p53 Affects Zeb1 Interactome of Breast Cancer Stem Cells
Abstract
:1. Introduction
2. Results
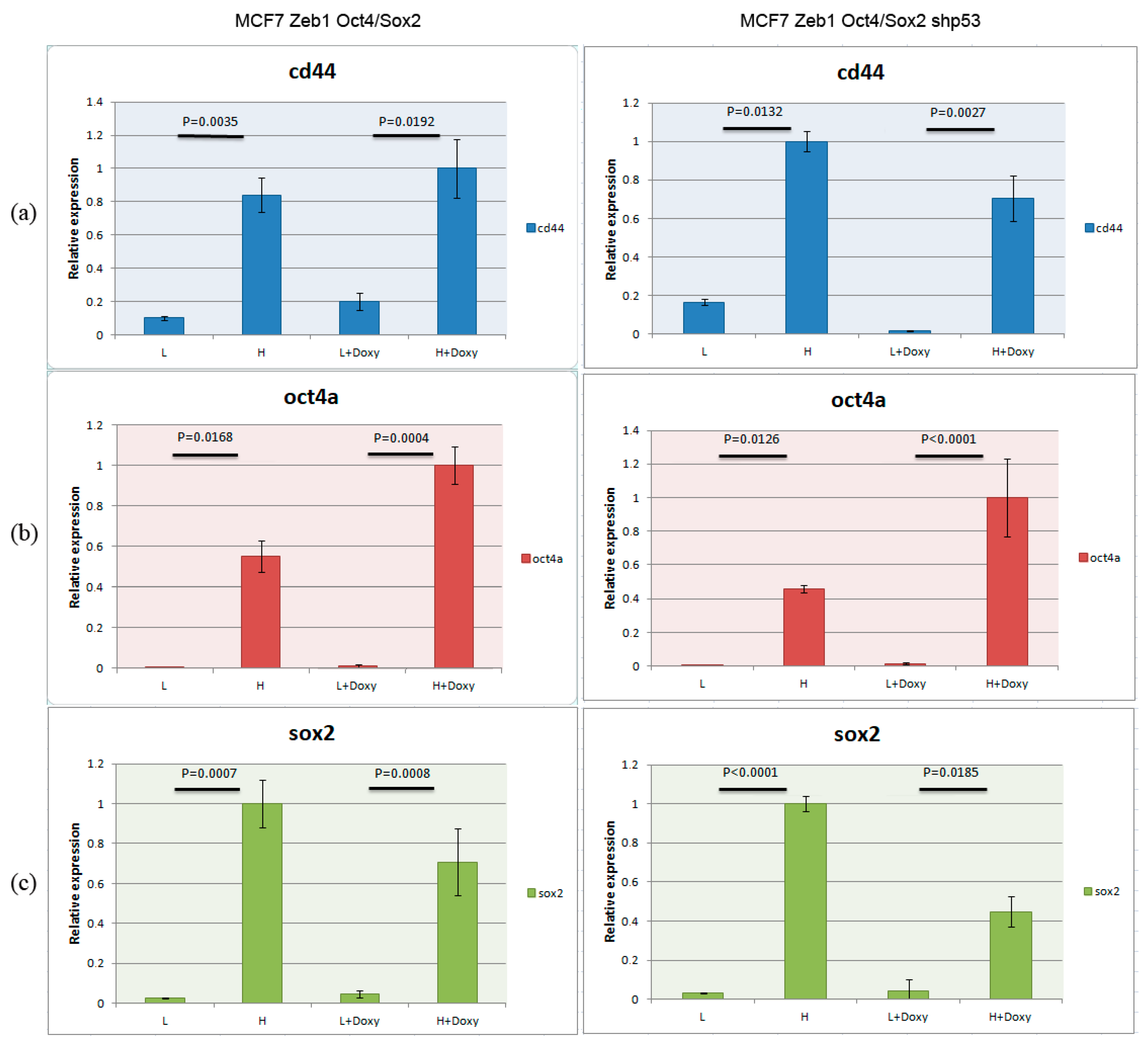
2.1. Cell Model Validation
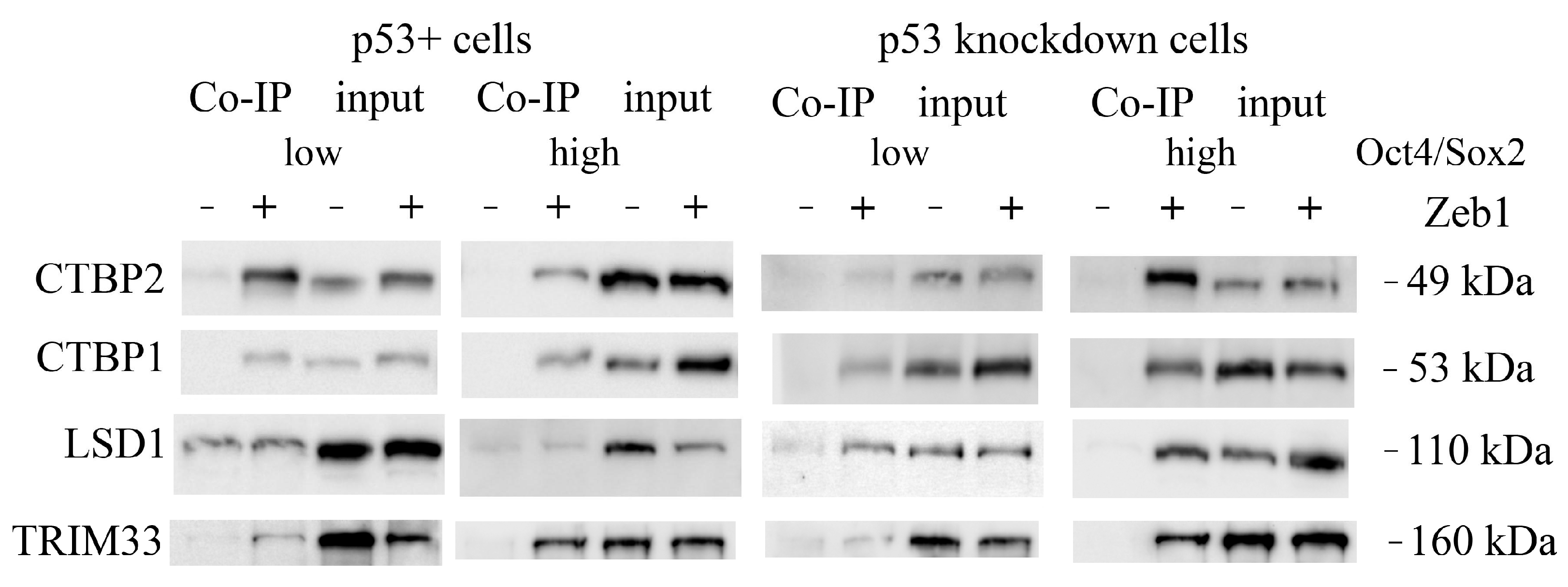
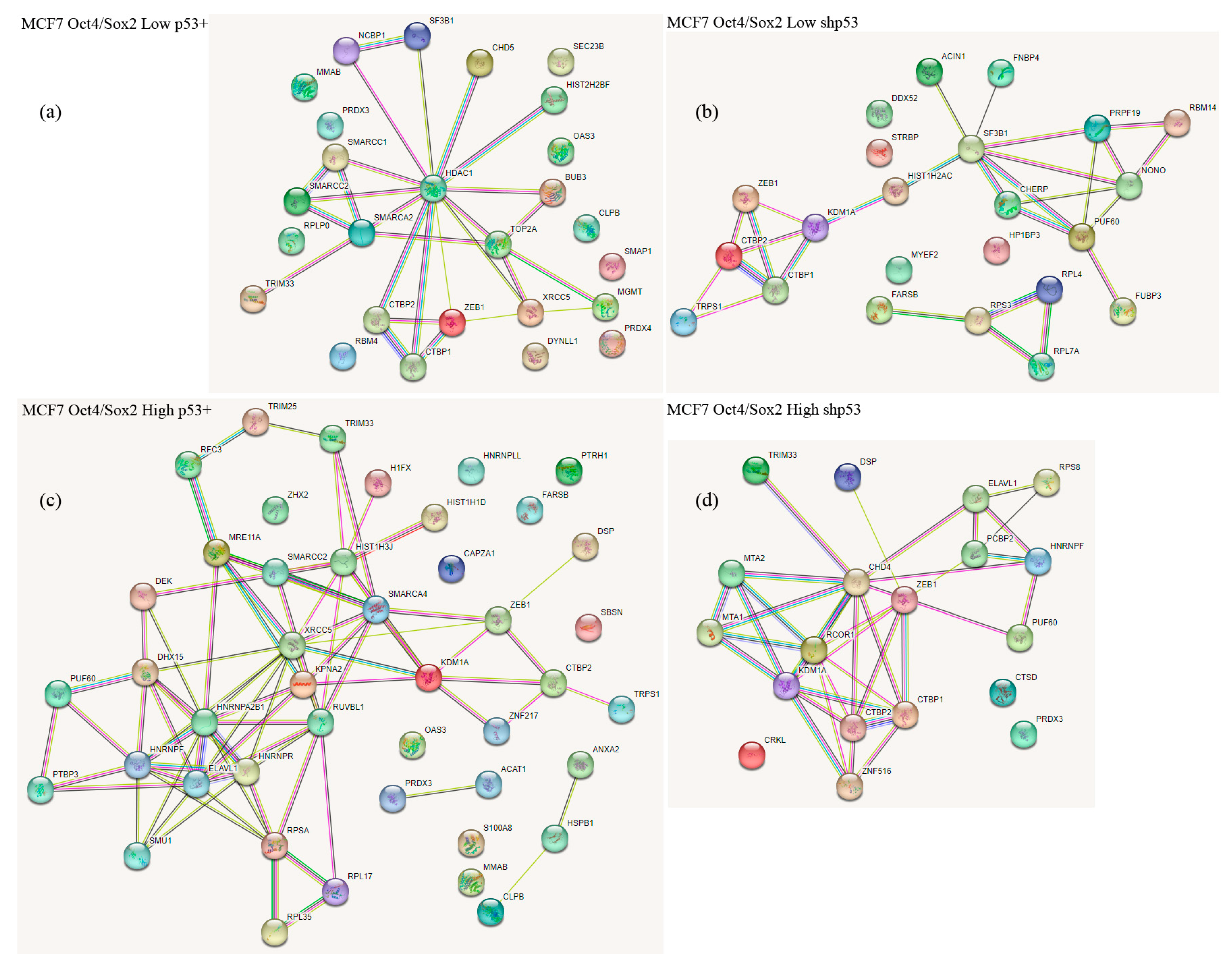
2.2. Co-Immunoprecipitation and Mass Spectrometric Identification of Zeb1 EMT-TF Interacting Proteins
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Co-Immunoprecipitation
4.3. Sample Preparation and LC-MALDI Mass Spectrometry
4.4. Western-Blotting
4.5. Real-Time Polymerase Chain Reaction
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCSCs | breast cancer stem cells |
| CHCA | α-cyanohydroxycinnamic acid |
| CMV | cytomegalovirus |
| CSCs | cancer stem cells |
| EMT | epithelial-to-mesenchymal transition |
| EMT-TF (TF-EMT) | transcription factors that govern EMT |
| ESRPs | epithelial splicing regulatory proteins |
| FDR | false discovery rate |
| GFP | green fluorescent protein |
| LC-MALDI TOF/TOF | liquid chromatography/matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry |
| OS-mC | Oct4/Sox2 mCherry reporter |
| SDS-PAGE | sodium dodecylsulphate polyacrylamide gel electrophoresis |
| SORE | Oct4/Sox2 responsive element |
References
- Liang, L.; Kaufmann, A.M. The Significance of Cancer Stem Cells and Epithelial–Mesenchymal Transition in Metastasis and Anti-Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 2555. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Pozdnyakov, D.Y.; Shuvalov, O.Y.; Barlev, N.A.; Mittenberg, A.G. Role of Zeb1 EMT-TF in metastasis and carcinogenesis. Tsitologiia 2019, 61, 915–925. (In Russian) [Google Scholar] [CrossRef]
- Pozdnyakov, D.Y.; Shuvalov, O.Y.; Barlev, N.A.; Mittenberg, A.G. Post-transcriptional regulation of Zeb1 EMT-TF activity in human breast carcinomae. Tsitologiia 2020, 62, 3–15. (In Russian) [Google Scholar] [CrossRef]
- Tevaarwerk, A.J.; Gray, R.J.; Schneider, B.P.; Smith, M.L.; Wagner, L.I.; Fetting, J.H.; Davidson, N.; Goldstein, L.J.; Miller, K.D.; Sparano, J.A. Survival in patients with metastatic recurrent breast cancer after adjuvant chemotherapy: Little evidence of improvement over the past 30 years. Cancer 2013, 119, 1140–1148. [Google Scholar] [CrossRef] [Green Version]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Morel, A.P.; Lièvre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [Green Version]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef]
- Tulchinsky, E.; Demidov, O.; Kriajevska, M.; Barlev, N.A.; Imyanitov, E. EMT: A mechanism for escape from EGFR-targeted therapy in lung cancer. BBA. Rev. Cancer 2019, 1871, 29–39. [Google Scholar] [CrossRef]
- Liu, S.; Dontu, G.; Mantle, I.D.; Patel, S.; Ahn, N.S.; Jackson, K.W.; Suri, P.; Wicha, M.S. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006, 66, 6063–6071. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Ghatak, D.; Das Ghosh, D.; Roychoudhury, S. Cancer Stemness: p53 at the Wheel. Front. Oncol. 2021, 10, 604124. [Google Scholar] [CrossRef]
- Parfenyev, S.; Singh, A.; Fedorova, O.; Daks, A.; Kulshreshtha, R.; Barlev, N.A. Interplay between p53 and non-coding RNAs in the regulation of EMT in breast cancer. Cell Death Dis. 2021, 12, 17. [Google Scholar] [CrossRef]
- Semenov, O.; Daks, A.; Fedorova, O.; Shuvalov, O.; Barlev, N.A. Opposing Roles of Wild-type and Mutant p53 in the Process of Epithelial to Mesenchymal Transition. Front. Mol. Biosci. 2022, 9, 928399. [Google Scholar] [CrossRef]
- Rizzotto, D.; Englmaier, L.; Villunger, A. At a Crossroads to Cancer: How p53-Induced Cell Fate Decisions Secure Genome Integrity. Int. J. Mol. Sci. 2021, 22, 10883. [Google Scholar] [CrossRef]
- Hu, S.; Ouyang, J.; Zheng, G.; Lu, Y.; Zhu, Q.; Wang, B.; Ye, L.; Zhu, C. Identification of mutant p53-specific proteins interaction network using TurboID-based proximity labeling. Biochem. Biophys. Res. Commun. 2022, 615, 163–171. [Google Scholar] [CrossRef]
- Moiseeva, T.N.; Bottrill, A.; Melino, G.; Barlev, N.A. DNA damage-induced ubiquitylation of proteasome controls its proteolytic activity. Oncotarget 2013, 4, 1338–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittenberg, A.G.; Moiseeva, T.N.; Barlev, N.A. Role of proteasomes in transcription and their regulation by covalent modifications. Front. Biosci. 2008, 13, 7184–7192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Ramirez, C.; Zhang, Z.; Warner, K.A.; Herzog, A.E.; Mantesso, A.; Zhang, Z.; Yoon, E.; Wang, S.; Wicha, M.S.; Nör, J.E. p53 Inhibits Bmi-1-driven Self-Renewal and Defines Salivary Gland Cancer Stemness. Clin. Cancer Res. 2022, 28, 4757–4770. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.; Miro, C.; Pezone, A.; Tramontano, A.; Di Cicco, E.; Sagliocchi, S.; Cicatiello, A.G.; Murolo, M.; Torabinejad, S.; Abbotto, E.; et al. Loss of p53 activates thyroid hormone via type 2 deiodinase and enhances DNA damage. Nat. Commun. 2023, 14, 1244. [Google Scholar] [CrossRef] [PubMed]
- Parfenyev, S.E.; Shabelnikov, S.V.; Pozdnyakov, D.Y.; Gnedina, O.O.; Adonin, L.S.; Barlev, N.A.; Mittenberg, A.G. Proteomic analysis of Zeb1 interactome in breast carcinoma cells. Molecules 2021, 26, 3143. [Google Scholar] [CrossRef]
- Pérez, G.; López-Moncada, F.; Indo, S.; Torres, M.J.; Castellón, E.A.; Contreras, H.R. Knockdown of ZEB1 reverses cancer stem cell properties in prostate cancer cells. Oncol. Rep. 2021, 45, 58. [Google Scholar] [CrossRef]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Gheldof, A.; Hulpiau, P.; van Roy, F.; De Craene, B.; Berx, G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell. Mol. Life Sci. CMLS 2012, 69, 2527–2541. [Google Scholar] [CrossRef]
- Zhou, C.; Jiang, H.; Zhang, Z.; Zhang, G.; Wang, H.; Zhang, Q.; Sun, P.; Xiang, R.; Yang, S. ZEB1 confers stem cell-like properties in breast cancer by targeting neurogenin-3. Oncotarget 2017, 8, 54388–54401. [Google Scholar] [CrossRef] [Green Version]
- Manshouri, R.; Coyaud, E.; Kundu, S.T.; Peng, D.H.; Stratton, S.A.; Alton, K.; Bajaj, R.; Fradette, J.J.; Minelli, R.; Peoples, M.D.; et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat. Commun. 2019, 10, 5125. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wei, Y.; Wang, L.; Debeb, B.G.; Yuan, Y.; Zhang, J.; Yuan, J.; Wang, M.; Chen, D.; Sun, Y.; et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014, 16, 864–875. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [Green Version]
- Davidovich, P.; Aksenova, V.; Petrova, V.; Tentler, D.; Orlova, D.; Smirnov, S.; Gurzhiy, V.; Okorokov, A.L.; Garabadzhiu, A.; Melino, G.; et al. Discovery of Novel Isatin-Based p53 Inducers. ACS Med. Chem. Lett. 2015, 6, 856–860. [Google Scholar] [CrossRef] [Green Version]
- Bulatov, E.; Sayarova, R.; Mingaleeva, R.; Miftakhova, R.; Gomzikova, M.; Ignatyev, Y.; Petukhov, A.; Davidovich, P.; Rizvanov, A.; Barlev, N.A. Isatin-Schiff base-copper (II) complex induces cell death in p53-positive tumors. Cell. Death Disc. 2018, 4, 103. [Google Scholar] [CrossRef] [Green Version]
- Fedorova, O.; Daks, A.; Petrova, V.; Petukhov, A.; Lezina, L.; Shuvalov, O.; Davidovich, P.; Kriger, D.; Lomert, E.; Tentler, D.; et al. Novel isatin-derived molecules activate p53 via interference with Mdm2 to promote apoptosis. Cell Cycle 2018, 17, 1917–1930. [Google Scholar] [CrossRef] [Green Version]
- Grinkevich, V.V.; Nikulenkov, F.; Shi, Y.; Enge, M.; Bao, W.; Maljukova, A.; Gluch, A.; Kel, A.; Sangfelt, O.; Selivanova, G. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell 2009, 15, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Scully, K.; Zhu, X.; Cai, L.; Zhang, J.; Prefontaine, G.G.; Krones, A.; Ohgi, K.A.; Zhu, P.; Garcia-Bassets, I.; et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007, 446, 882–887. [Google Scholar] [CrossRef]
- Shi, Y.; Sawada, J.; Sui, G.; Affar, B.; Whetstine, J.R.; Lan, F.; Ogawa, H.; Luke, M.P.; Nakatani, Y.; Shi, Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003, 422, 735–738. [Google Scholar] [CrossRef]
- Subramanian, T.; Chinnadurai, G. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 2003, 540, 255–258. [Google Scholar] [CrossRef] [Green Version]
- Kuppuswamy, M.; Vijayalingam, S.; Zhao, L.J.; Zhou, Y.; Subramanian, T.; Ryerse, J.; Chinnadurai, G. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol. Cell. Biol. 2008, 28, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lee, S.; Teh, C.E.; Bunting, K.; Ma, L.; Shannon, M.F. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Intl. Immunol. 2009, 21, 227–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolla, V.; Zhuang, T.; Higashi, M.; Naraparaju, K.; Brodeur, G.M. Role of CHD5 in human cancers: 10 years later. Cancer Res. 2014, 74, 652–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, J.K.; Webb, S.R.; Silva, A.P.; Saathoff, H.; Ryan, D.P.; Torrado, M.; Brofelth, M.; Parker, B.L.; Shepherd, N.E.; Mackay, J.P. CHD4 is a peripheral component of the nucleosome remodeling and deacetylase complex. J. Biol. Chem. 2016, 291, 15853–15866. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, X.; Zhang, H.; Liu, F.; Bi, Y.; Zhang, Y.; Cheng, C.; Liu, J. Knockdown of SF3B1 inhibits cell proliferation, invasion and migration triggering apoptosis in breast cancer via aberrant splicing. Breast Cancer 2020, 27, 464–476. [Google Scholar] [CrossRef]
- Daks, A.; Petukhov, A.; Fedorova, O.; Shuvalov, O.; Kizenko, A.; Tananykina, E.; Vasileva, E.; Semenov, O.; Bottrill, A.; Barlev, N. The RNA-binding protein HuR is a novel target of Pirh2 E3 ubiquitin ligase. Cell Death Dis. 2021, 12, 581. [Google Scholar] [CrossRef]
- Zhigalova, E.; Mittenberg, A. HuR prevents c-fos mRNA degradation by proteasome-associated ribonuclease in vitro. Porto Biomed. J. 2017, 2, 217–218. [Google Scholar] [CrossRef]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 2015, 34, 5729–5738. [Google Scholar] [CrossRef]
- Takayama, K.I.; Suzuki, T.; Tanaka, T.; Fujimura, T.; Takahashi, S.; Urano, T.; Ikeda, K.; Inoue, S. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene 2018, 37, 2165–2180. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wang, X.; Hu, W.; Feng, Z. Tumor suppressor p53 cross-talks with TRIM family proteins. Genes Dis. 2020, 8, 463–474. [Google Scholar] [CrossRef]
- Allton, K.; Jain, A.K.; Herz, H.M.; Tsai, W.W.; Jung, S.Y.; Qin, J.; Bergmann, A.; Johnson, R.L.; Barton, M.C. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 11612–11616. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Hatakeyama, S. TRIM proteins and diseases. J. Biochem. 2017, 161, 135–144. [Google Scholar] [CrossRef]
- Soule, H.D.; Vazguez, J.; Long, A.; Albert, S.; Brennan, M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973, 51, 1409–1416. [Google Scholar] [CrossRef]
- Tang, B.; Raviv, A.; Esposito, D.; Flanders, K.C.; Daniel, C.; Nghiem, B.T.; Garfield, S.; Lim, L.; Mannan, P.; Robles, A.I.; et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Rep. 2015, 4, 155–169. [Google Scholar] [CrossRef] [Green Version]


| Proteins Names | Number of Confident Peptides in MCF-7 Clones | |||
|---|---|---|---|---|
| LOW OS-mC p53+cells (L-P53WT) | HIGH OS-mC p53+cells (H-P53WT) | LOW OS-mC shp53 Cells (L-shP53) | HIGH OS-mC shp53 Cells (H-shP53) | |
| C-terminal-binding protein 2 (CTBP2) | 7 | 5 | 12 | 14 |
| 2′-5′-oligoadenylatesynthase 3 (OAS3) | 6 | 2 | 0 | 0 |
| Caseinolytic peptidase B protein homolog (CLPB) | 4 | 3 | 0 | 0 |
| Corrinoid adenosyltransferase (MMAB) | 2 | 2 | 0 | 0 |
| SWI/SNF complex sub-unit SMARCC2 (SMARCC2) | 6 | 6 | 0 | 0 |
| X-ray repair cross-complementing protein 5 (XRCC5) | 5 | 3 | 0 | 0 |
| Thioredoxin-dependent peroxide reductase, mitochondrial (PRDX3) | 6 | 5 | 0 | 4 |
| E3 ubiquitin-protein ligase TRIM33 (TRIM33) | 6 | 4 | 0 | 11 |
| C-terminal-binding protein 1 (CTBP1) | 6 | 0 | 11 | 10 |
| Splicing factor 3B sub-unit 1 (SF3B1) | 3 | 0 | 3 | 0 |
| Methylated-DNA--protein-cysteine methyltransferase (MGMT) | 5 | 0 | 0 | 3 |
| Heterogeneous nuclear ribonucleoprotein F (HNRNPF) | 0 | 9 | 0 | 4 |
| Poly(U)-binding-splicing factor PUF60 (PUF60) | 0 | 2 | 4 | 4 |
| Zinc finger transcription factor Trps1 (TRPS1) | 0 | 3 | 3 | 0 |
| Phenylalanine--tRNA ligase beta sub-unit (FARSB) | 0 | 4 | 4 | 0 |
| Desmoplakin (DSP) | 0 | 3 | 0 | 4 |
| ELAV-like protein 1 (ELAVL1) | 0 | 5 | 0 | 4 |
| Lysine-specific histone demethylase 1A (KDM1A) | 0 | 0 | 8 | 9 |
| Gene | 5′-3′ Primer Sequence |
|---|---|
| Oct4a | F: CTTCTGCTGATCCTGTCTGATG |
| R: TGCTGTGAAGGGAGATGTATTG | |
| Cd44 | F: GTCTCCTCTGACTTCAACAGCG |
| R: ACCACCCTGTTGCTGTAGCCAA | |
| Sox2 | F: GGCATACACCTACTCAACTACGG |
| R: TGGGCGGTGTAGAATCAGAGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parfenyev, S.E.; Shabelnikov, S.V.; Tolkunova, E.N.; Barlev, N.A.; Mittenberg, A.G. p53 Affects Zeb1 Interactome of Breast Cancer Stem Cells. Int. J. Mol. Sci. 2023, 24, 9806. https://doi.org/10.3390/ijms24129806
Parfenyev SE, Shabelnikov SV, Tolkunova EN, Barlev NA, Mittenberg AG. p53 Affects Zeb1 Interactome of Breast Cancer Stem Cells. International Journal of Molecular Sciences. 2023; 24(12):9806. https://doi.org/10.3390/ijms24129806
Chicago/Turabian StyleParfenyev, Sergey E., Sergey V. Shabelnikov, Elena N. Tolkunova, Nickolai A. Barlev, and Alexey G. Mittenberg. 2023. "p53 Affects Zeb1 Interactome of Breast Cancer Stem Cells" International Journal of Molecular Sciences 24, no. 12: 9806. https://doi.org/10.3390/ijms24129806







