Microcystin-LR-Induced Interaction between M2 Tumor-Associated Macrophage and Colorectal Cancer Cell Promotes Colorectal Cancer Cell Migration through Regulating the Expression of TGF-β1 and CST3
Abstract
:1. Introduction
2. Results
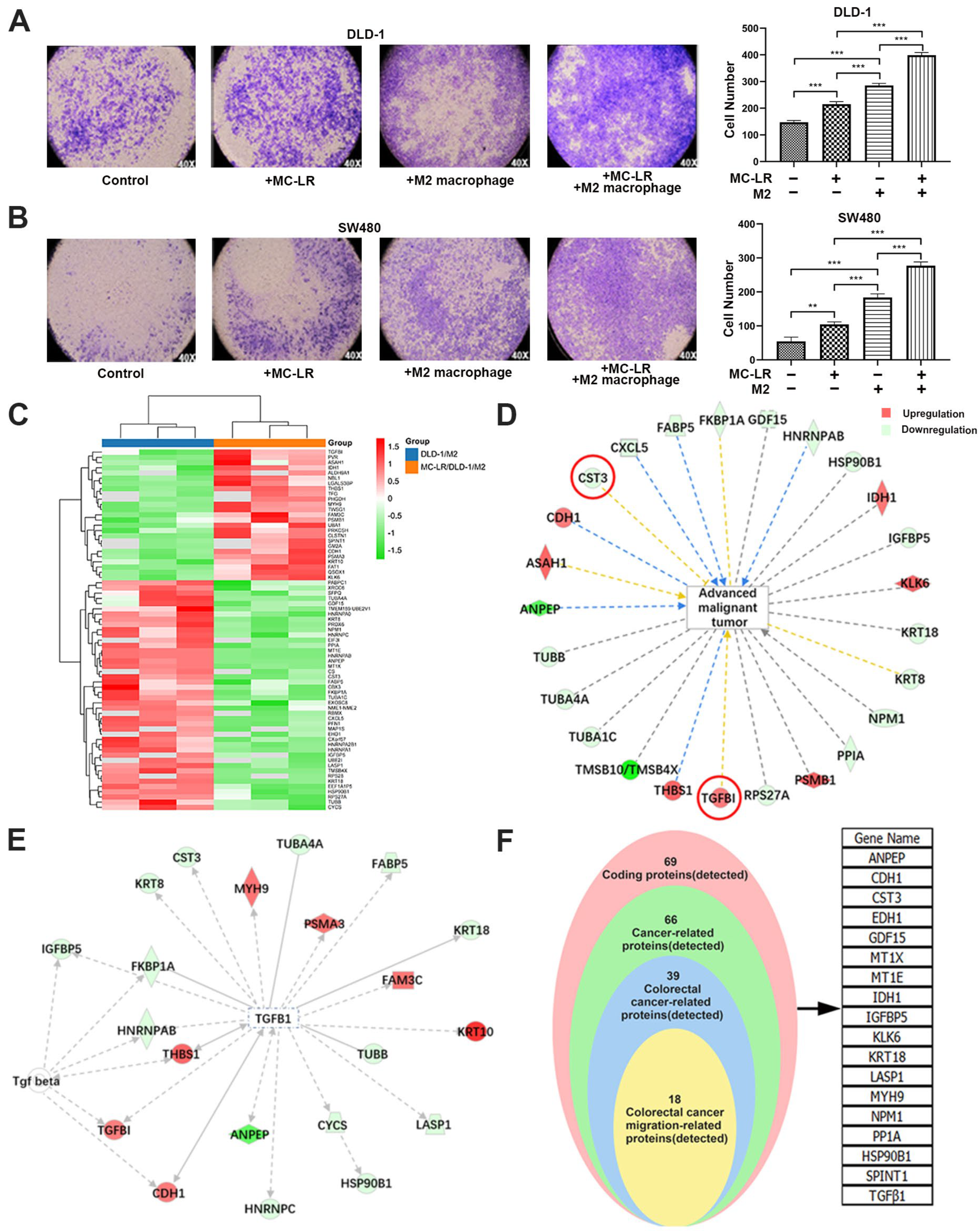
2.1. MC-LR Promoted the Migration of CRC Cells and the Change of Secreted Protein
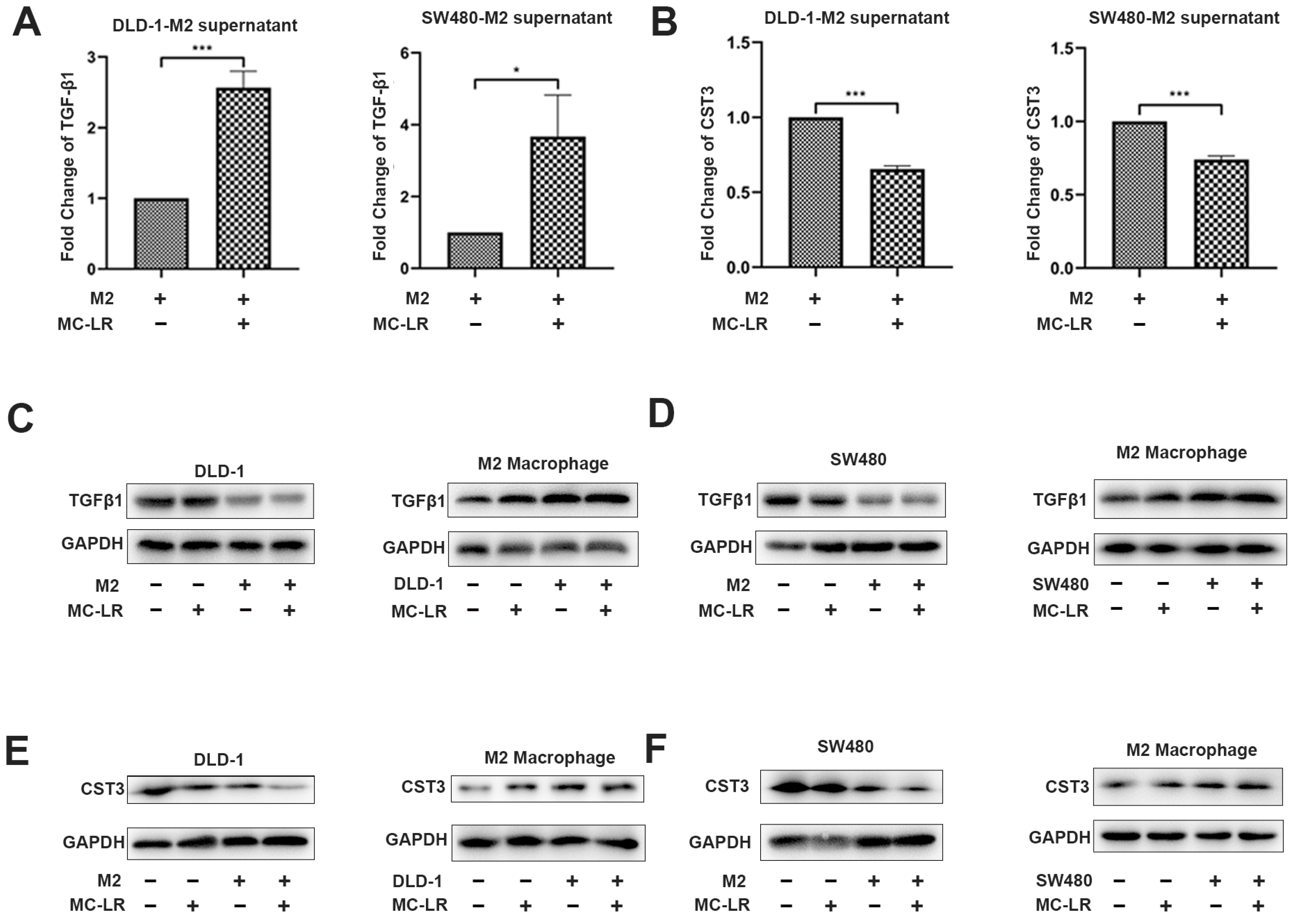
2.2. MC-LR Promoted the Expression of TGF-β1 and Reduced the Expression of CST3 in CRC Cell–M2 Macrophage Co-Culture System
2.3. The Expression of TGF-β1 and CST3 in Co-Culture System Regulated by MC-LR Prompted the Migration of CRC Cells
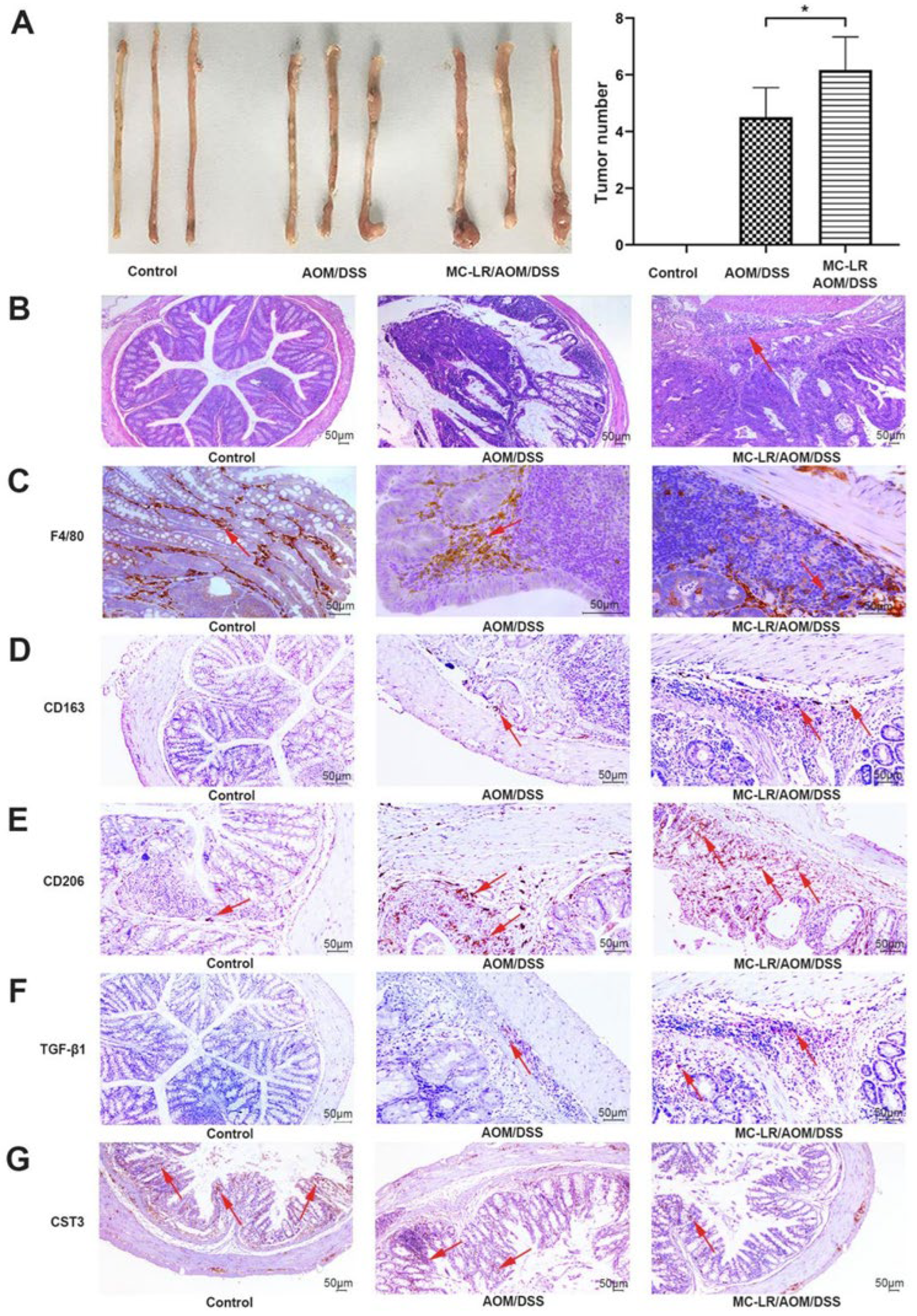
2.4. MC-LR Increased M2 Macrophage Infiltration and Decreased the Expression of CST3 to Exacerbate Tumor Progression in AOM/DSS-Model Mice
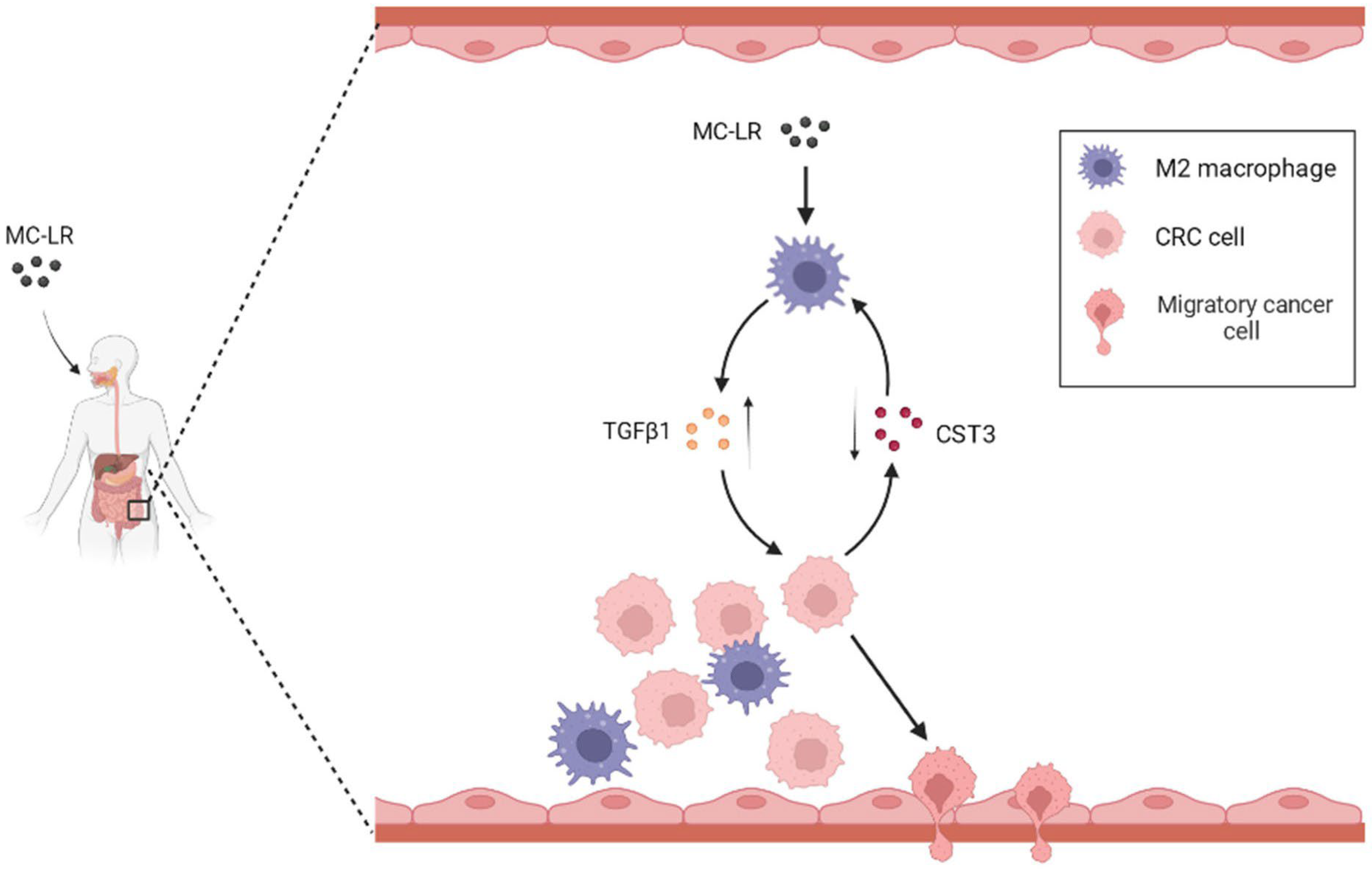
3. Discussion
4. Materials and Methods
4.1. Cell Culture and THP-1 Polarization
4.2. Co-Culture of CRC Cells and M2 Macrophages
4.3. Animal Experiments
4.4. Flow Cytometry
4.5. Western Blotting
4.6. Quantitative Real-Time Polymerase Chain Reaction
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Transfection In Vitro
4.9. Transwell Migration Assay
4.10. Hematoxylin & Eosin (H&E) Staining
4.11. Immunohistochemistry (IHC)
4.12. Isobaric Tags for Relative and Absolute Quantification (iTRAQ)
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, Q.; Liu, K.; Song, Z.; Sun, R.; Zhang, J.; Yin, L.; Pu, Y. Effects of Microcystin-LR on Metabolic Functions and Structure Succession of Sediment Bacterial Community under Anaerobic Conditions. Toxins 2020, 12, 183. [Google Scholar] [PubMed] [Green Version]
- MacKintosh, C.; Beattie, K.A.; Klumpp, S.; Cohen, P.; Codd, G.A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 1990, 264, 187–192. [Google Scholar] [PubMed] [Green Version]
- Laureano-Rosario, A.E.; McFarland, M.; Bradshaw, D.J.; Metz, J.; Brewton, R.A.; Pitts, T.; Perricone, C.; Schreiber, S.; Stockley, N.; Wang, G.; et al. Dynamics of microcystins and saxitoxin in the Indian River Lagoon, Florida. Harmful Algae 2021, 103, 102012. [Google Scholar] [PubMed]
- Xu, Q.; Chen, W.; Gao, G. Seasonal variations in microcystin concentrations in Lake Taihu, China. Environ. Monit. Assess. 2008, 145, 75–79. [Google Scholar]
- Lone, Y.; Koiri, R.K.; Bhide, M. An overview of the toxic effect of potential human carcinogen Microcystin-LR on testis. Toxicol. Rep. 2015, 2, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Zhang, L.; Chen, X.; Xiang, Z.; Li, D.; Han, X. miR-541 Contributes to Microcystin-LR-Induced Reproductive Toxicity through Regulating the Expression of p15 in Mice. Toxins 2016, 8, 260. [Google Scholar]
- Yi, X.; Xu, S.; Huang, F.; Wen, C.; Zheng, S.; Feng, H.; Guo, J.; Chen, J.; Feng, X.; Yang, F. Effects of Chronic Exposure to Microcystin-LR on Kidney in Mice. Int. J. Environ. Res. Public Health 2019, 16, 5030. [Google Scholar]
- Yuan, G.; Xie, P.; Zhang, X.; Tang, R.; Gao, Y.; Li, D.; Li, L. In vivo studies on the immunotoxic effects of microcystins on rabbit. Environ. Toxicol. 2010, 27, 83–89. [Google Scholar]
- Lone, Y.; Bhide, M.; Koiri, R.K. Microcystin-LR Induced Immunotoxicity in Mammals. J. Toxicol. 2016, 2016, 8048125. [Google Scholar] [CrossRef] [Green Version]
- Falconer, I.R.; Yeung, D.S.K. Cytoskeletal changes in hepatocytes induced by Microcystis toxins and their relation to hyperphosphorylation of cell proteins. Chem. Biol. Interact. 1992, 81, 181–196. [Google Scholar]
- Liu, W.; Wang, L.; Yang, X.; Zeng, H.; Zhang, R.; Pu, C.; Zheng, C.; Tan, Y.; Luo, Y.; Feng, X.; et al. Environmental Microcystin Exposure Increases Liver Injury Risk Induced by Hepatitis B Virus Combined with Aflatoxin: A Cross-Sectional Study in Southwest China. Environ. Sci. Technol. 2017, 51, 6367–6378. [Google Scholar]
- Hernández, J.M.; López-Rodas, V.; Costas, E. Microcystins from tap water could be a risk factor for liver and colorectal cancer: A risk intensified by global change. Med. Hypotheses 2009, 72, 539–540. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.-X.; Miao, C.; Fu, Z.; Li, Z.; Zhang, G.; Zheng, M.; Liu, Y.; Yang, L.; Wang, T. Promotion of Melanoma Cell Invasion and Tumor Metastasis by Microcystin-LR via Phosphatidylinositol 3-Kinase/AKT Pathway. Environ. Sci. Technol. 2013, 47, 8801–8808. [Google Scholar]
- Gan, N.; Sun, X.; Song, L. Activation of Nrf2 by Microcystin-LR Provides Advantages for Liver Cancer Cell Growth. Chem. Res. Toxicol. 2010, 23, 1477–1484. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Mayta, A.; Mrdjen, I.; Weghorst, C.; Knobloch, T. Microcystis toxin-mediated tumor promotion and toxicity lead to shifts in mouse gut microbiome. Ecotoxicol. Environ. Saf. 2020, 206, 111204. [Google Scholar]
- Ren, Y.; Yang, M.; Chen, M.; Zhu, Q.; Zhou, L.; Qin, W.; Wang, T. Microcystin-LR promotes epithelial-mesenchymal transition in colorectal cancer cells through PI3-K/AKT and SMAD2. Toxicol. Lett. 2017, 265, 53–60. [Google Scholar] [CrossRef]
- Ren, Y.; Yang, M.; Ma, R.; Gong, Y.; Zou, Y.; Wang, T.; Wu, J. Microcystin-LR promotes migration via the cooperation between microRNA-221/PTEN and STAT3 signal pathway in colon cancer cell line DLD-1. Ecotoxicol. Environ. Saf. 2019, 167, 107–113. [Google Scholar]
- Wang, Y.; Smith, W.; Hao, D.; He, B.; Kong, L. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int. Immunopharmacol. 2019, 70, 459–466. [Google Scholar]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 58. [Google Scholar]
- Li, R.; Zhou, R.; Wang, H.; Li, W.; Pan, M.; Yao, X.; Zhan, W.; Yang, S.; Xu, L.; Ding, Y.; et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019, 26, 2447–2463. [Google Scholar]
- Ren, B.; Cui, M.; Yang, G.; Wang, H.; Feng, M.; You, L.; Zhao, Y. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol. Cancer 2018, 17, 108. [Google Scholar] [PubMed] [Green Version]
- Zhao, J.; Li, H.; Zhao, S.; Wang, E.; Zhu, J.; Feng, D.; Zhu, Y.; Dou, W.; Fan, Q.; Hu, J.; et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer 2021, 20, 46. [Google Scholar] [PubMed]
- Chen, A. Chitinase-3-like 1 protein complexes modulate macrophage-mediated immune suppression in glioblastoma. J. Clin. Investig. 2021, 131, 209–214. [Google Scholar]
- Badawi, M.A.; Abouelfadl, D.M.; El-Sharkawy, S.L.; Abd El-Aal, W.E.; Abbas, N.F. Tumor-Associated Macrophage (TAM) and Angiogenesis in Human Colon Carcinoma. Open Access Maced. J. Med. Sci. 2015, 3, 209–214. [Google Scholar]
- Hwang, I.; Kim, J.W.; Ylaya, K.; Chung, E.J.; Kitano, H.; Perry, C.; Hanaoka, J.; Fukuoka, J.; Chung, J.-Y.; Hewitt, S.M. Tumor-associated macrophage, angiogenesis and lymphangiogenesis markers predict prognosis of non-small cell lung cancer patients. J. Transl. Med. 2020, 18, 443. [Google Scholar]
- Zeng, Y.-J.; Lai, W.; Wu, H.; Liu, L.; Xu, H.-Y.; Wang, J.; Chu, Z.-H. Neuroendocrine-like cells -derived CXCL10 and CXCL11 induce the infiltration of tumor-associated macrophage leading to the poor prognosis of colorectal cancer. Oncotarget 2016, 7, 27394–27407. [Google Scholar]
- Kaak, J.; de Sá, F.D.L.; Turner, J.R.; Schulzke, J.; Bücker, R. Unraveling the intestinal epithelial barrier in cyanotoxin microcystin-treated Caco-2 cell monolayers. Ann. N. Y. Acad. Sci. 2022, 1516, 188–196. [Google Scholar] [CrossRef]
- Miao, C.; Ren, Y.; Chen, M.; Wang, Z.; Wang, T. Microcystin-LR promotes migration and invasion of colorectal cancer through matrix metalloproteinase-13 up-regulation. Mol. Carcinog. 2015, 55, 514–524. [Google Scholar] [CrossRef]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar]
- Shi, J.; Deng, H.; Pan, H.; Xu, Y.; Zhang, M. Epigallocatechin-3-gallate attenuates microcystin-LR induced oxidative stress and inflammation in human umbilical vein endothelial cells. Chemosphere 2017, 168, 25–31. [Google Scholar]
- Chen, Y.; Wang, J.; Zhang, Q.; Xiang, Z.; Li, D.; Han, X. Microcystin-leucine arginine exhibits immunomodulatory roles in testicular cells resulting in orchitis. Environ. Pollut. 2017, 229, 964–975. [Google Scholar]
- Su, R.C.; Breidenbach, J.D.; Alganem, K.; Khalaf, F.K.; French, B.W.; Dube, P.; Malhotra, D.; McCullumsmith, R.; Presloid, J.B.; Wooten, R.M.; et al. Microcystin-LR (MC-LR) Triggers Inflammatory Responses in Macrophages. Int. J. Mol. Sci. 2021, 22, 9939. [Google Scholar] [CrossRef]
- Yang, K.; Xie, Y.; Xue, L.; Li, F.; Luo, C.; Liang, W.; Zhang, H.; Li, Y.; Ren, Y.; Zhao, M.; et al. M2 tumor-associated macrophage mediates the maintenance of stemness to promote cisplatin resistance by secreting TGF-beta1 in esophageal squamous cell carcinoma. J. Transl. Med. 2023, 21, 26. [Google Scholar]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming growth factor (TGF)-beta signaling in cardiac remodeling. J. Mol. Cell Cardiol. 2011, 51, 600–606. [Google Scholar]
- Clark, D.A.; Coker, R. Transforming growth factor-beta (TGF-beta). Int. J. Biochem. Cell. Biol. 1998, 30, 293–298. [Google Scholar] [CrossRef]
- Tian, M.; Neil, J.R.; Schiemann, W.P. Transforming growth factor-beta and the hallmarks of cancer. Cell Signal 2011, 23, 951–962. [Google Scholar]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [Green Version]
- David, C.J.; Massague, J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat. Rev. Mol. Cell. Biol. 2018, 19, 419–435. [Google Scholar]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.; Liu, X.; Chen, J.; Huang, S.; Wei, W.; Zou, Y.; Liu, X.; Deng, K.; Mo, S.; Chen, J.; et al. Anti-TGF-beta attenuates tumor growth via polarization of tumor associated neutrophils towards an anti-tumor phenotype in colorectal cancer. J. Cancer 2020, 11, 2580–2592. [Google Scholar] [CrossRef]
- Shen, X.; Hu, X.; Mao, J.; Wu, Y.; Liu, H.; Shen, J.; Yu, J.; Chen, W. The long noncoding RNA TUG1 is required for TGF-beta/TWIST1/EMT-mediated metastasis in colorectal cancer cells. Cell Death Dis. 2020, 11, 65. [Google Scholar]
- Costa, T.F.; Lima, A.P. Natural cysteine protease inhibitors in protozoa: Fifteen years of the chagasin family. Biochimie 2016, 122, 197–207. [Google Scholar] [PubMed]
- Turk, V.; Stoka, V.; Turk, D. Cystatins: Biochemical and structural properties, and medical relevance. Front. Biosci. 2008, 13, 5406–5420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Fu, Z.; Zhang, Y.; Wang, M.; Mao, C.; Ge, W. Serum proteomics analysis of candidate predictive biomarker panel for the diagnosis of trastuzumab-based therapy resistant breast cancer. Biomed. Pharmacother. 2020, 129, 110465. [Google Scholar] [PubMed]
- Indacochea, A.; Guerrero, S.; Ureña, M.; Araujo, F.; Coll, O.; Lleonart, M.E.; Gebauer, F. Cold-inducible RNA binding protein promotes breast cancer cell malignancy by regulating Cystatin C levels. RNA 2020, 27, 190–201. [Google Scholar] [CrossRef]
- Ahn, S.B.; Sharma, S.; Mohamedali, A.; Mahboob, S.; Redmond, W.J.; Pascovici, D.; Wu, J.X.; Zaw, T.; Adhikari, S.; Vaibhav, V.; et al. Potential early clinical stage colorectal cancer diagnosis using a proteomics blood test panel. Clin. Proteom. 2019, 16, 34. [Google Scholar]
- Tokarzewicz, A.; Guszcz, T.; Onopiuk, A.; Kozlowski, R.; Gorodkiewicz, E. Utility of cystatin C as a potential bladder tumour biomarker confirmed by surface plasmon resonance technique. Indian J. Med Res. 2018, 147, 46–50. [Google Scholar]
- Zhou, X.; Wang, X.; Huang, K.; Liao, X.; Yang, C.; Yu, T.; Liu, J.; Han, C.; Zhu, G.; Su, H.; et al. Investigation of the clinical significance and prospective molecular mechanisms of cystatin genes in patients with hepatitis B virus-related hepatocellular carcinoma. Oncol. Rep. 2019, 42, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Sokol, J.P.; Schiemann, W.P. Cystatin C antagonizes transforming growth factor beta signaling in normal and cancer cells. Mol. Cancer Res. 2004, 2, 183–195. [Google Scholar]
- Solem, M.; Rawson, C.; Lindburg, K.; Barnes, D. Transforming growth factor beta regulates cystatin C in serum-free mouse embryo (SFME) cells. Biochem. Biophys. Res. Commun. 1990, 172, 945–951. [Google Scholar] [CrossRef]
- Afonso, S.; Tovar, C.; Romagnano, L.; Babiarz, B. Control and expression of cystatin C by mouse decidual cultures. Mol. Reprod. Dev. 2002, 61, 155–163. [Google Scholar] [CrossRef]
- Nowak, G.; Schnellmann, R.G. Autocrine production and TGF-beta 1-mediated effects on metabolism and viability in renal cells. Am. J. Physiol. Content 1996, 271, F689–F697. [Google Scholar]
- Urata, K.; Kajihara, I.; Miyauchi, H.; Mijiddorj, T.; Otsuka-Maeda, S.; Sakamoto, R.; Sawamura, S.; Kanemaru, H.; Kanazawa-Yamada, S.; Makino, K. The Warburg effect and tumour immune microenvironment in extramammary Paget’s disease: Overexpression of lactate dehydrogenase A correlates with immune resistance. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1715–1721. [Google Scholar]
- Laurent-Matha, V.; Huesgen, P.F.; Masson, O.; Derocq, D.; Prébois, C.; Lecaille, F.; Rebière, B.; Meurice, G.; Oréar, C.; Hollingsworth, R.E.; et al. Proteolysis of cystatin C by cathepsin D in the breast cancer microenvironment. FASEB J. 2012, 26, 5172–5181. [Google Scholar]
- Al-Awadhi, F.H.; Law, B.K.; Paul, V.J.; Luesch, H. Grassystatins D–F, Potent Aspartic Protease Inhibitors from Marine Cyanobacteria as Potential Antimetastatic Agents Targeting Invasive Breast Cancer. J. Nat. Prod. 2017, 80, 2969–2986. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Zhang, H.; Zhang, H.; Wang, F.; Wang, X.; Ding, T.; Zhang, X.; Wang, T. Microcystin-LR-Induced Interaction between M2 Tumor-Associated Macrophage and Colorectal Cancer Cell Promotes Colorectal Cancer Cell Migration through Regulating the Expression of TGF-β1 and CST3. Int. J. Mol. Sci. 2023, 24, 10527. https://doi.org/10.3390/ijms241310527
Jiang X, Zhang H, Zhang H, Wang F, Wang X, Ding T, Zhang X, Wang T. Microcystin-LR-Induced Interaction between M2 Tumor-Associated Macrophage and Colorectal Cancer Cell Promotes Colorectal Cancer Cell Migration through Regulating the Expression of TGF-β1 and CST3. International Journal of Molecular Sciences. 2023; 24(13):10527. https://doi.org/10.3390/ijms241310527
Chicago/Turabian StyleJiang, Xinying, Hailing Zhang, Hengshuo Zhang, Fan Wang, Xiaochang Wang, Tong Ding, Xuxiang Zhang, and Ting Wang. 2023. "Microcystin-LR-Induced Interaction between M2 Tumor-Associated Macrophage and Colorectal Cancer Cell Promotes Colorectal Cancer Cell Migration through Regulating the Expression of TGF-β1 and CST3" International Journal of Molecular Sciences 24, no. 13: 10527. https://doi.org/10.3390/ijms241310527
APA StyleJiang, X., Zhang, H., Zhang, H., Wang, F., Wang, X., Ding, T., Zhang, X., & Wang, T. (2023). Microcystin-LR-Induced Interaction between M2 Tumor-Associated Macrophage and Colorectal Cancer Cell Promotes Colorectal Cancer Cell Migration through Regulating the Expression of TGF-β1 and CST3. International Journal of Molecular Sciences, 24(13), 10527. https://doi.org/10.3390/ijms241310527






