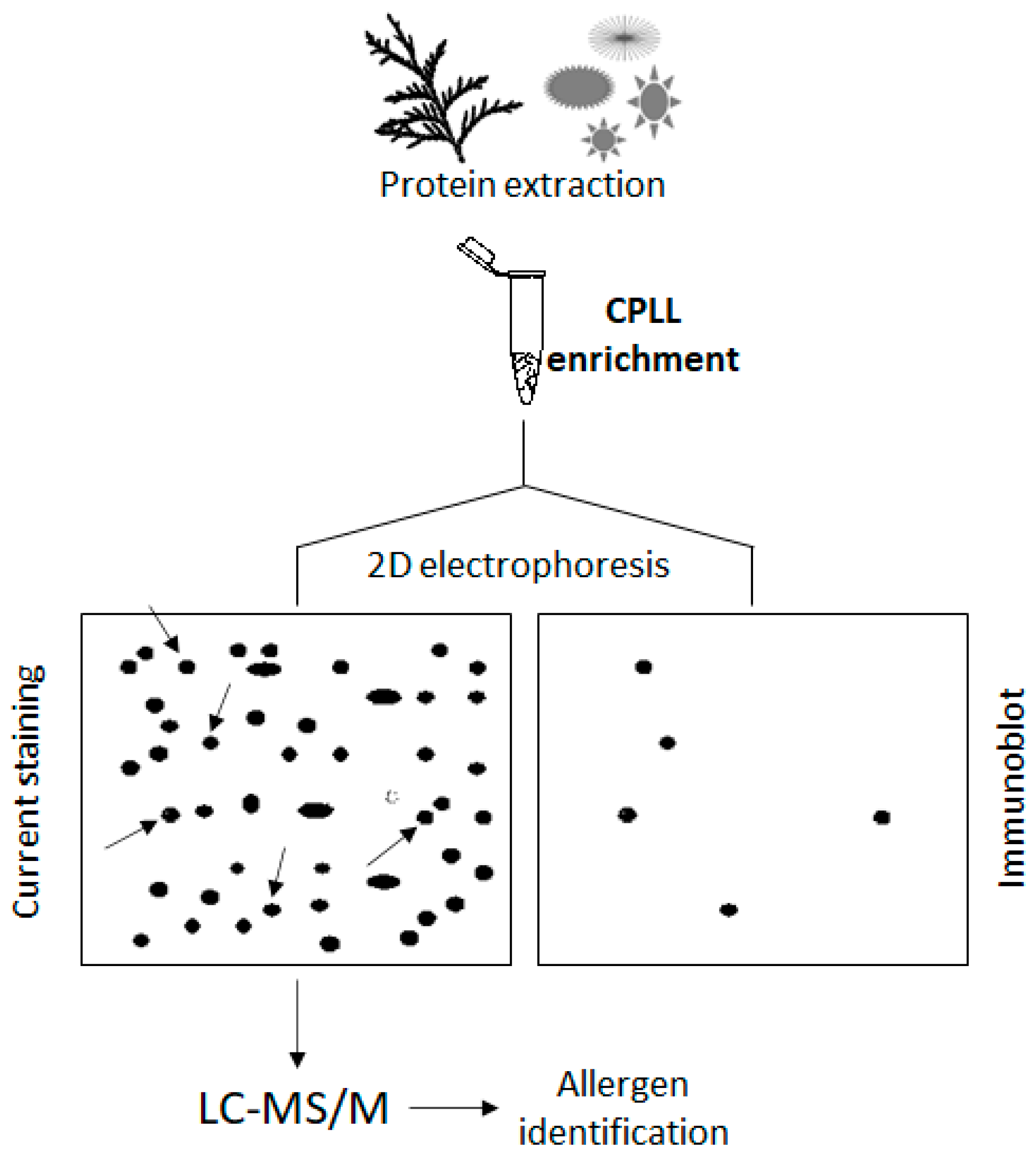
| Method | Principle | Advantages | Drawbacks |
|---|---|---|---|
| Fractionation | Chromatography | High binding capacity Cheap Various conditions | Fraction overlapping Non specific Large dilution |
| Precipitation | Differential solubility | Easy handling Cheap Large and small samples Large applications | Non specific Protein entrapping Rough method Fraction overlapping |
| Immunosubtraction | Antibodies against HAP | High specificity Easy handling Small samples | Restricted samples Large co-subtraction Large dilution Expensive Low binding capacity |
| Capture of LAP groups | Various affinity ligands | Group specific Large choice Concentration of LAP | Non-specific binding Restricted to protein groups |
| Reduction of dynamic range with CPLL | Multiple affinity-like overloading. | Concentration of LAP Reduction of HAP No sample restriction Possible fractionated harvesting | Large samples Expensive Single use |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boschetti, E.; Righetti, P.G. Low-Abundance Protein Enrichment for Medical Applications: The Involvement of Combinatorial Peptide Library Technique. Int. J. Mol. Sci. 2023, 24, 10329. https://doi.org/10.3390/ijms241210329
Boschetti E, Righetti PG. Low-Abundance Protein Enrichment for Medical Applications: The Involvement of Combinatorial Peptide Library Technique. International Journal of Molecular Sciences. 2023; 24(12):10329. https://doi.org/10.3390/ijms241210329
Chicago/Turabian StyleBoschetti, Egisto, and Pier Giorgio Righetti. 2023. "Low-Abundance Protein Enrichment for Medical Applications: The Involvement of Combinatorial Peptide Library Technique" International Journal of Molecular Sciences 24, no. 12: 10329. https://doi.org/10.3390/ijms241210329
APA StyleBoschetti, E., & Righetti, P. G. (2023). Low-Abundance Protein Enrichment for Medical Applications: The Involvement of Combinatorial Peptide Library Technique. International Journal of Molecular Sciences, 24(12), 10329. https://doi.org/10.3390/ijms241210329






