Identification of NRF2 Activation as a Prognostic Biomarker in T-Cell Acute Lymphoblastic Leukaemia
Abstract
1. Introduction
2. Results
2.1. High Expression of NFE2L2 Associates with Poor Prognosis in T-Cell Acute Lymphoblastic Leukaemia

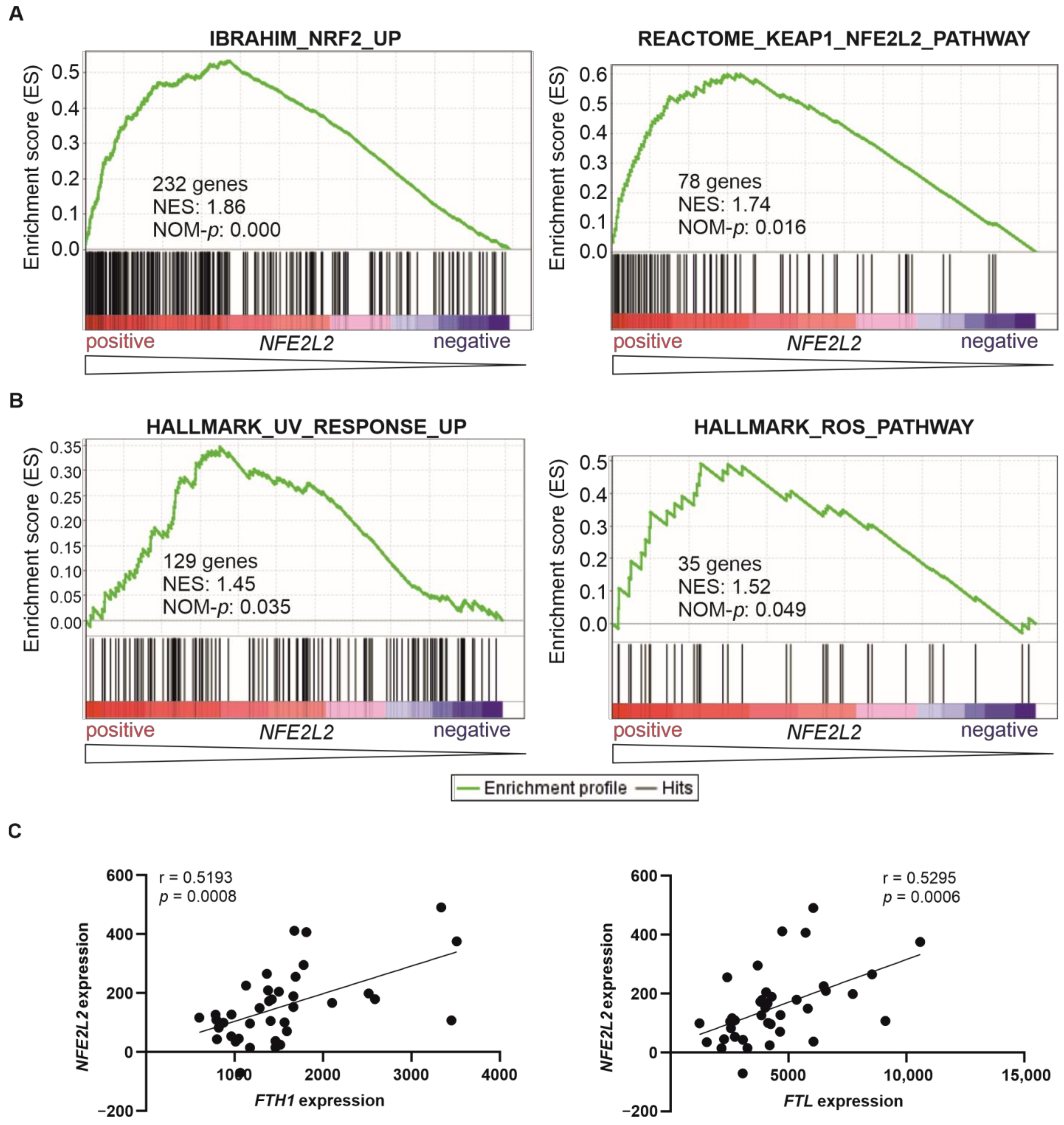
2.2. T-ALL Patients with High NFE2L2 Expression Exhibit a Genetic Program of NRF2-Induced Transcriptional Targets and Signalling
2.3. Aberrant Activation of the MAPK-ERK and PI3K-AKT-mTOR Signalling Pathways in T-ALL Patients with High NFE2L2
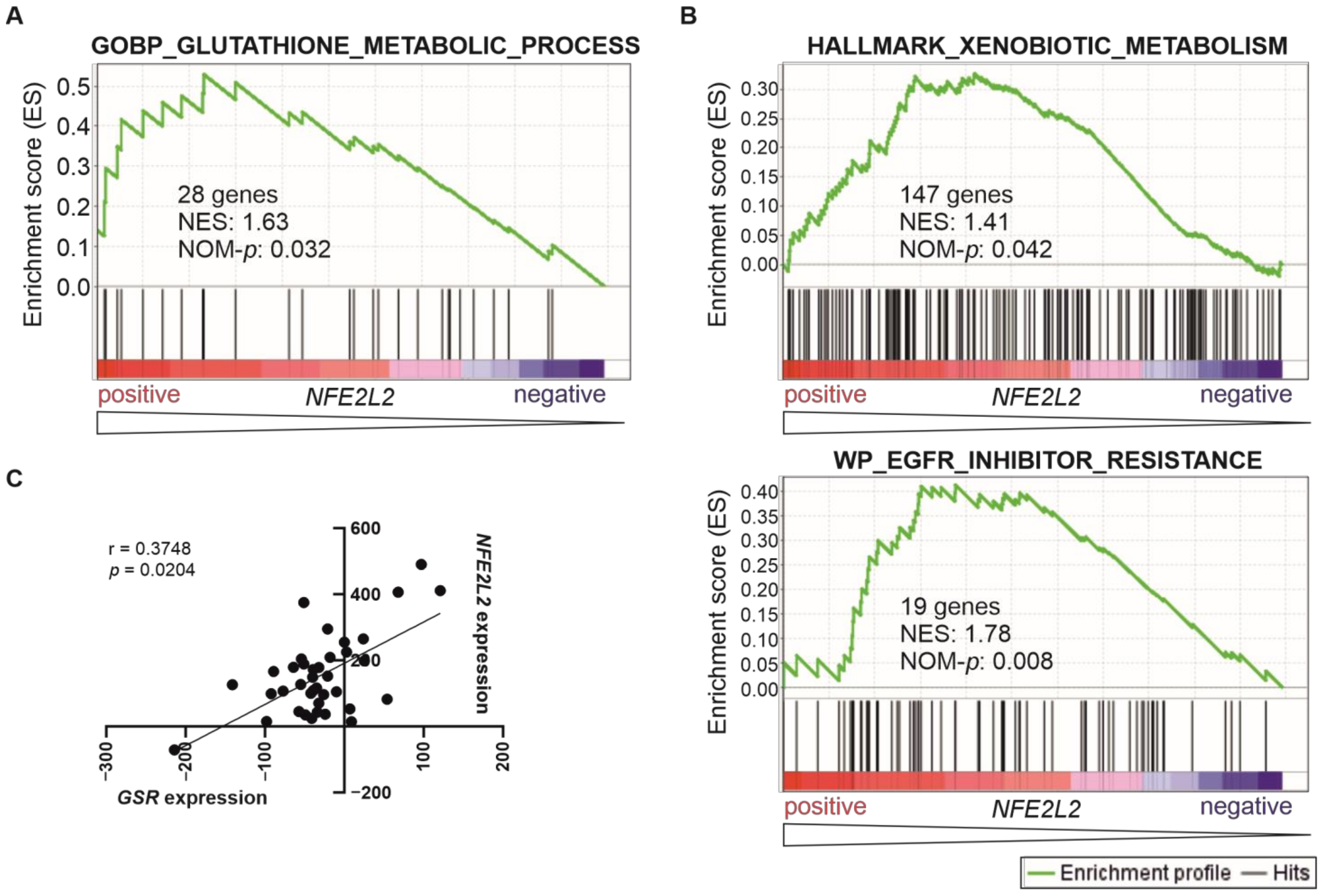
2.4. T-ALL Patients with High NFE2L2 Levels May Develop a Phenotype of Drug Resistance Provided by NRF2-Induced Biosynthesis of Glutathione
3. Discussion
4. Materials and Methods
4.1. Description of Patient Cohorts
4.2. In Silico Analysis of Published Datasets
4.3. Gene Set Enrichment Analysis (GSEA)
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Durinck, K.; Goossens, S.; Peirs, S.; Wallaert, A.; Van Loocke, W.; Matthijssens, F.; Pieters, T.; Milani, G.; Lammens, T.; Rondou, P.; et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp. Hematol. 2015, 43, 625–639. [Google Scholar] [CrossRef]
- Girardi, T.; Vicente, C.; Cools, J.; De Keersmaecker, K. The genetics and molecular biology of T-ALL. Blood 2017, 129, 1113–1123. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Spijkers-Hagelstein, J.A.P.; Buijs-Gladdines, J.G.C.A.M.; Uitdehaag, J.C.M.; Smits, W.; Van Der Zwet, J.; Buijsman, R.; Zaman, G.; Pieters, R.; Meijerink, J.P.P. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia 2016, 30, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Litzow, M.R.; Ferrando, A.A. How I treat T-cell acute lymphoblastic leukemia in adults. Blood 2015, 126, 833–841. [Google Scholar] [CrossRef]
- Lonetti, A.; Cappellini, A.; Bertaina, A.; Locatelli, F.; Pession, A.; Buontempo, F.; Evangelisti, C.; Evangelisti, C.; Orsini, E.; Zambonin, L.; et al. Improving nelarabine efficacy in T cell acute lymphoblastic leukemia by targeting aberrant PI3K/AKT/mTOR signaling pathway. J. Hematol. Oncol. 2016, 9, 114. [Google Scholar] [CrossRef]
- Fattizzo, B.; Rosa, J.; Giannotta, J.A.; Baldini, L.; Fracchiolla, N. The Physiopathology of T- Cell Acute Lymphoblastic Leukemia: Focus on Molecular Aspects. Front. Oncol. 2020, 10, 273. [Google Scholar] [CrossRef]
- Bortolozzi, R.; Bresolin, S.; Rampazzo, E.; Paganin, M.; Maule, F.; Mariotto, E.; Boso, D.; Minuzzo, S.; Agnusdei, V.; Viola, G.; et al. AKR1C enzymes sustain therapy resistance in paediatric T-ALL. Br. J. Cancer 2018, 118, 985–994. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Fahl, W.E. Functional Characterization of Transcription Regulators That Interact with the Electrophile Response Element. Biochem. Biophys. Res. Commun. 2001, 289, 212–219. [Google Scholar] [CrossRef]
- Motohashi, H.; Yamamoto, M. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004, 10, 549–557. [Google Scholar] [CrossRef] [PubMed]
- DeBlasi, J.M.; DeNicola, G.M. Dissecting the Crosstalk between NRF2 Signaling and Metabolic Processes in Cancer. Cancers 2020, 12, 3023. [Google Scholar] [CrossRef] [PubMed]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; McMahon, M.; Chowdhry, S.; Dinkova-Kostova, A.T.; Welsh, N.; Díaz-Alonso, J.; Paraíso-Luna, J.; Navarrete, C.; del Río, C.; Cantarero, I.; et al. Cancer Chemoprevention Mechanisms Mediated Through the Keap1–Nrf2 Pathway. Antioxid. Redox Signal. 2010, 13, 1713–1748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Peng, H.; Wang, H.; Xue, P.; Hou, Y.; Dong, J.; Zhou, T.; Qu, W.; Peng, S.; Carmichael, P.L.; et al. The Nrf2-Keap1-ARE Signaling Pathway: The Regulation and Dual Function of Nrf2 in Cancer. Antioxid. Redox Signal. 2010, 13, 1623–1626. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Yang, J.; Zhou, S.; Wang, Y.; Li, Y.; Tong, X. The Role of the Pentose Phosphate Pathway in Diabetes and Cancer. Front. Endocrinol. 2020, 11, 365. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Kitamura, H.; Motohashi, H. NRF2 addiction in cancer cells. Cancer Sci. 2018, 109, 900–911. [Google Scholar] [CrossRef]
- Almeida, M.; Soares, M.; Ramalhinho, A.C.; Moutinho, J.F.; Breitenfeld, L.; Pereira, L. The prognostic value of NRF2 in breast cancer patients: A systematic review with meta-analysis. Breast Cancer Res. Treat. 2020, 179, 523–532. [Google Scholar] [CrossRef]
- Bao, L.; Wu, J.; Dodson, M.; de la Vega, E.M.R.; Ning, Y.; Zhang, Z.; Yao, M.; Zhang, D.D.; Xu, C.; Yi, X. ABCF2, an Nrf2 target gene, contributes to cisplatin resistance in ovarian cancer cells. Mol. Carcinog. 2017, 56, 1543–1553. [Google Scholar] [CrossRef]
- Jeddi, F.; Soozangar, N.; Sadeghi, M.R.; Somi, M.H.; Shirmohamadi, M.; Eftekhar-Sadat, A.-T.; Samadi, N. Nrf2 overexpression is associated with P-glycoprotein upregulation in gastric cancer. Biomed. Pharmacother. 2018, 97, 286–292. [Google Scholar] [CrossRef]
- Roh, J.-L.; Jang, H.; Kim, E.H.; Shin, D. Targeting of the Glutathione, Thioredoxin, and Nrf2 Antioxidant Systems in Head and Neck Cancer. Antioxidants Redox Signal. 2017, 27, 106–114. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, N.; Zhou, S.; Ye, W.; Jing, G.; Zhang, M. Propofol induces proliferation and invasion of gallbladder cancer cells through activation of Nrf2. J. Exp. Clin. Cancer Res. 2012, 31, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Zhong, L.; Dan, W.; Wang, X.; Zhang, Z.; Liu, Z.; Lu, Y.; Shao, X.; Zhou, Z.; Chen, S.; et al. DNMT3A R882H mutation drives daunorubicin resistance in acute myeloid leukemia via regulating NRF2/NQO1 pathway. Cell Commun. Signal. 2022, 20, 168. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Ma, D.; Wang, P.; Pan, C.; Fang, Q.; Wang, J. Nrf2 overexpression increases risk of high tumor mutation burden in acute myeloid leukemia by inhibiting MSH2. Cell Death Dis. 2021, 12, 20. [Google Scholar] [CrossRef]
- Shang, Q.; Pan, C.; Zhang, X.; Yang, T.; Hu, T.; Zheng, L.; Cao, S.; Feng, C.; Hu, X.; Chai, X.; et al. Nuclear factor Nrf2 promotes glycosidase OGG1 expression by activating the AKT pathway to enhance leukemia cell resistance to cytarabine. J. Biol. Chem. 2023, 299, 102798. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Aziz, A.; MacEwan, D.J.; Bowles, K.M.; Rushworth, S.A. Oxidative Stress Responses and NRF2 in Human Leukaemia. Oxidative Med. Cell. Longev. 2015, 2015, 4546591. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X.; Kang, Q.; Pan, C.; Zhang, T.; Feng, C.; Chen, L.; Wei, S.; Wang, J. Nrf2 Overexpression Decreases Vincristine Chemotherapy Sensitivity Through the PI3K-AKT Pathway in Adult B-Cell Acute Lymphoblastic Leukemia. Front. Oncol. 2022, 12, 876556. [Google Scholar] [CrossRef]
- Chu, X.; Zhong, L.; Dan, W.; Wang, X.; Zhang, Z.; Liu, Z.; Lu, Y.; Shao, X.; Zhou, Z.; Chen, S.; et al. DNMT3A R882H mutation promotes acute leukemic cell survival by regulating glycolysis through the NRF2/NQO1 axis. Cell. Signal. 2023, 105, 110626. [Google Scholar] [CrossRef]
- Rushworth, S.A.; Bowles, K.M.; MacEwan, D.J. High Basal Nuclear Levels of Nrf2 in Acute Myeloid Leukemia Reduces Sensitivity to Proteasome Inhibitors. Cancer Res. 2011, 71, 1999–2009. [Google Scholar] [CrossRef]
- Rushworth, S.A.; Zaitseva, L.; Murray, M.Y.; Shah, N.M.; Bowles, K.M.; MacEwan, D.J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-kappaB and underlies its chemo-resistance. Blood 2012, 120, 5188–5198. [Google Scholar] [CrossRef]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef]
- Ferrando, A.A.; Neuberg, D.S.; Staunton, J.; Loh, M.L.; Huard, C.; Raimondi, S.C.; Behm, F.G.; Pui, C.-H.; Downing, J.R.; Gilliland, D.; et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 2002, 1, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Basak, P.; Sadhukhan, P.; Sarkar, P.; Sil, P.C. Perspectives of the Nrf-2 signaling pathway in cancer progression and therapy. Toxicol. Rep. 2017, 4, 306–318. [Google Scholar] [CrossRef]
- Zimta, A.-A.; Cenariu, D.; Irimie, A.; Magdo, L.; Nabavi, S.M.; Atanasov, A.G.; Berindan-Neagoe, I. The Role of Nrf2 Activity in Cancer Development and Progression. Cancers 2019, 11, 1755. [Google Scholar] [CrossRef]
- Den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.-F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E.; et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- Lien, E.C.; Lyssiotis, C.A.; Juvekar, A.; Hu, H.; Asara, J.M.; Cantley, L.C.; Toker, A. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol. 2016, 18, 572–578. [Google Scholar] [CrossRef]
- Jasek-Gajda, E.; Jurkowska, H.; Jasińska, M.; Lis, G.J. Targeting the MAPK/ERK and PI3K/AKT Signaling Pathways Affects NRF2, Trx and GSH Antioxidant Systems in Leukemia Cells. Antioxidants 2020, 9, 633. [Google Scholar] [CrossRef]
- Jorge, J.; Magalhães, N.; Alves, R.; Lapa, B.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B. Antitumor Effect of Brusatol in Acute Lymphoblastic Leukemia Models Is Triggered by Reactive Oxygen Species Accumulation. Biomedicines 2022, 10, 2207. [Google Scholar] [CrossRef]
- Silic-Benussi, M.; Sharova, E.; Ciccarese, F.; Cavallari, I.; Raimondi, V.; Urso, L.; Corradin, A.; Kotler, H.; Scattolin, G.; Buldini, B.; et al. mTOR inhibition downregulates glucose-6-phosphate dehydrogenase and induces ROS-dependent death in T-cell acute lymphoblastic leukemia cells. Redox Biol. 2022, 51, 102268. [Google Scholar] [CrossRef] [PubMed]
- Akın-Balı, D.F.; Aktas, S.H.; Unal, M.A.; Kankılıc, T. Identification of novel Nrf2/Keap1 pathway mutations in pediatric acute lymphoblastic leukemia. Pediatr. Hematol. Oncol. 2020, 37, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/β-TrCP Promotes Glycogen Synthase Kinase 3-Dependent Degradation of the Nrf2 Transcription Factor in a Keap1-Independent Manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef]
- Tung, M.-C.; Lin, P.-L.; Wang, Y.-C.; He, T.-Y.; Lee, M.-C.; Yeh, S.-D.; Chen, C.-Y.; Lee, H. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 2015, 6, 41692–41705. [Google Scholar] [CrossRef]
- Hayes, J.D.; Chowdhry, S.; Dinkova-Kostova, A.T.; Sutherland, C. Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of β-TrCP and GSK-3. Biochem. Soc. Trans. 2015, 43, 611–620. [Google Scholar] [CrossRef]
- Taguchi, K.; Hirano, I.; Itoh, T.; Tanaka, M.; Miyajima, A.; Suzuki, A.; Motohashi, H.; Yamamoto, M. Nrf2 Enhances Cholangiocyte Expansion in Pten-Deficient Livers. Mol. Cell. Biol. 2014, 34, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and Consequences of Cysteine S-Glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [Google Scholar] [CrossRef]
- Calvert, P.; Yao, K.-S.; Hamilton, T.C.; O’dwyer, P.J. Clinical studies of reversal of drug resistance based on glutathione. Chem. Interact. 1998, 111, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, A.M.; Rolfo, M.; Mangerini, R.; Gaetani, G.F. Increased glutathione in chronic lymphocytic leukemia lymphocytes. Am. J. Hematol. 1994, 47, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Maung, Z.T.; Hogarth, L.; Reid, M.M.; Proctor, S.J.; Hamilton, P.J.; Hall, A.G. Raised intracellular glutathione levels correlate with in vitro resistance to cytotoxic drugs in leukaemic cells from patients with acute lymphoblastic leukemia. Leukemia 1994, 8, 1487–1491. [Google Scholar] [PubMed]
- Ghalaut, V.S.; Kharb, S.; Ghalaut, P.; Rawal, A. Lymphocyte glutathione levels in acute leukemia. Clin. Chim. Acta 1999, 285, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]

| Patient | Gene * | Mutation | Location | Variation Type | Consequence | HGVS **_cDNA | HGVS **_Protein |
|---|---|---|---|---|---|---|---|
| 1 | PTEN | -/CT | 10:89,717,712 | insertion | frameshift_variant | c.737_738insTC | p.Leu247ArgfsTer10 |
| 2 | PTEN | G/T | 10:89,692,905 | SNV | missense_variant | c.389G>T | p.Arg130Leu |
| 3 | PTEN | -/G | 10:89,711,893 | insertion | frameshift_variant | c.510_511insG | p.Gln171AlafsTer9 |
| PTEN | -/CTCACTC | 10:89,711,895 | insertion | frameshift_variant | c.512_513insCTCACTC | p.Gln171HisfsTer11 | |
| 4 | KRAS | C/T | 12:25,398,284 | SNV | missense_variant | c.35G>A | p.Gly12Asp |
| 5 | KRAS | C/A | 12:25,398,284 | SNV | missense_variant | c.35G>T | p.Gly12Val |
| 6 | PTEN | -/A | 10:89,717,671 | insertion | frameshift_variant | c.696dup | p.Arg233ThrfsTer10 |
| 7 | KRAS | C/G | 12:25,398,284 | SNV | missense_variant | c.35G>C | p.Gly12Ala |
| 8 | PTEN | -/G | 10:89,717,676 | insertion | frameshift_variant | c.703dup | p.Glu235GlyfsTer8 |
| 9 | PTEN | C/T | 10:89,720,741 | SNV | stop_gained | c.892C>T | p.Gln298Ter |
| 10 | PTEN | T/A | 10:89,717,739 | SNV | missense_variant | c.764T>A | p.Val255Glu |
| 11 | PTEN | -/G | 10:89,717,699 | insertion | frameshift_variant | c.724dup | p.Glu242GlyfsTer11 |
| 12 | PTEN | A/G | 10:89,711,903 | SNV | missense_variant | c.521A>G | p.Tyr174Cys |
| PTEN | -/G | 10:89,717,672 | insertion | frameshift_variant | c.696_697insG | p.Arg233AlafsTer10 | |
| PTEN | C/T | 10:89,717,672 | SNV | stop_gained | c.697C>T | p.Arg233Ter | |
| 13 | PTEN | -/ CAGCCGCCGCTTTTGGAGGG | 10:89,717,672 | insertion | frameshift_variant | c.697_ 698insAGCCGCCGCTTTTGGAGGGC | p.Arg233GlnfsTer30 |
| 14 | KRAS | C/A | 12:25,398,284 | SNV | missense_variant | c.35G>T | p.Gly12Val |
| 15 | PTEN | -/GGTGT | 10:89,624,297 | insertion | frameshift_variant | c.70_71insGGTGT | p.Asp24GlyfsTer4 |
| PTEN | -/GT | 10:89,624,298 | insertion | frameshift_variant | c.71_72insGT | p.Asp24GlufsTer3 | |
| 16 | PTEN | -/T | 10:89,717,672 | insertion | frameshift_variant | c.696_697insT | p.Arg233SerfsTer10 |
| PTEN | C/T | 10:89,717,672 | SNV | stop_gained | c.697C>T | p.Arg233Ter | |
| 17 | KRAS | C/T | 12:25,398,281 | SNV | missense_variant | c.38G>A | p.Gly13Asp |
| 18 | PTEN | C/T | 10:89,692,904 | SNV | stop_gained | c.388C>T | p.Arg130Ter |
| PTEN | C/T | 10:89,720,852 | SNV | stop_gained | c.1003C>T | p.Arg335Ter | |
| 19 | KRAS | G/C | 12:25,398,218 | SNV | missense_variant | c.101C>G | p.Pro34Arg |
| 20 | KRAS | C/T | 12:25,398,284 | SNV | missense_variant | c.35G>A | p.Gly12Asp |
| PTEN | -/AA | 10:89,692,954 | insertion | frameshift_variant | c.439_440dup | p.Ala148ArgfsTer6 | |
| 21 | PTEN | C/T | 10:89,692,805 | SNV | stop_gained | c.289C>T | p.Gln97Ter |
| 22 | KRAS | C/T | 12:25,398,284 | SNV | missense_variant | c.35G>A | p.Gly12Asp |
| PTEN | A/T | 10:89,717,636 | SNV | stop_gained | c.661A>T | p.Lys221Ter | |
| 23 | PTEN | ATAG/- | 10:89,725,147 | deletion | frameshift_variant | c.1133_1136del | p.Arg378IlefsTer37 |
| 24 | TP53 | G/A | 17:7,577,094 | SNV | missense_variant | c.844C>T | p.Arg282Trp |
| TP53 | G/A | 17:7,577,094 | SNV | missense_variant | c.844C>T | p.Arg282Trp | |
| 25 | KRAS | C/A | 12:25,398,284 | SNV | missense_variant | c.35G>T | p.Gly12Val |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villa-Morales, M.; Pérez-Gómez, L.; Pérez-Gómez, E.; López-Nieva, P.; Fernández-Navarro, P.; Santos, J. Identification of NRF2 Activation as a Prognostic Biomarker in T-Cell Acute Lymphoblastic Leukaemia. Int. J. Mol. Sci. 2023, 24, 10350. https://doi.org/10.3390/ijms241210350
Villa-Morales M, Pérez-Gómez L, Pérez-Gómez E, López-Nieva P, Fernández-Navarro P, Santos J. Identification of NRF2 Activation as a Prognostic Biomarker in T-Cell Acute Lymphoblastic Leukaemia. International Journal of Molecular Sciences. 2023; 24(12):10350. https://doi.org/10.3390/ijms241210350
Chicago/Turabian StyleVilla-Morales, María, Laura Pérez-Gómez, Eduardo Pérez-Gómez, Pilar López-Nieva, Pablo Fernández-Navarro, and Javier Santos. 2023. "Identification of NRF2 Activation as a Prognostic Biomarker in T-Cell Acute Lymphoblastic Leukaemia" International Journal of Molecular Sciences 24, no. 12: 10350. https://doi.org/10.3390/ijms241210350
APA StyleVilla-Morales, M., Pérez-Gómez, L., Pérez-Gómez, E., López-Nieva, P., Fernández-Navarro, P., & Santos, J. (2023). Identification of NRF2 Activation as a Prognostic Biomarker in T-Cell Acute Lymphoblastic Leukaemia. International Journal of Molecular Sciences, 24(12), 10350. https://doi.org/10.3390/ijms241210350







