StackTHPred: Identifying Tumor-Homing Peptides through GBDT-Based Feature Selection with Stacking Ensemble Architecture
Abstract
:1. Introduction
2. Results and Discussion
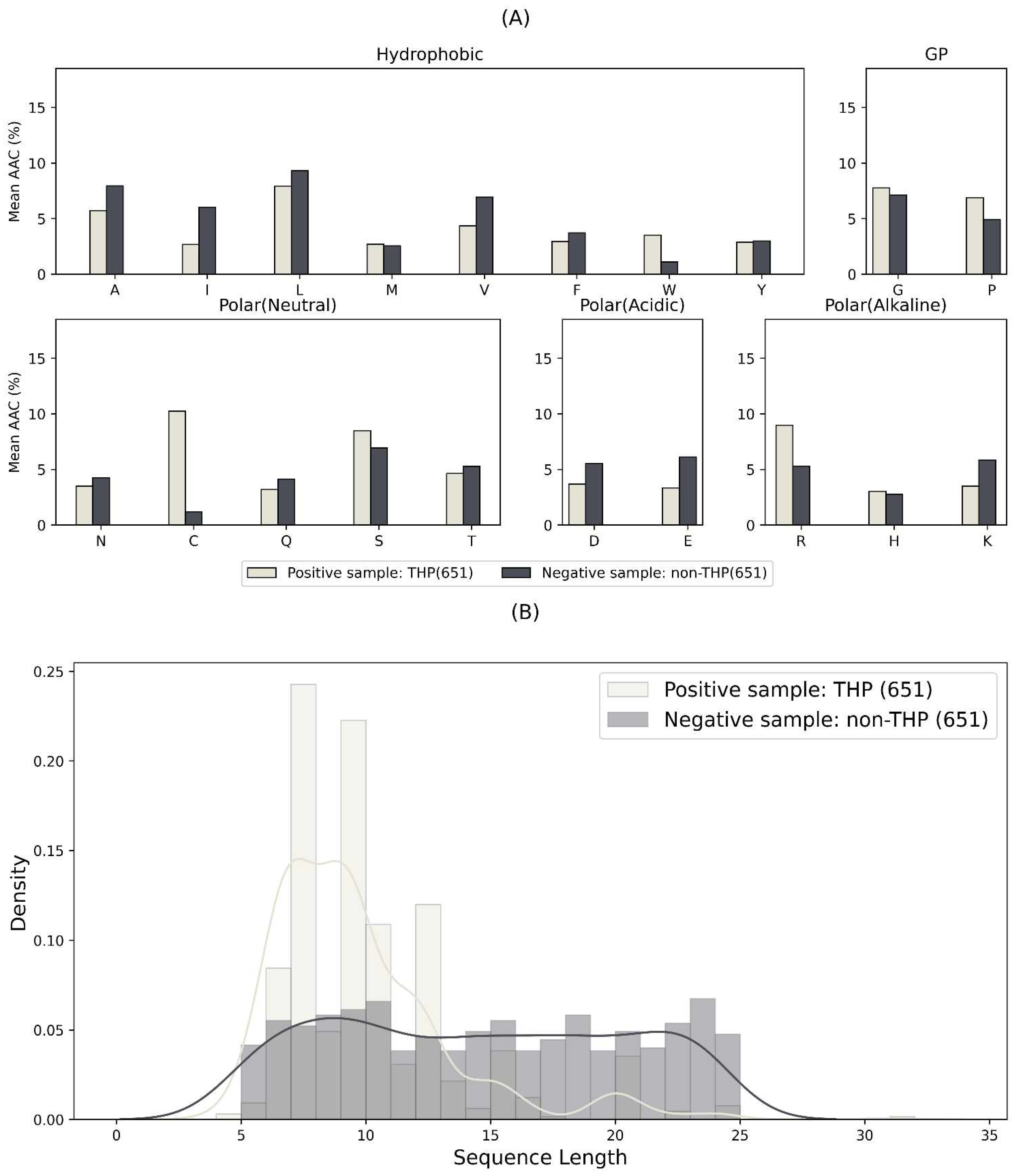
2.1. Overview of the THP and Non-THP Data
2.2. Evaluation Metrics
2.3. Analysis and Comparison of Feature Selection
2.4. Performance Comparison with Other Existing Methods
2.5. Effectiveness Analysis of the Stacking Architecture
2.6. Case Study
2.7. Peptide Features Importance Analysis
3. Materials and Methods
3.1. Dataset Preparation
3.2. Feature Extraction
3.2.1. Amino Acid Composition
3.2.2. Pseudo-Amino Acid Composition
3.2.3. Physicochemical Properties
3.2.4. BLOSUM62
3.2.5. Z-Scale
3.3. Feature Selection
3.4. Stacking Architecture
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Jhong, J.H.; Chen, Q.; Huang, K.Y.; Strittmatter, K.; Kreuzer, J.; DeRan, M.; Wu, X.; Lee, T.Y.; Slavov, N.; et al. Global characterization of macrophage polarization mechanisms and identification of M2-type polarization inhibitors. Cell Rep. 2021, 37, 109955. [Google Scholar] [CrossRef]
- Lee, T.Y.; Huang, K.Y.; Chuang, C.H.; Lee, C.Y.; Chang, T.H. Incorporating deep learning and multi-omics autoencoding for analysis of lung adenocarcinoma prognostication. Comput. Biol. Chem. 2020, 87, 107277. [Google Scholar] [CrossRef]
- Bretana, N.A.; Lu, C.T.; Chiang, C.Y.; Su, M.G.; Huang, K.Y.; Lee, T.Y.; Weng, S.L. Identifying protein phosphorylation sites with kinase substrate specificity on human viruses. PLoS ONE 2012, 7, e40694. [Google Scholar] [CrossRef]
- Bui, V.M.; Weng, S.L.; Lu, C.T.; Chang, T.H.; Weng, J.T.Y.; Lee, T.Y. SOHSite: Incorporating Evolutionary Information and Physicochemical Properties to Identify Protein S-sulfenylation Sites. In BMC Genomics; BioMed Central: London, UK, 2016; Volume 17, pp. 59–70. [Google Scholar]
- Svensen, N.; Walton, J.G.; Bradley, M. Peptides for cell-selective drug delivery. Trends Pharmacol. Sci. 2012, 33, 186–192. [Google Scholar] [CrossRef]
- Khongorzul, P.; Ling, C.J.; Khan, F.U.; Ihsan, A.U.; Zhang, J. Antibody–Drug Conjugates: A Comprehensive ReviewAntibody–Drug Conjugates in Cancer Immunotherapy. Mol. Cancer Res. 2020, 18, 3–19. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.; Kapoor, P.; Chaudhary, K.; Kumar, R.; Raghava, G.; Source Drug Discovery Consortium. Tumor homing peptides as molecular probes for cancer therapeutics, diagnostics and theranostics. Curr. Med. Chem. 2014, 21, 2367–2391. [Google Scholar] [CrossRef]
- Pasqualini, R.; Koivunen, E.; Kain, R.; Lahdenranta, J.; Sakamoto, M.; Stryhn, A.; Ashmun, R.A.; Shapiro, L.H.; Arap, W.; Ruoslahti, E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000, 60, 722–727. [Google Scholar]
- Kondo, E.; Iioka, H.; Saito, K. Tumor-homing peptide and its utility for advanced cancer medicine. Cancer Sci. 2021, 112, 2118–2125. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Lingasamy, P.; Tobi, A.; Kurm, K.; Kopanchuk, S.; Sudakov, A.; Salumäe, M.; Rätsep, T.; Asser, T.; Bjerkvig, R.; Teesalu, T. Tumor-penetrating peptide for systemic targeting of Tenascin-C. Sci. Rep. 2020, 10, 5809. [Google Scholar] [CrossRef] [Green Version]
- Laakkonen, P.; Åkerman, M.E.; Biliran, H.; Yang, M.; Ferrer, F.; Karpanen, T.; Hoffman, R.M.; Ruoslahti, E. Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9381–9386. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, P.; Singh, H.; Gautam, A.; Chaudhary, K.; Kumar, R.; Raghava, G.P. TumorHoPe: A database of tumor homing peptides. PLoS ONE 2012, 7, e35187. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef] [Green Version]
- Shoombuatong, W.; Schaduangrat, N.; Pratiwi, R.; Nantasenamat, C. THPep: A machine learning-based approach for predicting tumor homing peptides. Comput. Biol. Chem. 2019, 80, 441–451. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Chiangjong, W.; Nantasenamat, C.; Moni, M.A.; Lio’, P.; Manavalan, B.; Shoombuatong, W. SCMTHP: A new approach for identifying and characterizing of tumor-homing peptides using estimated propensity scores of amino acids. Pharmaceutics 2022, 14, 122. [Google Scholar] [CrossRef]
- He, W.; Jiang, Y.; Jin, J.; Li, Z.; Zhao, J.; Manavalan, B.; Su, R.; Gao, X.; Wei, L. Accelerating bioactive peptide discovery via mutual information-based meta-learning. Brief. Bioinform. 2022, 23, bbab499. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Schaduangrat, N.; Moni, M.A.; Manavalan, B.; Shoombuatong, W.; Lio, P. NEPTUNE: A novel computational approach for accurate and large-scale identification of tumor homing peptides. Comput. Biol. Med. 2022, 148, 105700. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.C.; You, Z.H.; Zhou, X.; Cheng, L.; Li, X.; Jiang, T.H.; Chen, Z.H. ACP-DL: A deep learning long short-term memory model to predict anticancer peptides using high-efficiency feature representation. Mol. Ther. Nucleic Acids 2019, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ghulam, A.; Ali, F.; Sikander, R.; Ahmad, A.; Ahmed, A.; Patil, S. ACP-2DCNN: Deep learning-based model for improving prediction of anticancer peptides using two-dimensional convolutional neural network. Chemom. Intell. Lab. Syst. 2022, 226, 104589. [Google Scholar] [CrossRef]
- Chung, C.R.; Kuo, T.R.; Wu, L.C.; Lee, T.Y.; Horng, J.T. Characterization and identification of antimicrobial peptides with different functional activities. Brief. Bioinform. 2020, 21, 1098–1114. [Google Scholar] [CrossRef]
- Yao, L.; Li, W.; Zhang, Y.; Deng, J.; Pang, Y.; Huang, Y.; Chung, C.R.; Yu, J.; Chiang, Y.C.; Lee, T.Y. Accelerating the Discovery of Anticancer Peptides through Deep Forest Architecture with Deep Graphical Representation. Int. J. Mol. Sci. 2023, 24, 4328. [Google Scholar] [CrossRef]
- Chen, X.; Huang, J.; He, B. AntiDMPpred: A web service for identifying anti-diabetic peptides. PeerJ 2022, 10, e13581. [Google Scholar] [CrossRef]
- Chang, K.Y.; Yang, J.R. Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS ONE 2013, 8, e70166. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-based prediction of anti-inflammatory peptides using random forest. Front. Pharmacol. 2018, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, B.; Basith, S.; Shin, T.H.; Wei, L.; Lee, G. AtbPpred: A robust sequence-based prediction of anti-tubercular peptides using extremely randomized trees. Comput. Struct. Biotechnol. J. 2019, 17, 972–981. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. iGHBP: Computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput. Struct. Biotechnol. J. 2018, 16, 412–420. [Google Scholar] [CrossRef]
- Arif, M.; Ahmad, S.; Ali, F.; Fang, G.; Li, M.; Yu, D.J. TargetCPP: Accurate prediction of cell-penetrating peptides from optimized multi-scale features using gradient boost decision tree. J. Comput. Aided Mol. Des. 2020, 34, 841–856. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, X. iACP-GE: Accurate identification of anticancer peptides by using gradient boosting decision tree and extra tree. SAR QSAR Environ. Res. 2023, 34, 1–19. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [Green Version]
- Sugahara, K.N.; Braun, G.B.; de Mendoza, T.H.; Kotamraju, V.R.; French, R.P.; Lowy, A.M.; Teesalu, T.; Ruoslahti, E. Tumor-Penetrating iRGD Peptide Inhibits MetastasisiRGD Inhibits Metastasis and Repels Tumor Cells. Mol. Cancer Ther. 2015, 14, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Dai, R.; Yan, W.; Zhang, W.; Bin, Y.; Xia, E.; Xia, J. Identifying multi-functional bioactive peptide functions using multi-label deep learning. Brief. Bioinform. 2022, 23, bbab414. [Google Scholar] [CrossRef]
- Jhong, J.H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. dbAMP 2.0: Updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief. Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Khatun, M.; Hasan, M.; Kurata, H. PreAIP: Computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front. Genet. 2019, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, B.; Basith, S.; Shin, T.H.; Wei, L.; Lee, G. mAHTPred: A sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 2019, 35, 2757–2765. [Google Scholar] [CrossRef]
- Teke, A.; Kavzoglu, T. Determination of effective predisposing factors using Random Forest-Based Gini Index in landslide susceptibility mapping. Intercont. Geoinf. Days 2021, 2, 198–201. [Google Scholar]
- Boulesteix, A.L.; Bender, A.; Lorenzo Bermejo, J.; Strobl, C. Random forest Gini importance favours SNPs with large minor allele frequency: Impact, sources and recommendations. Brief. Bioinform. 2012, 13, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Sandri, M.; Zuccolotto, P. A bias correction algorithm for the Gini variable importance measure in classification trees. J. Comput. Graph. Stat. 2008, 17, 611–628. [Google Scholar] [CrossRef]
- Pang, Y.; Yao, L.; Jhong, J.H.; Wang, Z.; Lee, T.Y. AVPIden: A new scheme for identification and functional prediction of antiviral peptides based on machine learning approaches. Brief. Bioinform. 2021, 22, bbab263. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Bhasin, M.; Raghava, G.P. Classification of nuclear receptors based on amino acid composition and dipeptide composition. J. Biol. Chem. 2004, 279, 23262–23266. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.S.; Zhang, T.L. Using Chou’s pseudo amino acid composition to predict subcellular localization of apoptosis proteins: An approach with immune genetic algorithm-based ensemble classifier. Pattern Recognit. Lett. 2008, 29, 1887–1892. [Google Scholar] [CrossRef]
- Han, Y.; Lin, Z.; Zhou, J.; Yun, G.; Guo, R.; Richardson, J.J.; Caruso, F. Polyphenol-mediated assembly of proteins for engineering functional materials. Angew. Chem. Int. Ed. 2020, 59, 15618–15625. [Google Scholar] [CrossRef]
- Azzarito, V.; Long, K.; Murphy, N.S.; Wilson, A.J. Inhibition of α-helix-mediated protein–protein interactions using designed molecules. Nat. Chem. 2013, 5, 161–173. [Google Scholar] [CrossRef]
- Singh, A.; Orsat, V.; Raghavan, V. Soybean hydrophobic protein response to external electric field: A molecular modeling approach. Biomolecules 2013, 3, 168–179. [Google Scholar] [CrossRef] [Green Version]
- Danoff, E.J.; Fleming, K.G. The soluble, periplasmic domain of OmpA folds as an independent unit and displays chaperone activity by reducing the self-association propensity of the unfolded OmpA transmembrane β-barrel. Biophys. Chem. 2011, 159, 194–204. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Huang, L.; Zhang, C.; Deng, Y.; Xie, P.; Liu, L.; Cheng, J. Research advances in chemical modifications of starch for hydrophobicity and its applications: A review. Carbohydr. Polym. 2020, 240, 116292. [Google Scholar] [CrossRef]
- Boman, H.; Wade, D.; Boman, I.; Wåhlin, B.; Merrifield, R. Antibacterial and antimalarial properties of peptides that are cecropin-melittin hybrids. FEBS Lett. 1989, 259, 103–106. [Google Scholar] [CrossRef] [Green Version]
- Pihlasalo, S.; Auranen, L.; Hanninen, P.; Harma, H. Method for estimation of protein isoelectric point. Anal. Chem. 2012, 84, 8253–8258. [Google Scholar] [CrossRef]
- Hayat, M.; Khan, A. WRF-TMH: Predicting transmembrane helix by fusing composition index and physicochemical properties of amino acids. Amino Acids 2013, 44, 1317–1328. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, M.; Eriksson, L.; Jonsson, J.; Sjöström, M.; Wold, S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem. 1998, 41, 2481–2491. [Google Scholar] [CrossRef]
- Tang, H.; Su, Z.D.; Wei, H.H.; Chen, W.; Lin, H. Prediction of cell-penetrating peptides with feature selection techniques. Biochem. Biophys. Res. Commun. 2016, 477, 150–154. [Google Scholar] [CrossRef]
- Wang, P.; Hu, L.; Liu, G.; Jiang, N.; Chen, X.; Xu, J.; Zheng, W.; Li, L.; Tan, M.; Chen, Z.; et al. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS ONE 2011, 6, e18476. [Google Scholar] [CrossRef]
- Rao, H.; Shi, X.; Rodrigue, A.K.; Feng, J.; Xia, Y.; Elhoseny, M.; Yuan, X.; Gu, L. Feature selection based on artificial bee colony and gradient boosting decision tree. Appl. Soft Comput. 2019, 74, 634–642. [Google Scholar] [CrossRef]
- Xu, Z.; Huang, G.; Weinberger, K.Q.; Zheng, A.X. Gradient boosted feature selection. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 522–531. [Google Scholar]
- Upadhyay, D.; Manero, J.; Zaman, M.; Sampalli, S. Gradient boosting feature selection with machine learning classifiers for intrusion detection on power grids. IEEE Trans. Netw. Serv. Manag. 2020, 18, 1104–1116. [Google Scholar] [CrossRef]
- Yu, B.; Wang, X.; Zhang, Y.; Gao, H.; Wang, Y.; Liu, Y.; Gao, X. RPI-MDLStack: Predicting RNA–protein interactions through deep learning with stacking strategy and LASSO. Appl. Soft Comput. 2022, 120, 108676. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Chiangjong, W.; Nantasenamat, C.; Hasan, M.M.; Manavalan, B.; Shoombuatong, W. StackIL6: A stacking ensemble model for improving the prediction of IL-6 inducing peptides. Brief. Bioinform. 2021, 22, bbab172. [Google Scholar] [CrossRef]
- Gattani, S.; Mishra, A.; Hoque, M.T. StackCBPred: A stacking based prediction of protein-carbohydrate binding sites from sequence. Carbohydr. Res. 2019, 486, 107857. [Google Scholar] [CrossRef]
- Pavlyshenko, B. Using stacking approaches for machine learning models. In Proceedings of the 2018 IEEE Second International Conference on Data Stream Mining & Processing (DSMP), Lviv, Ukraine, 21–25 August 2018; pp. 255–258. [Google Scholar]



| Dataset | Method | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| main dataset | THPep | 0.846 | 0.792 | 0.900 | 0.696 |
| SCMTHP | 0.827 | 0.869 | 0.785 | 0.656 | |
| MIMML | 0.885 | 0.876 | 0.894 | 0.770 | |
| NEPTUNE | 0.885 | 0.900 | 0.869 | 0.770 | |
| StackTHPred | 0.915 | 0.915 | 0.915 | 0.831 | |
| small dataset | THPep | 0.798 | 0.862 | 0.734 | 0.601 |
| SCMTHP | 0.798 | 0.766 | 0.830 | 0.597 | |
| MIMML | 0.840 | 0.807 | 0.874 | 0.682 | |
| NEPTUNE | 0.856 | 0.830 | 0.883 | 0.714 | |
| StackTHPred | 0.883 | 0.862 | 0.904 | 0.767 |
| Dataset | Model | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| main dataset | Only-ET | 0.896 | 0.908 | 0.885 | 0.793 |
| Only-RF | 0.889 | 0.915 | 0.862 | 0.778 | |
| Only-GBDT | 0.885 | 0.915 | 0.854 | 0.771 | |
| Stacking model | 0.915 | 0.915 | 0.915 | 0.831 | |
| small dataset | Only-ET | 0.851 | 0.819 | 0.883 | 0.704 |
| Only-RF | 0.846 | 0.809 | 0.883 | 0.695 | |
| Only-GBDT | 0.824 | 0.830 | 0.819 | 0.649 | |
| Stacking model | 0.883 | 0.862 | 0.904 | 0.767 |
| Method | Accuracy | Sensitivity | Specificity | MCC |
|---|---|---|---|---|
| SCMTHP | 0.827 | 0.869 | 0.785 | 0.656 |
| NEPTUNE-main | 0.832 | 0.901 | 0.763 | 0.670 |
| NEPTUNE-small | 0.844 | 0.817 | 0.870 | 0.688 |
| StackTHPred | 0.924 | 0.924 | 0.924 | 0.847 |
| Positive | Negative | Total | Max Length | Min Length | ||
|---|---|---|---|---|---|---|
| main dataset | Train Set | 521 | 521 | 1042 | 31 | 4 |
| Test Set | 130 | 130 | 260 | 24 | 5 | |
| small dataset | Train Set | 375 | 375 | 750 | 10 | 4 |
| Test Set | 94 | 94 | 188 | 10 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, J.; Yao, L.; Chung, C.-R.; Chiang, Y.-C.; Lee, T.-Y. StackTHPred: Identifying Tumor-Homing Peptides through GBDT-Based Feature Selection with Stacking Ensemble Architecture. Int. J. Mol. Sci. 2023, 24, 10348. https://doi.org/10.3390/ijms241210348
Guan J, Yao L, Chung C-R, Chiang Y-C, Lee T-Y. StackTHPred: Identifying Tumor-Homing Peptides through GBDT-Based Feature Selection with Stacking Ensemble Architecture. International Journal of Molecular Sciences. 2023; 24(12):10348. https://doi.org/10.3390/ijms241210348
Chicago/Turabian StyleGuan, Jiahui, Lantian Yao, Chia-Ru Chung, Ying-Chih Chiang, and Tzong-Yi Lee. 2023. "StackTHPred: Identifying Tumor-Homing Peptides through GBDT-Based Feature Selection with Stacking Ensemble Architecture" International Journal of Molecular Sciences 24, no. 12: 10348. https://doi.org/10.3390/ijms241210348
APA StyleGuan, J., Yao, L., Chung, C.-R., Chiang, Y.-C., & Lee, T.-Y. (2023). StackTHPred: Identifying Tumor-Homing Peptides through GBDT-Based Feature Selection with Stacking Ensemble Architecture. International Journal of Molecular Sciences, 24(12), 10348. https://doi.org/10.3390/ijms241210348








