Features of DNA–Montmorillonite Binding Visualized by Atomic Force Microscopy
Abstract
1. Introduction
2. Results and Discussion
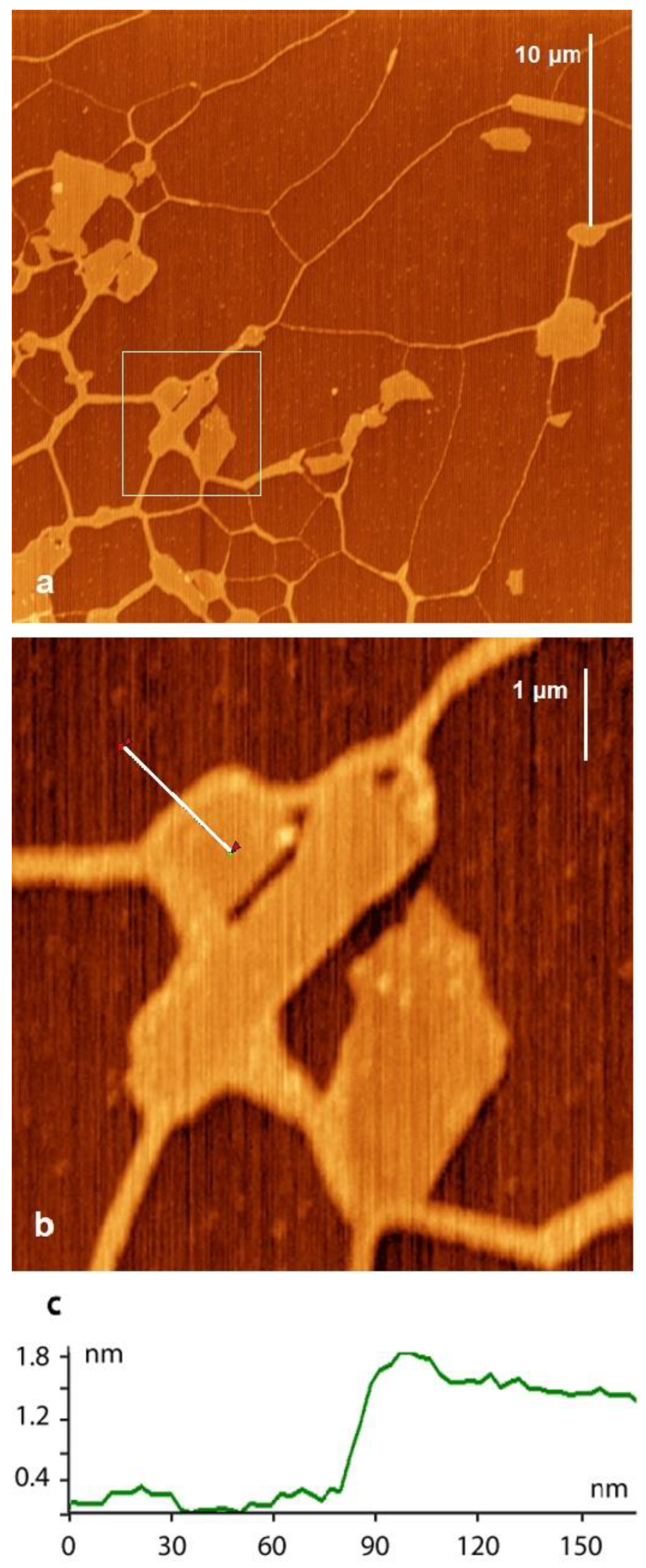
2.1. DNA and Mt in Distilled Water
2.2. DNA–Mt Interaction in a Mg2+ Solution
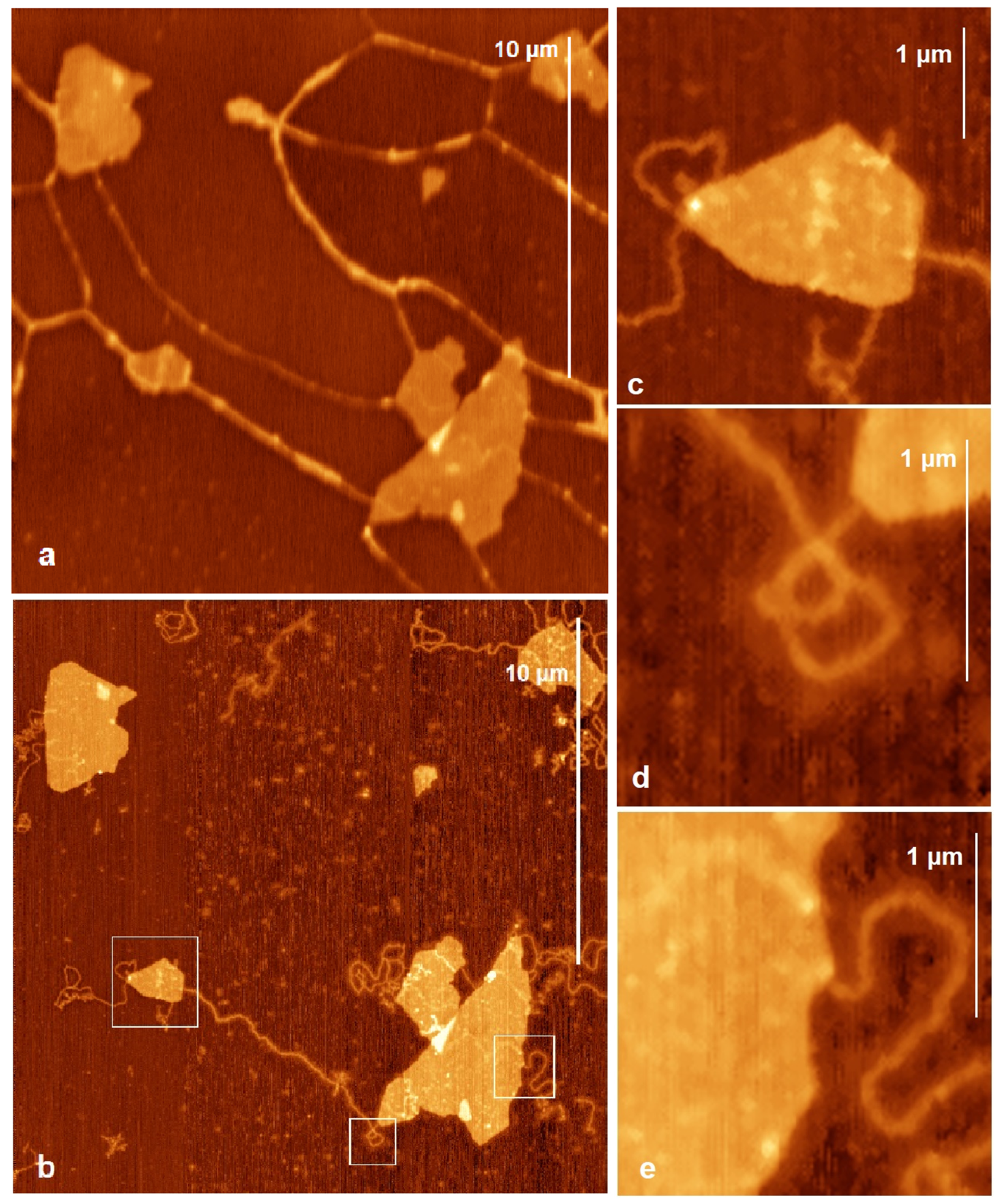
2.2.1. Rinsed DNA and Mt Samples with Mg2+ Solution
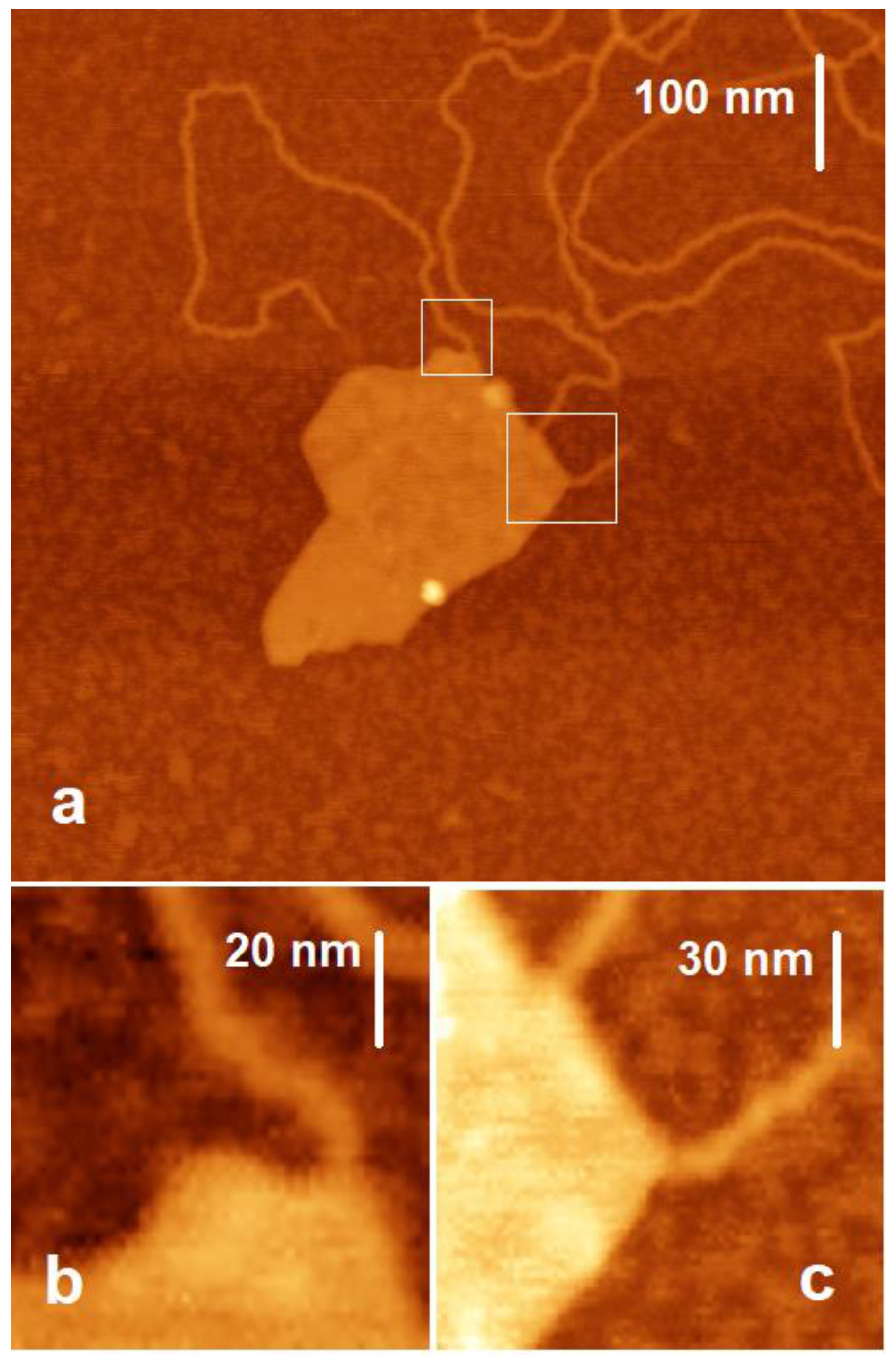
2.2.2. DNA and Mt Mixed in Mg2+ Solution
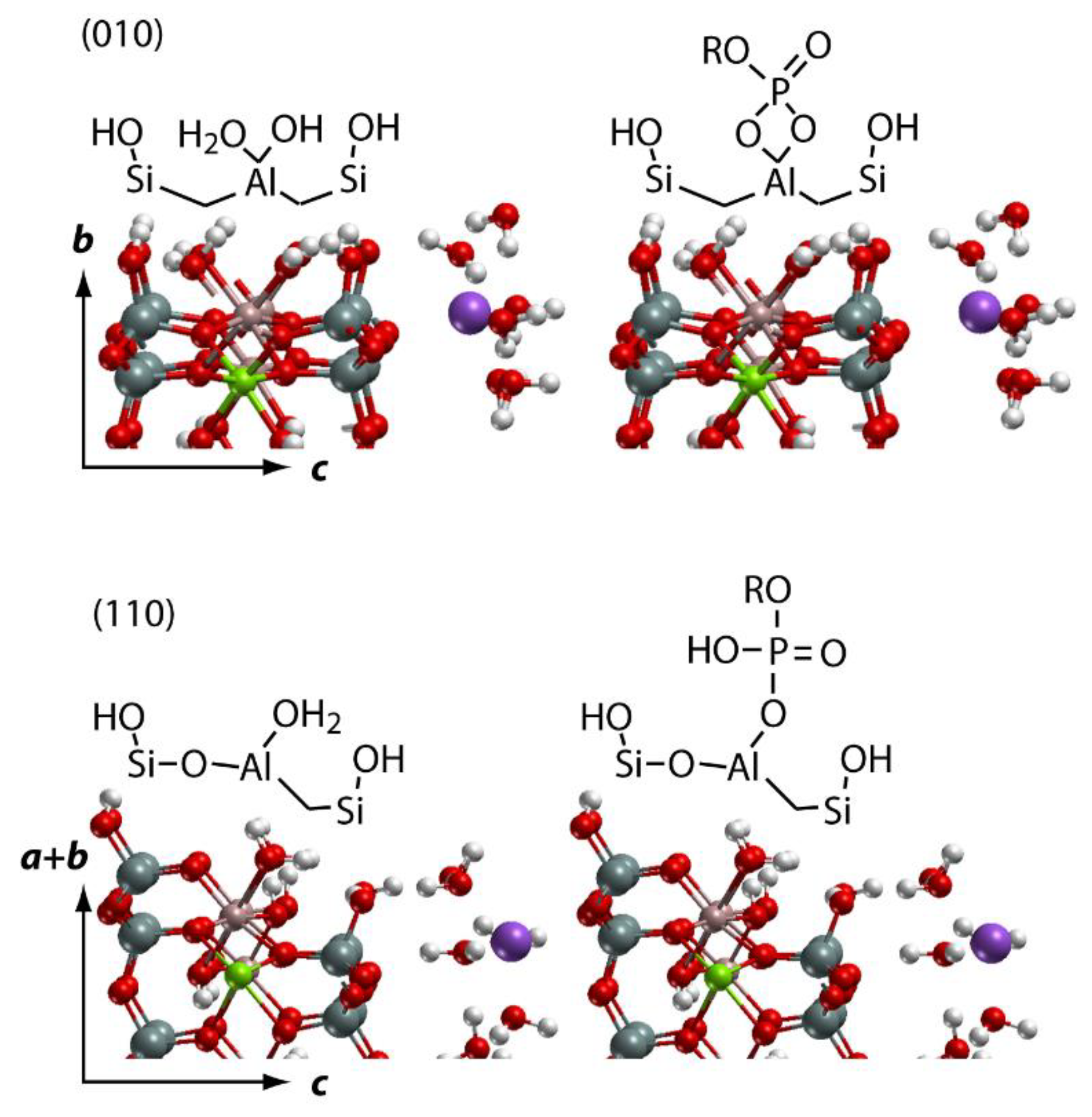
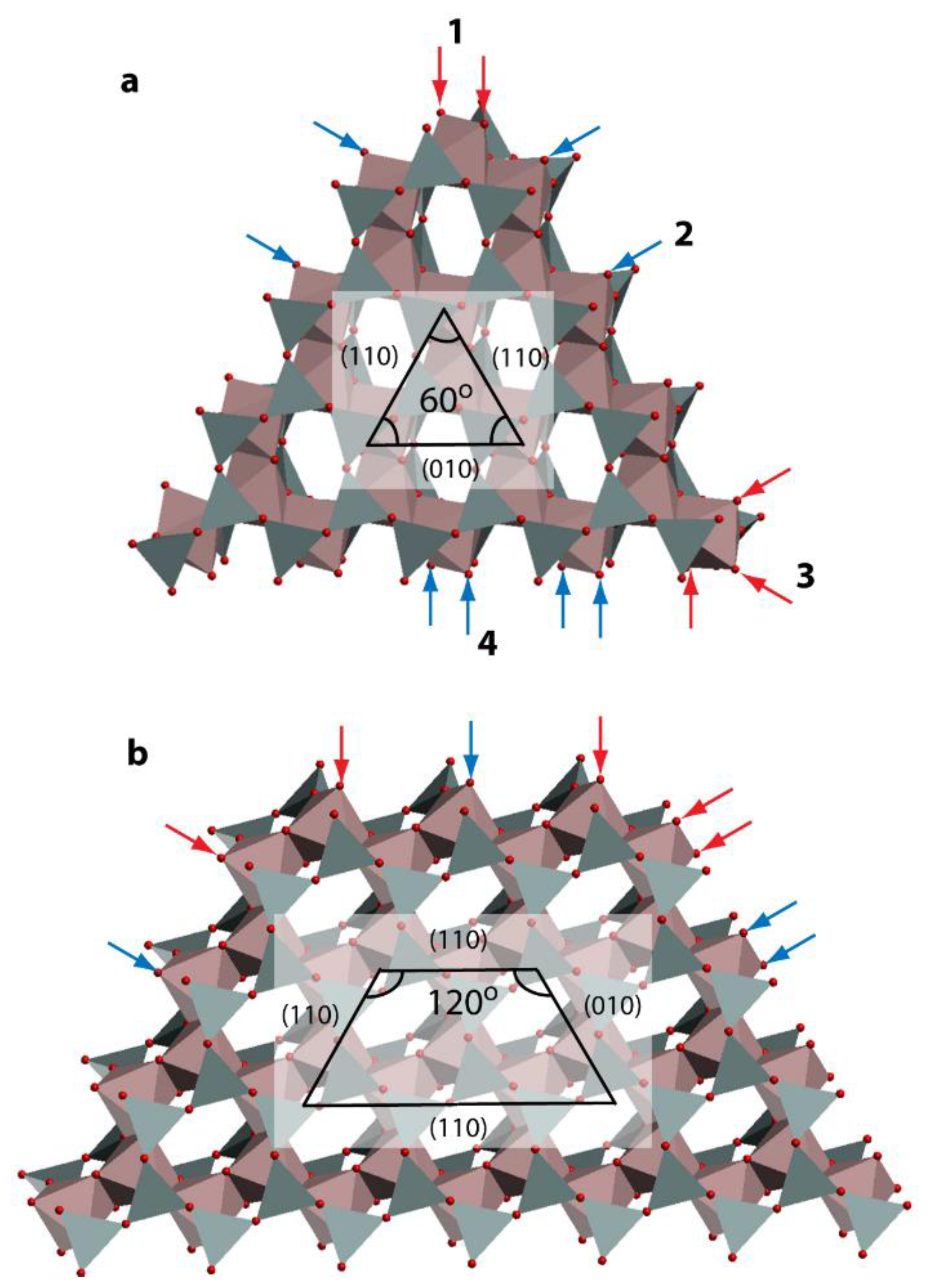
2.3. DFT-Based Interpretation of Edge Joint Reactivity
3. Materials and Methods
3.1. Sample Preparation
3.2. AFM
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on Nanoparticles and Nanostructured Materials: History, Sources, Toxicity and Regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef]
- Cavallaro, G.; Fakhrullin, R.; Pasbakhsh, P. Clay Nanoparticles: Properties and Applications, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Jeon, I.-Y.; Baek, J.-B. Nanocomposites Derived from Polymers and Inorganic Nanoparticles. Materials 2010, 3, 3654–3674. [Google Scholar] [CrossRef]
- Mousa, M.; Evans, N.D.; Oreffo, R.O.C.; Dawson, J.I. Clay Nanoparticles for Regenerative Medicine and Biomaterial Design: A Review of Clay Bioactivity. Biomaterials 2018, 159, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Gaharwar, A.K.; Mihaila, S.M.; Swami, A.; Patel, A.; Sant, S.; Reis, R.L.; Marques, A.P.; Gomes, M.E.; Khademhosseini, A. Bioactive Silicate Nanoplatelets for Osteogenic Differentiation of Human Mesenchymal Stem Cells. Adv. Mater. 2013, 25, 3329–3336. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C.; Sleiman, H.F. DNA Nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological Mass Production of DNA Origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]
- Bernal, J.D. The Physical Basis of Life. Proc. Phys. Soc. B 1949, 62, 752. [Google Scholar] [CrossRef]
- Huang, W.; Ferris, J.P. One-Step, Regioselective Synthesis of up to 50-Mers of RNA Oligomers by Montmorillonite Catalysis. J. Am. Chem. Soc. 2006, 128, 8914–8919. [Google Scholar] [CrossRef]
- Subramaniam, A.B.; Wan, J.; Gopinath, A.; Stone, H.A. Semi-Permeable Vesicles Composed of Natural Clay. Soft Matter 2011, 7, 2600–2612. [Google Scholar] [CrossRef]
- Saladino, R.; Crestini, C.; Ciambecchini, U.; Ciciriello, F.; Costanzo, G.; Di Mauro, E. Synthesis and Degradation of Nucleobases and Nucleic Acids by Formamide in the Presence of Montmorillonites. ChemBioChem 2004, 5, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Khanna, M.; Stotzky, G. Transformation of Bacillus Subtilis by DNA Bound on Montmorillonite and Effect of DNase on the Transforming Ability of Bound DNA. Appl. Environ. Microbiol. 1992, 58, 1930–1939. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Huang, Q.-Y.; Zhang, X.-W. Interactions of DNA with Clay Minerals and Soil Colloidal Particles and Protection against Degradation by DNase. Environ. Sci. Technol. 2006, 40, 2971–2976. [Google Scholar] [CrossRef] [PubMed]
- Pietramellara, G.; Ascher, J.; Ceccherini, M.T.; Nannipieri, P.; Wenderoth, D. Adsorption of Pure and Dirty Bacterial DNA on Clay Minerals and Their Transformation Frequency. Biol. Fertil. Soils 2007, 43, 731–739. [Google Scholar] [CrossRef]
- Kawase, M.; Hayashi, Y.; Kinoshita, F.; Yamato, E.; Miyazaki, J.; Yamakawa, J.; Ishida, T.; Tamura, M.; Yagi, K. Protective Effect of Montmorillonite on Plasmid DNA in Oral Gene Delivery into Small Intestine. Biol. Pharm. Bull. 2004, 27, 2049–2051. [Google Scholar] [CrossRef]
- Sironmani, T.A. Comparison of Nanocarriers for Gene Delivery and Nanosensing Using Montmorillonite, Silver Nanoparticles and Multiwalled Carbon Nanotubes. Appl. Clay Sci. 2015, 103, 55–61. [Google Scholar] [CrossRef]
- Jayrajsinh, S.; Shankar, G.; Agrawal, Y.K.; Bakre, L. Montmorillonite Nanoclay as a Multifaceted Drug-Delivery Carrier: A Review. J. Drug Deliv. Sci. Technol. 2017, 39, 200–209. [Google Scholar] [CrossRef]
- Repalli, S.K.; Geda, C.K.; Rao, G.J.N. MMT, a High Promising, Cost Effective Micro-Carrier for Gene Delivery. J. Microb. Biochem. Technol. 2019, 11, 417. [Google Scholar]
- Franchi, M.; Gallori, E. A Surface-Mediated Origin of the RNA World: Biogenic Activities of Clay-Adsorbed RNA Molecules. Gene 2005, 346, 205–214. [Google Scholar] [CrossRef]
- Pedreira-Segade, U.; Feuillie, C.; Pelletier, M.; Michot, L.J.; Daniel, I. Adsorption of Nucleotides onto Ferromagnesian Phyllosilicates: Significance for the Origin of Life. Geochim. Cosmochim. Acta 2016, 176, 81–95. [Google Scholar] [CrossRef]
- Pedreira-Segade, U.; Hao, J.; Razafitianamaharavo, A.; Pelletier, M.; Marry, V.; Le Crom, S.; Michot, L.J.; Daniel, I. How Do Nucleotides Adsorb Onto Clays? Life 2018, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Mignon, P.; Corbin, G.; Le Crom, S.; Marry, V.; Hao, J.; Daniel, I. Adsorption of Nucleotides on Clay Surfaces: Effects of Mineral Composition, PH and Solution Salts. Appl. Clay Sci. 2020, 190, 105544. [Google Scholar] [CrossRef]
- Khanna, M.; Yoder, M.; Calamai, L.; Stotzky, G. X-ray Diffractometry and Electron Microscopy of DNA from Bacillus Subtilis Bound on Clay Minerals. Sci. Soils 1998, 3, 1–10. [Google Scholar] [CrossRef]
- Franchi, M.; Bramanti, E.; Morassi Bonzi, L.; Luigi Orioli, P.; Vettori, C.; Gallori, E. Clay-Nucleic Acid Complexes: Characteristics and Implications for the Preservation of Genetic Material in Primeval Habitats. Orig. Life Evol. Biosph. 1999, 29, 297–315. [Google Scholar] [CrossRef] [PubMed]
- Delain, E.; Fourcade, A.; Poulin, J.-C.; Barbin, A.; Coulaud, D.; Cam, E.L.; Paris, E. Comparative Observations of Biological Specimens, Especially DNA and Filamentous Actin Molecules in Atomic Force, Tunnelling and Electron Microscopes. Microsc. Microanal. Microstruct. 1992, 3, 457–470. [Google Scholar] [CrossRef]
- Bezanilla, M.; Manne, S.; Laney, D.E.; Lyubchenko, Y.L.; Hansma, H.G. Adsorption of DNA to Mica, Silylated Mica, and Minerals: Characterization by Atomic Force Microscopy. Langmuir 1995, 11, 655–659. [Google Scholar] [CrossRef]
- Hansma, H.G.; Laney, D.E.; Bezanilla, M.; Sinsheimer, R.L.; Hansma, P.K. Applications for Atomic Force Microscopy of DNA. Biophys. J. 1995, 68, 1672–1677. [Google Scholar] [CrossRef]
- Allen, M.J.; Bradbury, E.M.; Balhorn, R. AFM Analysis of DNA-Protamine Complexes Bound to Mica. Nucleic. Acids. Res. 1997, 25, 2221–2226. [Google Scholar] [CrossRef]
- Sanchez-Sevilla, A.; Thimonier, J.; Marilley, M.; Rocca-Serra, J.; Barbet, J. Accuracy of AFM Measurements of the Contour Length of DNA Fragments Adsorbed on Mica in Air and in Aqueous Buffer. Ultramicroscopy 2002, 92, 151–158. [Google Scholar] [CrossRef]
- Sushko, M.L.; Shluger, A.L.; Rivetti, C. Simple Model for DNA Adsorption onto a Mica Surface in 1:1 and 2:1 Electrolyte Solutions. Langmuir 2006, 22, 7678–7688. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S. AFM for Analysis of Structure and Dynamics of DNA and Protein–DNA Complexes. Methods 2009, 47, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Jelavić, S.; Tobler, D.J.; Hassenkam, T.; Yoreo, J.J.D.; Stipp, S.L.S.; Sand, K.K. Prebiotic RNA Polymerisation: Energetics of Nucleotide Adsorption and Polymerisation on Clay Mineral Surfaces. Chem. Commun. 2017, 53, 12700–12703. [Google Scholar] [CrossRef] [PubMed]
- Pietramellara, G.; Franchi, M.; Gallori, E.; Nannipieri, P. Effect of Molecular Characteristics of DNA on Its Adsorption and Binding on Homoionic Montmorillonite and Kaolinite. Biol. Fertil. Soils 2001, 33, 402–409. [Google Scholar] [CrossRef]
- Paget, E.; Simonet, P. On the Track of Natural Transformation in Soil. FEMS Microbiol. Ecol. 1994, 15, 109–117. [Google Scholar] [CrossRef]
- Cai, P.; Huang, Q.; Zhang, X.; Chen, H. Adsorption of DNA on Clay Minerals and Various Colloidal Particles from an Alfisol. Soil Biol. Biochem. 2006, 38, 471–476. [Google Scholar] [CrossRef]
- Jenne, E. Adsorption of Metals by Geomedia; Elsevier: Amsterdam, The Netherlands, 1998; ISBN 978-0-12-384245-9. [Google Scholar]
- Churakov, S.V. Ab Initio Study of Sorption on Pyrophyllite: Structure and Acidity of the Edge Sites. J. Phys. Chem. B 2006, 110, 4135–4146. [Google Scholar] [CrossRef]
- Churakov, S.V. Structure and Dynamics of the Water Films Confined between Edges of Pyrophyllite: A First Principle Study. Geochim. Cosmochim. Acta 2007, 71, 1130–1144. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Wang, R.; Zhou, H.; Xu, S. Surface Complexes of Acetate on Edge Surfaces of 2:1 Type Phyllosilicate: Insights from Density Functional Theory Calculation. Geochim. Cosmochim. Acta 2008, 72, 5896–5907. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Meijer, E.J.; Wang, R.; Zhou, H. Atomic-Scale Structures of Interfaces between Phyllosilicate Edges and Water. Geochim. Cosmochim. Acta 2012, 81, 56–68. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Sprik, M.; Cheng, J.; Meijer, E.J.; Wang, R. Acidity of Edge Surface Sites of Montmorillonite and Kaolinite. Geochim. Cosmochim. Acta 2013, 117, 180–190. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J.; Sprik, M.; Lu, X.; Wang, R. Surface Acidity of 2:1-Type Dioctahedral Clay Minerals from First Principles Molecular Dynamics Simulations. Geochim. Cosmochim. Acta 2014, 140, 410–417. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J.; Sprik, M.; Lu, X.; Wang, R. Interfacial Structures and Acidity of Edge Surfaces of Ferruginous Smectites. Geochim. Cosmochim. Acta 2015, 168, 293–301. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Cheng, J.; Sprik, M.; Wang, R. Temperature Dependence of Interfacial Structures and Acidity of Clay Edge Surfaces. Geochim. Cosmochim. Acta 2015, 160, 91–99. [Google Scholar] [CrossRef]
- Churakov, S.V.; Dähn, R. Zinc Adsorption on Clays Inferred from Atomistic Simulations and EXAFS Spectroscopy. Environ. Sci. Technol. 2012, 46, 5713–5719. [Google Scholar] [CrossRef]
- Kremleva, A.; Krüger, S.; Rösch, N. Uranyl Adsorption at Solvated Edge Surfaces of 2:1 Smectites. A Density Functional Study. Phys. Chem. Chem. Phys. 2015, 17, 13757–13768. [Google Scholar] [CrossRef]
- Kremleva, A.; Krüger, S.; Rösch, N. Toward a Reliable Energetics of Adsorption at Solvated Mineral Surfaces: A Computational Study of Uranyl(VI) on 2:1 Clay Minerals. J. Phys. Chem. C 2016, 120, 324–335. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, X.; Lu, X.; He, M.; Jan Meijer, E.; Wang, R. Surface Complexation of Heavy Metal Cations on Clay Edges: Insights from First Principles Molecular Dynamics Simulation of Ni(II). Geochim. Cosmochim. Acta 2017, 203, 54–68. [Google Scholar] [CrossRef]
- Gao, P.; Liu, X.; Guo, Z.; Tournassat, C. Acid–Base Properties of Cis-Vacant Montmorillonite Edge Surfaces: A Combined First-Principles Molecular Dynamics and Surface Complexation Modeling Approach. Environ. Sci. Technol. 2023, 57, 1342–1352. [Google Scholar] [CrossRef]
- Pedreira-Segade, U.; Michot, L.J.; Daniel, I. Effects of Salinity on the Adsorption of Nucleotides onto Phyllosilicates. Phys. Chem. Chem. Phys. 2018, 20, 1938–1952. [Google Scholar] [CrossRef]
- Prescott, L.; Harley, J.; Klein, D. Microbiology, 3rd ed.; Wm.C. Brown Publishers: Dubuque, IA, USA, 1996. [Google Scholar]
- Tang, J.; Li, J.; Wang, C.; Bai, C. Enhancement of Resolution of DNA on Silylated Mica Using Atomic Force Microscopy. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. Process. Meas. Phenom. 2000, 18, 1858–1860. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Anraku, S.; Paineau, E.; Safinya, C.R.; Davidson, P.; Michot, L.J.; Miyamoto, N. Swelling Inhibition of Liquid Crystalline Colloidal Montmorillonite and Beidellite Clays by DNA. Sci. Rep. 2018, 8, 4367. [Google Scholar] [CrossRef]
- Bergaya, F.; Theng, B.K.G.; Lagaly, G. Handbook of Clay Science, Volume 1; Developments in Clay Science; Elsevier Science: Amsterdam, The Netherlands, 2006; Volume 1. [Google Scholar]
- Hartman, P.; Perdok, W.G. On the Relations between Structure and Morphology of Crystals. II. Acta Cryst. 1955, 8, 521–524. [Google Scholar] [CrossRef]
- Hartman, P.; Perdok, W.G. On the Relations between Structure and Morphology of Crystals. III. Acta Cryst. 1955, 8, 525–529. [Google Scholar] [CrossRef]
- Hartman, P.; Perdok, W.G. IUCr On the Relations between Structure and Morphology of Crystals. I. Available online: http://scripts.iucr.org/cgi-bin/paper?S0365110X55000121 (accessed on 14 November 2019).
- White, G.N.; Zelazny, L.W. Analysis and Implications of the Edge Structure of Dioctahedral Phyllosilicates. Clays Clay Miner. 1988, 36, 141–146. [Google Scholar] [CrossRef]
- Kuwahara, Y. In-Situ AFM Study of Smectite Dissolution under Alkaline Conditions at Room Temperature. Am. Mineral. 2006, 91, 1142–1149. [Google Scholar] [CrossRef]
- Kraevsky, S.V.; Tournassat, C.; Vayer, M.; Warmont, F.; Grangeon, S.; Ngouana Wakou, B.F.; Kalinichev, A.G. Identification of Montmorillonite Particle Edge Orientations by Atomic-Force Microscopy. Appl. Clay Sci. 2020, 186, 105442. [Google Scholar] [CrossRef]
- Churakov, S.V.; Liu, X. 3-Quantum-Chemical Modelling of Clay Mineral Surfaces and Clay Mineral–Surface–Adsorbate Interactions. In Developments in Clay Science; Schoonheydt, R., Johnston, C.T., Bergaya, F., Eds.; Surface and Interface Chemistry of Clay Minerals; Elsevier: Amsterdam, The Netherlands, 2018; Volume 9, pp. 49–87. [Google Scholar]
- Kresse, G.; Hafner, J. Ab Initio Molecular Dynamics for Liquid Metals. Phys. Rev. B 1993, 47, 558–561. [Google Scholar] [CrossRef]
- Kresse, G.; Hafner, J. Ab Initio Molecular-Dynamics Simulation of the Liquid-Metal--Amorphous-Semiconductor Transition in Germanium. Phys. Rev. B 1994, 49, 14251–14269. [Google Scholar] [CrossRef]
- Kresse, G.; Furthmüller, J. Efficiency of Ab-Initio Total Energy Calculations for Metals and Semiconductors Using a Plane-Wave Basis Set. Comput. Mater. Sci. 1996, 6, 15–50. [Google Scholar] [CrossRef]
- Kresse, G.; Furthmüller, J. Efficient Iterative Schemes for Ab Initio Total-Energy Calculations Using a Plane-Wave Basis Set. Phys. Rev. B 1996, 54, 11169–11186. [Google Scholar] [CrossRef]
- Napierska, D.; Thomassen, L.C.; Lison, D.; Martens, J.A.; Hoet, P.H. The Nanosilica Hazard: Another Variable Entity. Part Fibre. Toxicol. 2010, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-K.; Huang, P.-K.; Law, W.-C.; Chu, C.-H.; Chen, N.-T.; Lo, L.-W. Biodegradable Polymers for Gene-Delivery Applications. IJN 2020, 15, 2131–2150. [Google Scholar] [CrossRef] [PubMed]
- Tachi, Y.; Yotsuji, K. Diffusion and Sorption of Cs+, Na+, I− and HTO in Compacted Sodium Montmorillonite as a Function of Porewater Salinity: Integrated Sorption and Diffusion Model. Geochim. Cosmochim. Acta 2014, 132, 75–93. [Google Scholar] [CrossRef]
- Orucoglu, E.; Tournassat, C.; Robinet, J.-C.; Madé, B.; Lundy, M. From Experimental Variability to the Sorption Related Retention Parameters Necessary for Performance Assessment Models for Nuclear Waste Disposal Systems: The Example of Pb Adsorption on Clay Minerals. Appl. Clay Sci. 2018, 163, 20–32. [Google Scholar] [CrossRef]
- Klinov, D.; Magonov, S. True Molecular Resolution in Tapping-Mode Atomic Force Microscopy with High-Resolution Probes. Appl. Phys. Lett. 2004, 84, 2697–2699. [Google Scholar] [CrossRef]
- Prokhorov, V.V.; Klinov, D.V.; Chinarev, A.A.; Tuzikov, A.B.; Gorokhova, I.V.; Bovin, N.V. High-Resolution Atomic Force Microscopy Study of Hexaglycylamide Epitaxial Structures on Graphite. Langmuir 2011, 27, 5879–5890. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868, Erratum in Phys. Rev. Lett. 1997, 78, 1396. [Google Scholar] [CrossRef]
- Blöchl, P.E. Projector Augmented-Wave Method. Phys. Rev. B 1994, 50, 17953–17979. [Google Scholar] [CrossRef]
- Kresse, G.; Joubert, D. From Ultrasoft Pseudopotentials to the Projector Augmented-Wave Method. Phys. Rev. B 1999, 59, 1758–1775. [Google Scholar] [CrossRef]
- Ahlrichs, R.; Bär, M.; Häser, M.; Horn, H.; Kölmel, C. Electronic Structure Calculations on Workstation Computers: The Program System Turbomole. Chem. Phys. Lett. 1989, 162, 165–169. [Google Scholar] [CrossRef]
- Treutler, O.; Ahlrichs, R. Efficient Molecular Numerical Integration Schemes. J. Chem. Phys. 1995, 102, 346–354. [Google Scholar] [CrossRef]
- Klamt, A.; Schüürmann, G. COSMO: A New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient. J. Chem. Soc. Perkin Trans. 2 1993, 799–805. [Google Scholar] [CrossRef]
- Eichkorn, K.; Treutler, O.; Öhm, H.; Häser, M.; Ahlrichs, R. Auxiliary Basis Sets to Approximate Coulomb Potentials. Chem. Phys. Lett. 1995, 240, 283–290. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully Optimized Contracted Gaussian Basis Sets for Atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Dillen, P.M.W.; Noordaa, J. van der Rapid and Simple Method for Purification of Nucleic Acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraevsky, S.V.; Barinov, N.A.; Morozova, O.V.; Palyulin, V.V.; Kremleva, A.V.; Klinov, D.V. Features of DNA–Montmorillonite Binding Visualized by Atomic Force Microscopy. Int. J. Mol. Sci. 2023, 24, 9827. https://doi.org/10.3390/ijms24129827
Kraevsky SV, Barinov NA, Morozova OV, Palyulin VV, Kremleva AV, Klinov DV. Features of DNA–Montmorillonite Binding Visualized by Atomic Force Microscopy. International Journal of Molecular Sciences. 2023; 24(12):9827. https://doi.org/10.3390/ijms24129827
Chicago/Turabian StyleKraevsky, Sergey V., Nikolay A. Barinov, Olga V. Morozova, Vladimir V. Palyulin, Alena V. Kremleva, and Dmitry V. Klinov. 2023. "Features of DNA–Montmorillonite Binding Visualized by Atomic Force Microscopy" International Journal of Molecular Sciences 24, no. 12: 9827. https://doi.org/10.3390/ijms24129827
APA StyleKraevsky, S. V., Barinov, N. A., Morozova, O. V., Palyulin, V. V., Kremleva, A. V., & Klinov, D. V. (2023). Features of DNA–Montmorillonite Binding Visualized by Atomic Force Microscopy. International Journal of Molecular Sciences, 24(12), 9827. https://doi.org/10.3390/ijms24129827





