Comparative Transcriptome Profiling Unfolds a Complex Defense and Secondary Metabolite Networks Imparting Corynespora cassiicola Resistance in Soybean (Glycine max (L.) Merrill)
Abstract
:1. Introduction
2. Results
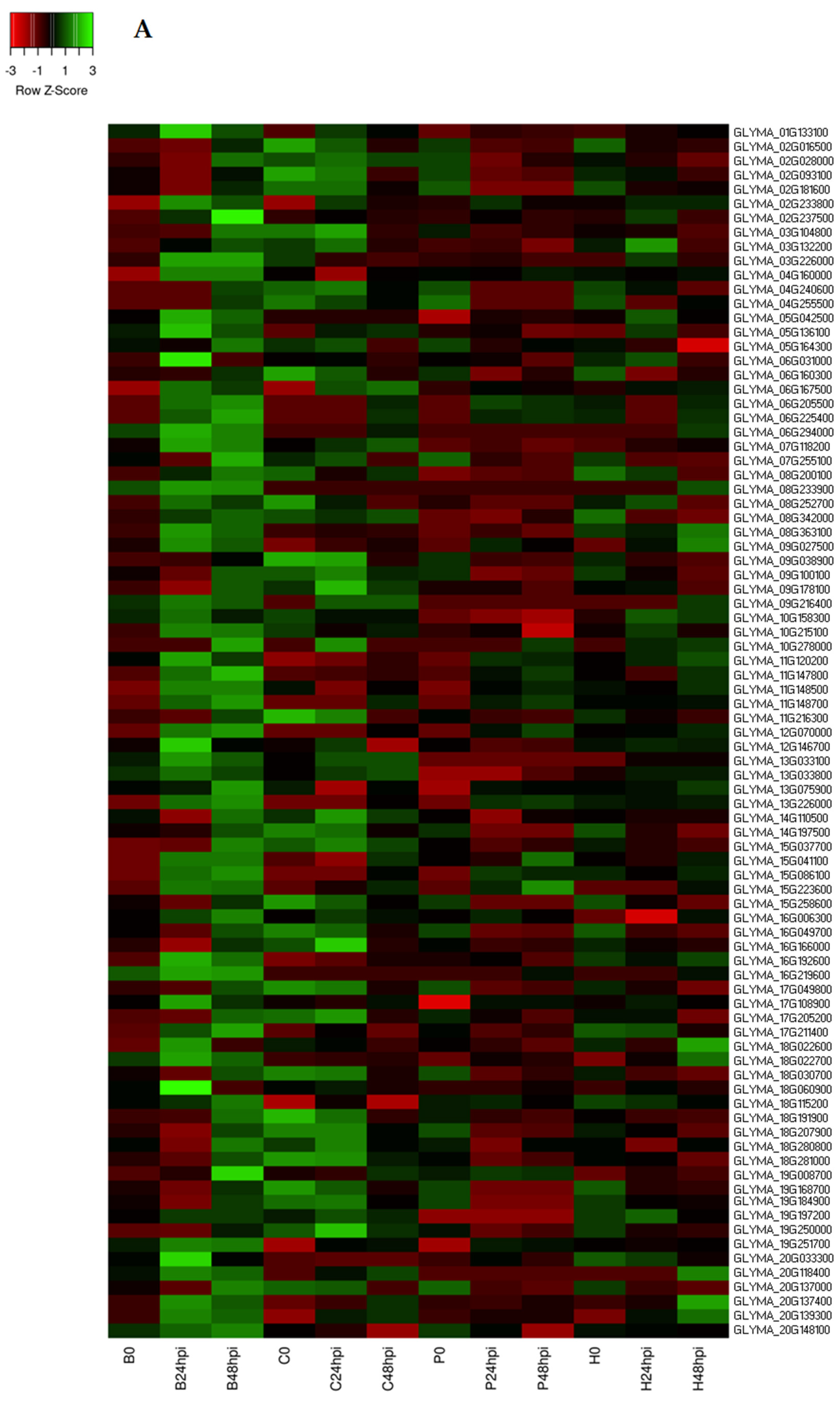
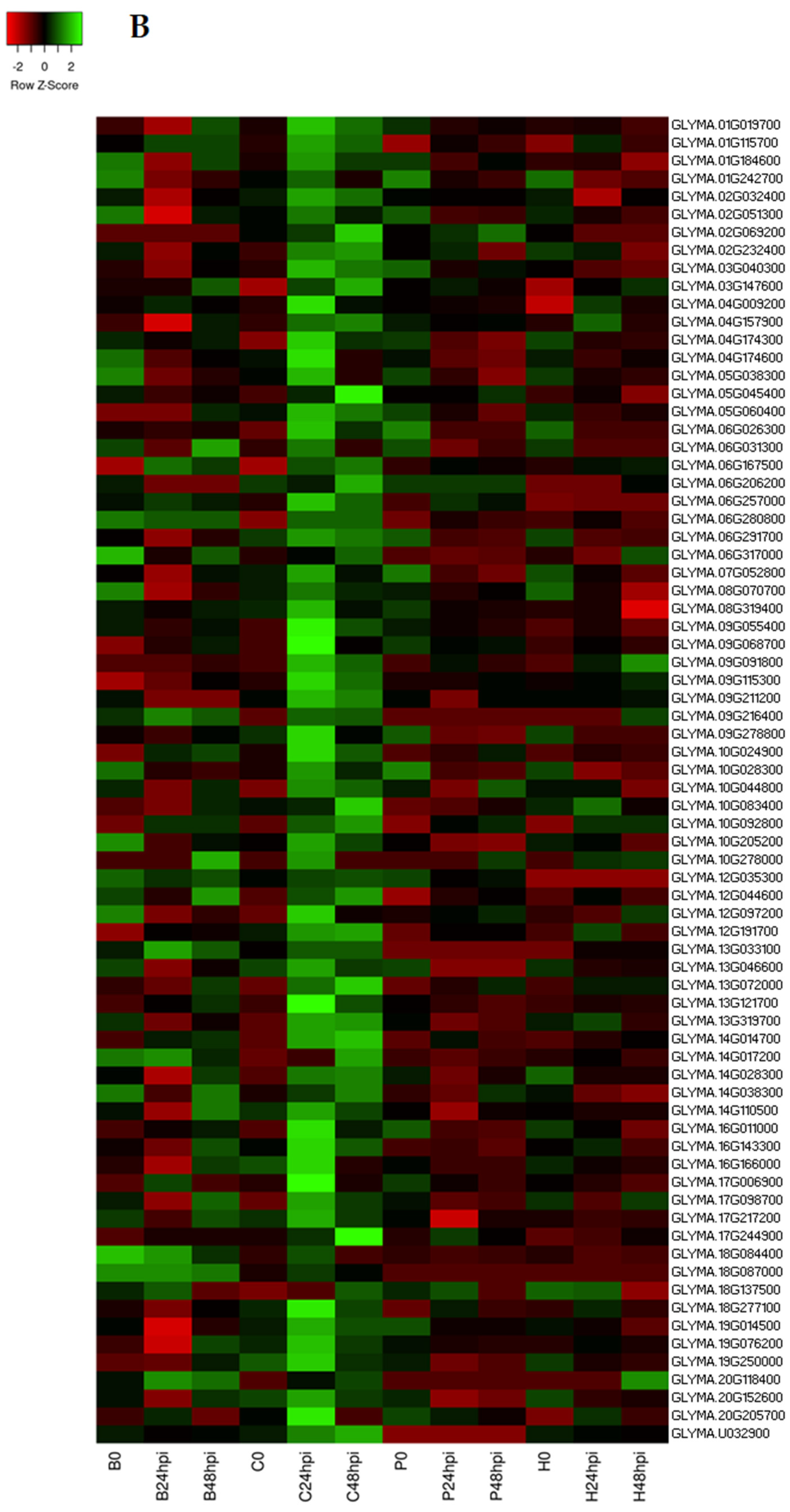
2.1. RNA Sequencing Data Analysis and DEGs in Response to C. cassiicola
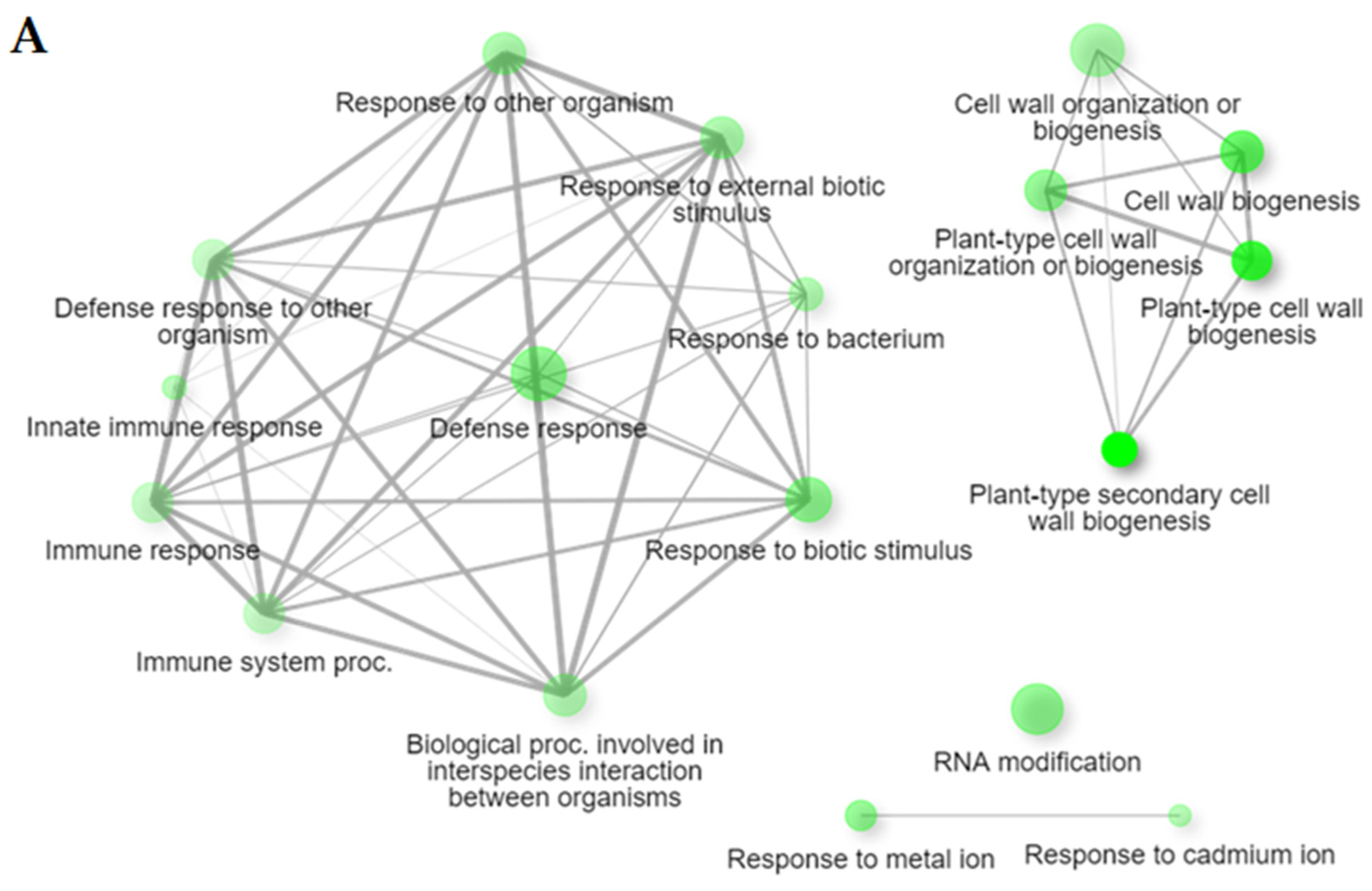
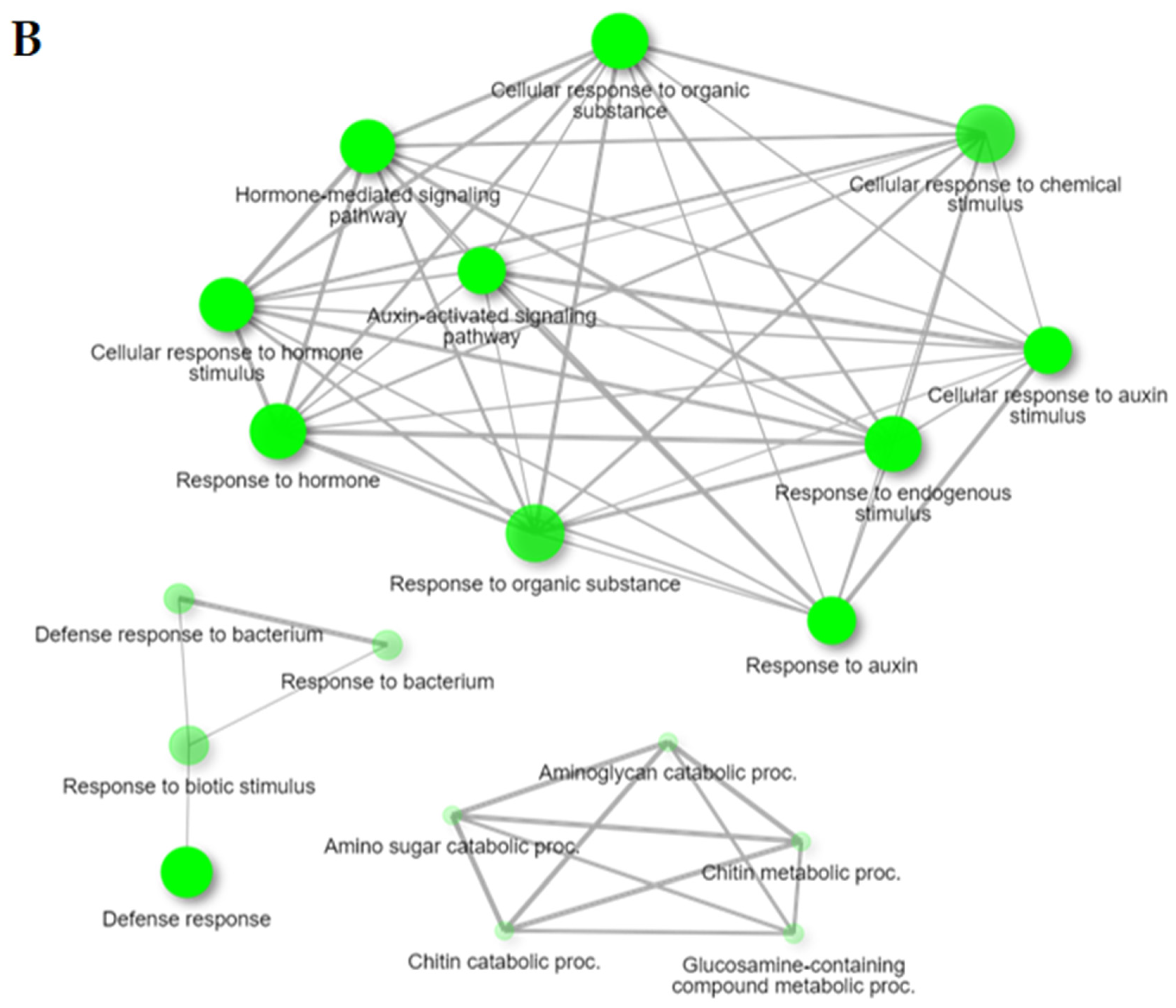
2.2. Gene Ontology (GO) Enrichment Analysis of DEGs
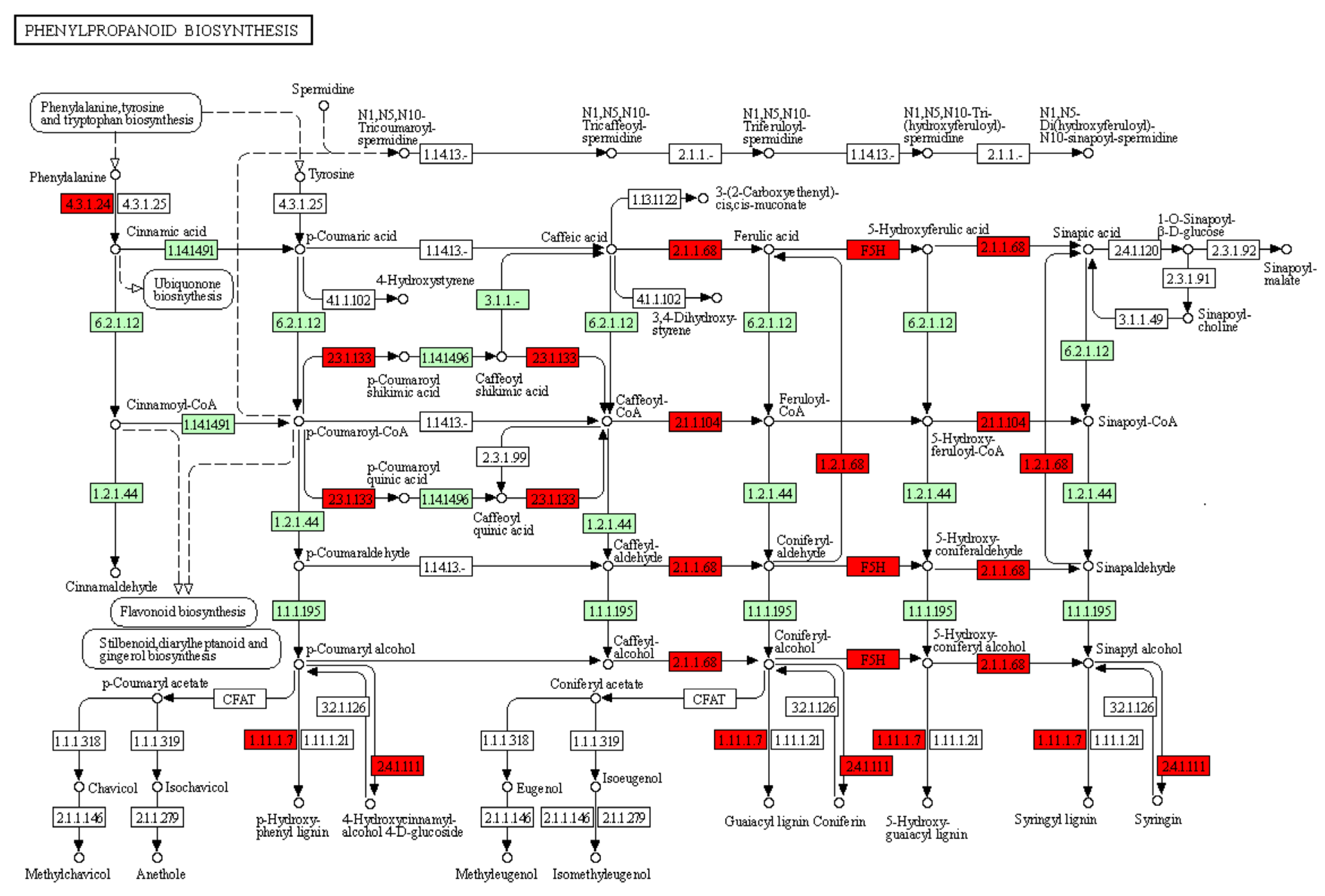
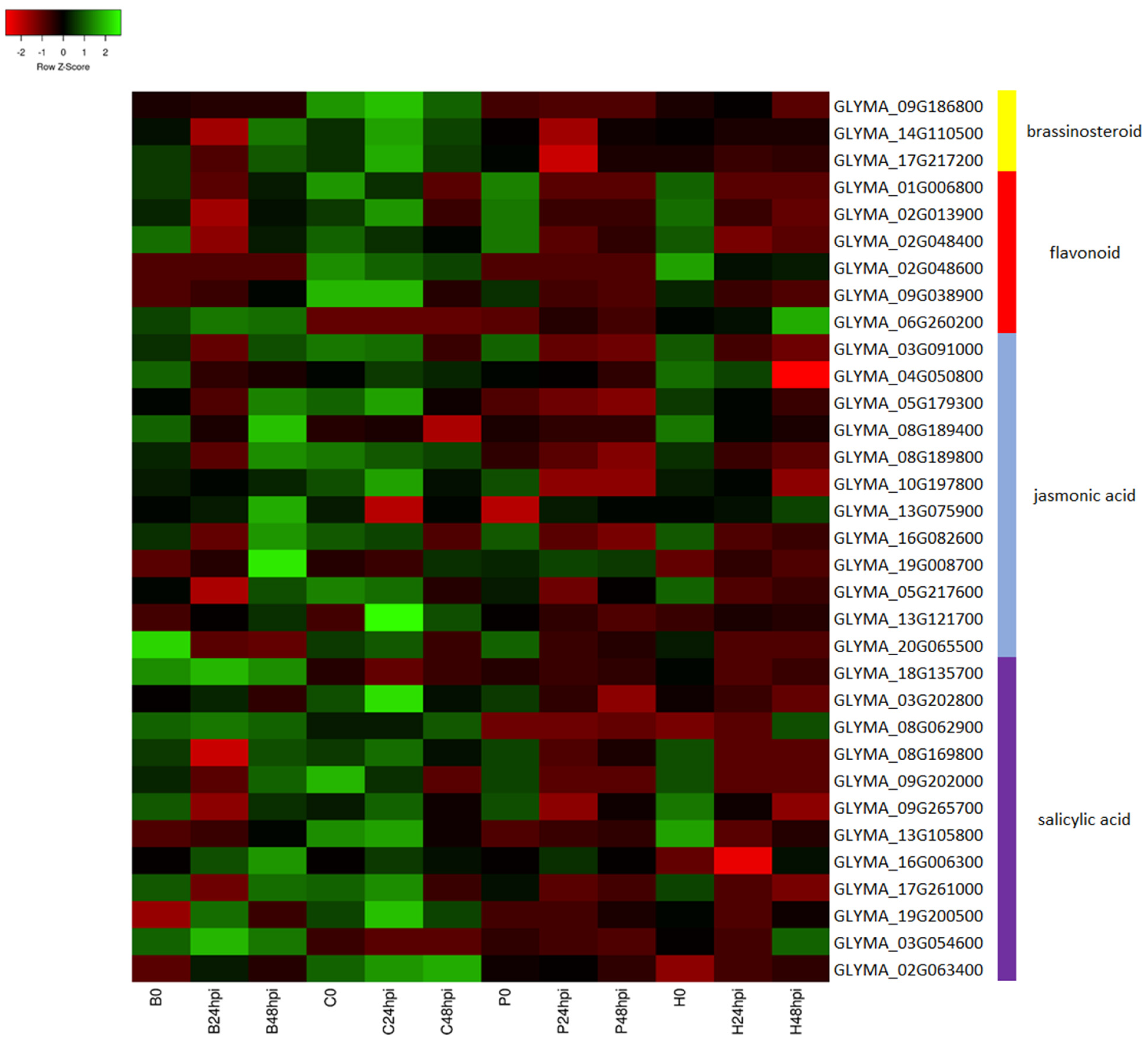
2.3. KEGG Pathway Analysis
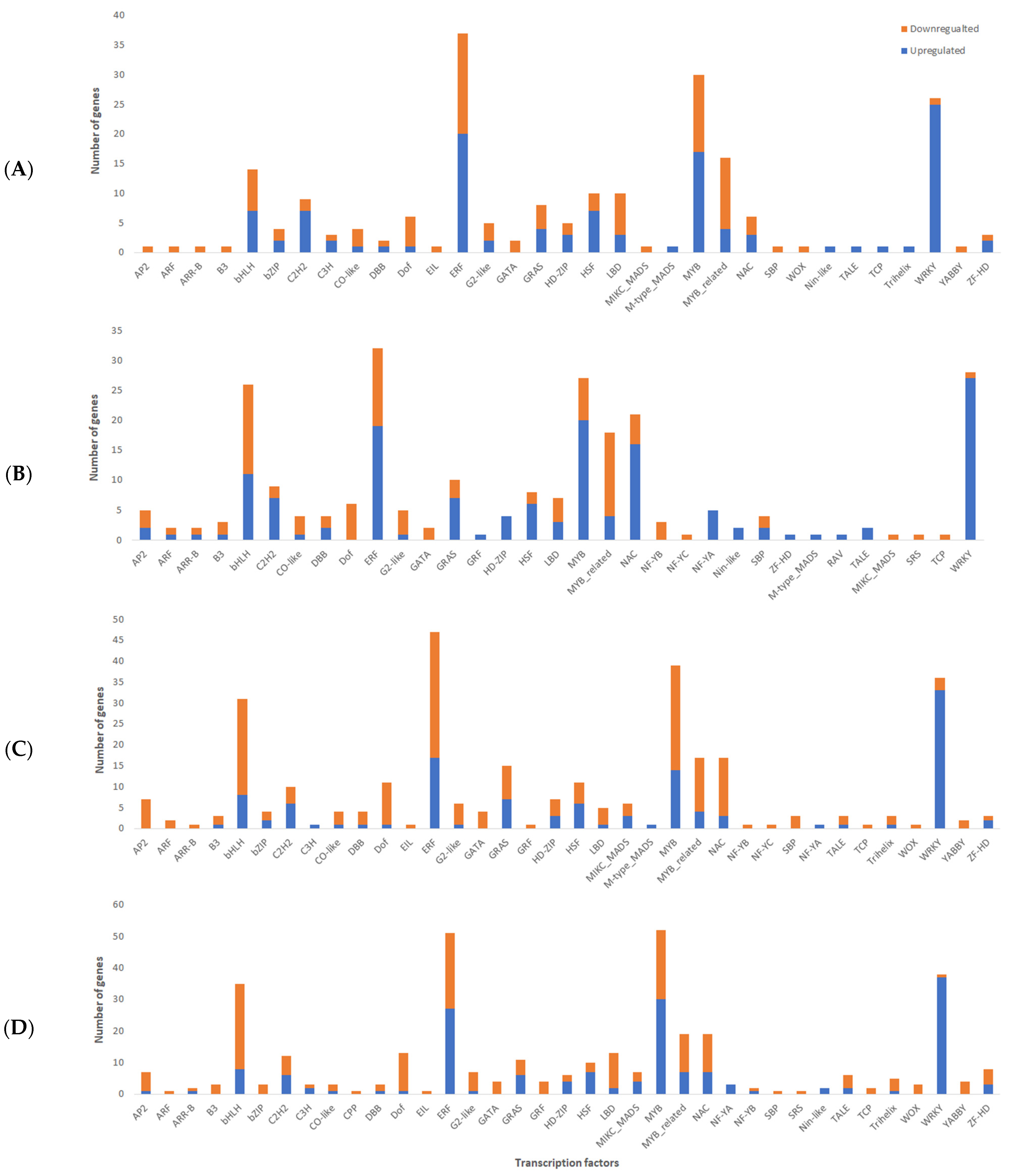
2.4. Identification of Differentially Expressed Transcription Factors (TFs)
2.5. Quantitative Real-Time Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Inoculation
4.2. RNA Extraction, Library Preparation, and Illumina Sequencing
4.3. RNA-Seq Data Analysis
4.4. GO Enrichment and KEGG Pathway Analysis
4.5. Identification of Transcription Factors (TFs)
4.6. Quantitative Real-Time PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- USDA. World Agricultural Production. Available online: https://apps.fas.usda.gov/psdonline/circulars/production.pdf (accessed on 30 July 2021).
- Hartman, G.L.; Rupe, J.C.; Sikora, E.J.; Domier, L.L.; Davis, J.A.; Steffey, K.L. (Eds.) Compendium of Soybean Diseases and Pests; Am Phytopath Society: St. Paul, MN, USA, 2015. [Google Scholar]
- Godoy, C.V. Target spot. In Compendium of Soybean Diseases and Pests; Hartman, G.L., Rupe, J.C., Sikora, E.J., Domier, L.L., Davis, J.A., Steffey, K.L., Eds.; Am Phytopath Society: St. Paul, MN, USA, 2015; pp. 62–63. [Google Scholar]
- Faske, T. Arkansas Soybeans: Target Spot—What Do We Know? Available online: https://agfax.com/2016/11/02/arkansas-soybeans-target-spot-what-do-we-know/ (accessed on 12 March 2019).
- Koenning, S.; Creswell, T.; Dunphy, E.; Sikora, E.; Mueller, J. Increased occurrence of target spot of soybean caused by Corynespora cassiicola in the Southeastern United States. Plant Dis. 2006, 90, 974. [Google Scholar] [CrossRef]
- Duan, Y.; Xin, W.; Lu, F.; Li, T.; Li, M.; Wu, J.; Wang, J.; Zhou, M. Benzimidazole-and QoI-resistance in Corynespora cassiicola populations from greenhouse-cultivated cucumber: An emerging problem in China. Pestic. Biochem. Physiol. 2019, 153, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Xavier, S.A.; Canteri, M.G.; Barros, D.; Godoy, C.V. Sensitivity of Corynespora cassiicola from soybean to carbendazim and prothioconazole. Trop. Plant Pathol. 2013, 38, 431–435. [Google Scholar] [CrossRef] [Green Version]
- de Mello, F.E.; Lopes-Caitar, V.S.; Prudente, H.; Xavier-Valencio, S.A.; Franzenburg, S.; Mehl, A.; Marcelino-Guimaraes, F.C.; Verreet, J.-A.; Balbi-Peña, M.I.; Godoy, C.V. Sensitivity of Cercospora spp. from soybean to quinone outside inhibitors and methyl benzimidazole carbamate fungicides in Brazil. Trop. Plant Pathol. 2021, 46, 69–80. [Google Scholar] [CrossRef]
- Patel, S.; Bowen, K.; Patel, J.; Koebernick, J. Evaluating target spot (Corynespora cassiicola) resistance in soybean (Glycine max (L.) Merrill) in a controlled environment. Crop Prot. 2022, 159, 106018. [Google Scholar] [CrossRef]
- Amaral, D.O.J.d.; Lima, M.M.d.A.; Resende, L.V.; Silva, M.V.d. Differential gene expression, induced by salicylic acid and Fusarium oxysporum f. sp. lycopersici infection, in tomato. Pesqui. Agropecuária Bras. 2008, 43, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Hickman, R.; Pieterse, C.M.; Van Wees, S.C. RNA-Seq: Revelation of the messengers. Trends Plant Sci. 2013, 18, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Miraeiz, E.; Chaiprom, U.; Afsharifar, A.; Karegar, A.; Drnevich, J.M.; Hudson, M.E. Early transcriptional responses to soybean cyst nematode HG Type 0 show genetic differences among resistant and susceptible soybeans. Theor. Appl. Genet. 2020, 133, 87–102. [Google Scholar] [CrossRef]
- Kofsky, J.; Zhang, H.; Song, B.-H. Novel resistance strategies to soybean cyst nematode (SCN) in wild soybean. Sci. Rep. 2021, 11, 7967. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Zhao, M.; Baumann, D.D.; Ping, J.; Sun, L.; Liu, Y.; Zhang, B.; Tang, Z.; Hughes, E.; Doerge, R.W. Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genom. 2014, 15, 18. [Google Scholar] [CrossRef] [Green Version]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Kang, Y.J.; Kim, D.H.; Yoon, M.Y.; Moon, J.-K.; Kim, M.Y.; Van, K.; Lee, S.-H. RNA-Seq analysis of a soybean near-isogenic line carrying bacterial leaf pustule-resistant and-susceptible alleles. DNA Res. 2011, 18, 483–497. [Google Scholar] [CrossRef] [Green Version]
- DeMers, L.C.; Redekar, N.R.; Kachroo, A.; Tolin, S.A.; Li, S.; Saghai Maroof, M. A transcriptional regulatory network of Rsv3-mediated extreme resistance against Soybean mosaic virus. PLoS ONE 2020, 15, e0231658. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Shi, S.; Zhang, C.; Zhu, S.; Li, M.; Tan, J.; Yu, Y.; Lin, L.; Jia, S.; Wang, X. Transcriptomic analysis of genes in soybean in response to Peronospora manshurica infection. BMC Genom. 2018, 19, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCabe, C.E.; Cianzio, S.R.; O’Rourke, J.A.; Graham, M.A. Leveraging RNA-Seq to characterize resistance to Brown stem rot and the Rbs3 locus in soybean. Mol. Plant-Microbe Interact. 2018, 31, 1083–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chee, K.H. Studies of sporulation, pathogenicity and epidemiology of Corynespora cassiicola on Hevea rubber. J. Not. Rubb. Res. 1988, 3, 21–29. [Google Scholar]
- Conner, K.; Hagan, A.; Zhang, L. First report of Corynespora cassiicola-incited target spot on cotton in Alabama. Plant Dis. 2013, 97, 1379. [Google Scholar] [CrossRef]
- Blazquez, C. Corynespora leaf spot of cucumber. Proc. Fla. State Hortic. Soc. 1967, 80, 177. [Google Scholar]
- Wang, X.; Zhang, D.; Cui, N.; Yu, Y.; Yu, G.; Fan, H. Transcriptome and miRNA analyses of the response to Corynespora cassiicola in cucumber. Sci. Rep. 2018, 8, 7798. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xin, M.; Zhou, X.; Wang, C.; Zhang, Y.; Qin, Z. Expression and functional analysis of the transcription factor-encoding Gene CsERF004 in cucumber during Pseudoperonospora cubensis and Corynespora cassiicola infection. BMC Plant Biol. 2017, 17, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, S.; Label, P.; Garcia, D.; Montoro, P.; Pujade-Renaud, V. Transcriptome profiling in susceptible and tolerant rubber tree clones in response to cassiicolin Cas1, a necrotrophic effector from Corynespora cassiicola. PLoS ONE 2021, 16, e0254541. [Google Scholar] [CrossRef]
- Roy, C.B.; Liu, H.; Rajamani, A.; Saha, T. Transcriptome profiling reveals genetic basis of disease resistance against Corynespora cassiicola in rubber tree (Hevea brasiliensis). Curr. Plant Biol. 2019, 17, 2–16. [Google Scholar] [CrossRef]
- Fortunato, A.A.; Debona, D.; Bernardeli, A.M.; Rodrigues, F.A. Defence-related enzymes in soybean resistance to target spot. J. Phytopathol. 2015, 163, 731–742. [Google Scholar] [CrossRef] [Green Version]
- Fortunato, A.A.; Araujo, L.; Rodrigues, F.Á. Association of the production of phenylpropanoid compounds at the infection sites of Corynespora cassiicola with soybean resistance against target spot. J. Phytopathol. 2017, 165, 131–142. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.-M.; Park, O.K. The Arabidopsis cysteine-rich receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Bosamia, T.C.; Dodia, S.M.; Mishra, G.P.; Ahmad, S.; Joshi, B.; Thirumalaisamy, P.P.; Kumar, N.; Rathnakumar, A.L.; Sangh, C.; Kumar, A.; et al. Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea L.) using comparative RNA-Seq analysis of resistant and susceptible genotypes. PLoS ONE 2020, 15, e0236823. [Google Scholar] [CrossRef]
- Gao, S.; Wang, F.; Niran, J.; Li, N.; Yin, Y.; Yu, C.; Jiao, C.; Yao, M. Transcriptome analysis reveals defense-related genes and pathways against Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum L.). PLoS ONE 2021, 16, e0240279. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, Y.; Yang, J.; Yang, W. Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Front. Plant Sci. 2015, 6, 1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor Kinases in Plant-Pathogen Interactions: More Than Pattern Recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Staal, J.; Kaliff, M.; Bohman, S.; Dixelius, C. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 2006, 46, 218–230. [Google Scholar] [CrossRef]
- Staal, J.; Kaliff, M.; Dewaele, E.; Persson, M.; Dixelius, C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008, 55, 188–200. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Elmore, J.M.; Creer, A.Y.; Feng, B.; Franco, J.Y.; Rufian, J.S.; He, P.; Phinney, B.; Coaker, G. A Cysteine-Rich Protein Kinase Associates with a Membrane Immune Complex and the Cysteine Residues Are Required for Cell Death. Plant Physiol 2017, 173, 771–787. [Google Scholar] [CrossRef] [Green Version]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojärvi, J.; Rayapuram, C.; Idänheimo, N.; Hunter, K.; Kimura, S.; Merilo, E. Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 2015, 11, e1005373. [Google Scholar] [CrossRef] [Green Version]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant–fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Toufiq, N.; Tabassum, B.; Bhatti, M.U.; Khan, A.; Tariq, M.; Shahid, N.; Nasir, I.A.; Husnain, T. Improved antifungal activity of barley derived chitinase I gene that overexpress a 32 kDa recombinant chitinase in Escherichia coli host. Braz. J. Microbiol. 2018, 49, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Schlumbaum, A.; Mauch, F.; Vögeli, U.; Boller, T. Plant chitinases are potent inhibitors of fungal growth. Nature 1986, 324, 365–367. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Yadav, M.; Chawade, A.; Vivekanand, V.; Pareek, N. Chitinases—Potential candidates for enhanced plant resistance towards fungal pathogens. Agriculture 2018, 8, 88. [Google Scholar] [CrossRef] [Green Version]
- Kabir, S.R.; Rahman, M.M.; Tasnim, S.; Karim, M.R.; Khatun, N.; Hasan, I.; Amin, R.; Islam, S.S.; Nurujjaman, M.; Kabir, A.H. Purification and characterization of a novel chitinase from Trichosanthes dioica seed with antifungal activity. Int. J. Biol. Macromol. 2016, 84, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Corrado, G.; Arciello, S.; Fanti, P.; Fiandra, L.; Garonna, A.; Digilio, M.C.; Lorito, M.; Giordana, B.; Pennacchio, F.; Rao, R. The Chitinase A from the baculovirus AcMNPV enhances resistance to both fungi and herbivorous pests in tobacco. Transgenic Res. 2008, 17, 557–571. [Google Scholar] [CrossRef]
- Reuber, T.L.; Ausubel, F.M. Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 1996, 8, 241–249. [Google Scholar]
- Wang, J.; Tian, W.; Tao, F.; Wang, J.; Shang, H.; Chen, X.; Xu, X.; Hu, X. TaRPM1 positively regulates wheat high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2020, 10, 1679. [Google Scholar] [CrossRef] [Green Version]
- Afzal, M.; Alghamdi, S.S.; Nawaz, H.; Migdadi, H.H.; Altaf, M.; El-Harty, E.; Al-Fifi, S.A.; Sohaib, M. Genome-wide identification and expression analysis of CC-NB-ARC-LRR (NB-ARC) disease-resistant family members from soybean (Glycine max L.) reveal their response to biotic stress. J. King Saud Univ.-Sci. 2022, 34, 101758. [Google Scholar] [CrossRef]
- Whaley, A.; Sheridan, J.; Safari, S.; Burton, A.; Burkey, K.; Schlueter, J. RNA-seq analysis reveals genetic response and tolerance mechanisms to ozone exposure in soybean. BMC Genom. 2015, 16, 426. [Google Scholar] [CrossRef] [Green Version]
- Parkhi, V.; Kumar, V.; Campbell, L.M.; Bell, A.A.; Shah, J.; Rathore, K.S. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 2010, 19, 959–975. [Google Scholar] [CrossRef]
- Stein, E.; Molitor, A.; Kogel, K.-H.; Waller, F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 2008, 49, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mir, Z.A.; Tyagi, A.; Mehari, H.; Meena, R.P.; Bhat, J.A.; Yadav, P.; Papalou, P.; Rawat, S.; Grover, A. Overexpression of NPR1 in Brassica juncea confers broad spectrum resistance to fungal pathogens. Front. Plant Sci. 2017, 8, 1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Ma, H.; Hong, H.; Yao, W.; Xie, W.; Xiao, J.; Li, X.; Wang, S. Transcriptome-based analysis of mitogen-activated protein kinase cascades in the rice response to Xanthomonas oryzae infection. Rice 2015, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Qi, S.D.; Qi, F.; Wu, C.A.; Yang, G.D.; Zheng, C.C. NtKTI1, a Kunitz trypsin inhibitor with antifungal activity from Nicotiana tabacum, plays an important role in tobacco’s defense response. FEBS J. 2010, 277, 4076–4088. [Google Scholar] [CrossRef] [PubMed]
- Sarowar, S.; Alam, S.T.; Makandar, R.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Targeting the pattern-triggered immunity pathway to enhance resistance to Fusarium graminearum. Mol. Plant Pathol. 2019, 20, 626–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashi, K.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet. Genom. 2008, 279, 303–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicko, M.H.; Gruppen, H.; Barro, C.; Traoré, A.S.; van Berkel, W.J.; Voragen, A.G. Impact of phenolic compounds and related enzymes in sorghum varieties for resistance and susceptibility to biotic and abiotic stresses. J. Chem. Ecol. 2005, 31, 2671–2688. [Google Scholar] [CrossRef]
- Lozovaya, V.V.; Lygin, A.V.; Zernova, O.V.; Ulanov, A.V.; Li, S.; Hartman, G.L.; Widholm, J.M. Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 2007, 225, 665–679. [Google Scholar] [CrossRef]
- Mitchell, H.J.; Hall, S.A.; Stratford, R.; Hall, J.L.; Barber, M.S. Differential induction of cinnamyl alcohol dehydrogenase during defensive lignification in wheat (Triticum aestivum L.): Characterisation of the major inducible form. Planta 1999, 208, 31–37. [Google Scholar] [CrossRef]
- Hamberger, B.; Ellis, M.; Friedmann, M.; de Azevedo Souza, C.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene families. Botany 2007, 85, 1182–1201. [Google Scholar]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dao, T.T.; Linthorst, H.J.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Algar, E.; Gutierrez-Mañero, F.J.; Garcia-Villaraco, A.; García-Seco, D.; Lucas, J.A.; Ramos-Solano, B. The role of isoflavone metabolism in plant protection depends on the rhizobacterial MAMP that triggers systemic resistance against Xanthomonas axonopodis pv. glycines in Glycine max (L.) Merr. cv. Osumi. Plant Physiol. Biochem. 2014, 82, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fan, R.; Guo, S.; Wang, P.; Zhu, X.; Fan, Y.; Chen, Y.; He, K.; Kumar, A.; Shi, J. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 2019, 166, 103807. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef]
- Mahajan, M.; Yadav, S.K. Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol. Biol. 2014, 85, 551–573. [Google Scholar] [CrossRef]
- Jan, R.; Aaqil Khan, M.; Asaf, S.; Park, J.-R.; Lee, I.-J.; Kim, K.-M. Flavonone 3-hydroxylase relieves bacterial leaf blight stress in rice via overaccumulation of antioxidant flavonoids and induction of defense genes and hormones. Int. J. Mol. Sci. 2021, 22, 6152. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Chen, W.; Muehlbauer, F.J. Constitutive expression of the Flavanone 3-hydroxylase gene related to pathotype-specific ascochyta blight resistance in Cicer arietinum L. Physiol. Mol. Plant Pathol. 2005, 67, 100–107. [Google Scholar] [CrossRef]
- Verhage, A.; van Wees, S.C.; Pieterse, C.M. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef] [Green Version]
- Kohli, S.K.; Bali, S.; Khanna, K.; Bakshi, P.; Sharma, P.; Sharma, A.; Verma, V.; Ohri, P.; Mir, B.A.; Kaur, R. A current scenario on role of brassinosteroids in plant defense triggered in response to biotic challenges. In Brassinosteroids: Plant Growth and Development; Springer: Berlin/Heidelberg, Germany, 2019; pp. 367–388. [Google Scholar]
- Tariq, R.; Wang, C.; Qin, T.; Xu, F.; Tang, Y.; Gao, Y.; Ji, Z.; Zhao, K. Comparative transcriptome profiling of rice near-isogenic line carrying Xa23 under infection of Xanthomonas oryzae pv. oryzae. Int. J. Mol. Sci. 2018, 19, 717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, U.; Mishra, G.P.; Dikshit, H.K.; Mishra, D.C.; Bosamia, T.; Roy, A.; Bhati, J.; Aski, M.; Kumar, R.R.; Singh, A.K. Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]. PLoS ONE 2021, 16, e0244593. [Google Scholar] [CrossRef] [PubMed]
- French, E.; Kim, B.S.; Iyer-Pascuzzi, A.S. Mechanisms of quantitative disease resistance in plants. Semin. Cell Dev. Biol. 2016, 56, 201–208. [Google Scholar] [CrossRef]
- Poland, J.A.; Balint-Kurti, P.J.; Wisser, R.J.; Pratt, R.C.; Nelson, R.J. Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 2009, 14, 21–29. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Mizukami, Y.; Koga, H.; Yamanouchi, U.; Yoshioka, Y.; Hayashi, N.; Ebana, K.; Mizobuchi, R.; Yano, M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015, 5, 7773. [Google Scholar] [CrossRef] [Green Version]
- Richardson, K.; Vales, M.; Kling, J.; Mundt, C.; Hayes, P. Pyramiding and dissecting disease resistance QTL to barley stripe rust. Theor. Appl. Genet. 2006, 113, 485–495. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Samples | Raw Reads | Clean Reads | Map Reads | Raw Data (G) | Clean Data (G) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| Bedford—0 hpi | 26,390,793 | 25,793,638 | 23,582,978 | 7.9 | 7.8 | 93.39 | 45.04 |
| Council—0 hpi | 23,089,787 | 22,563,156 | 20,640,957 | 6.9 | 6.8 | 93.53 | 45.19 |
| Henderson—0 hpi | 22,838,228 | 22,381,404 | 20,209,026 | 6.9 | 6.7 | 93.19 | 44.79 |
| Pembina—0 hpi | 25,554,633 | 25,099,735 | 22,617,422 | 7.7 | 7.5 | 93.39 | 44.72 |
| Bedford—24 hpi | 28,423,021 | 28,174,656 | 24,664,671 | 8.6 | 8.5 | 93.19 | 44.69 |
| Council—24 hpi | 26,514,264 | 26,147,721 | 23,551,179 | 8.0 | 7.8 | 93.17 | 45.18 |
| Henderson—24 hpi | 27,493,410 | 27,128,017 | 23,310,773 | 8.2 | 8.1 | 93.05 | 44.69 |
| Pembina—24 hpi | 33,485,280 | 33,067,807 | 29,065,093 | 10.1 | 9.9 | 93.18 | 44.42 |
| Bedford—48 hpi | 25,860,451 | 25,641,661 | 22,746,284 | 7.8 | 7.7 | 93.37 | 44.94 |
| Council—48 hpi | 23,417,306 | 23,237,271 | 20,529,754 | 7.0 | 7.0 | 93.31 | 45.03 |
| Henderson—48 hpi | 25,075,023 | 24,887,851 | 21,578,946 | 7.5 | 7.4 | 93.09 | 44.53 |
| Pembina—48 hpi | 24,925,859 | 24,478,453 | 21,424,984 | 7.5 | 7.3 | 93.26 | 44.54 |
| Term | ID | Input Number | Background Number | p-Value | Corrected p-Value |
|---|---|---|---|---|---|
| Biosynthesis of secondary metabolites | gmx01110 | 129 | 2121 | 1.83 × 10−19 | 1.79 × 10−17 |
| Metabolic pathways | gmx01100 | 180 | 4144 | 6.06 × 10−13 | 2.97 × 10−11 |
| Phenylpropanoid biosynthesis | gmx00940 | 35 | 349 | 2.87 × 10−11 | 9.37 × 10−10 |
| Circadian rhythm—plant | gmx04712 | 18 | 106 | 1.29 × 10−9 | 3.17 × 10−8 |
| Cyanoamino acid metabolism | gmx00460 | 15 | 87 | 2.53 × 10−8 | 4.97 × 10−7 |
| Flavonoid biosynthesis | gmx00941 | 15 | 94 | 6.30 × 10−8 | 1.03 × 10−6 |
| Isoflavonoid biosynthesis | gmx00943 | 10 | 42 | 4.13 × 10−7 | 5.78 × 10−6 |
| Starch and sucrose metabolism | gmx00500 | 23 | 299 | 4.73 × 10−6 | 5.80 × 10−5 |
| Thiamine metabolism | gmx00730 | 7 | 43 | 0.000191 | 0.00199 |
| Ubiquinone and other terpenoid-quinone biosynthesis | gmx00130 | 10 | 93 | 0.000203 | 0.00199 |
| Fatty acid elongation | gmx00062 | 7 | 49 | 0.000391 | 0.003485 |
| MAPK signaling pathway—plant | gmx04016 | 19 | 307 | 0.00043 | 0.003513 |
| Plant hormone signal transduction | gmx04075 | 30 | 675 | 0.00233 | 0.017564 |
| Ascorbate and aldarate metabolism | gmx00053 | 8 | 90 | 0.002635 | 0.018442 |
| Cutin, suberine, and wax biosynthesis | gmx00073 | 6 | 54 | 0.003245 | 0.021197 |
| Nitrogen metabolism | gmx00910 | 6 | 65 | 0.007429 | 0.043641 |
| Carotenoid biosynthesis | gmx00906 | 6 | 66 | 0.007942 | 0.043641 |
| Glycerolipid metabolism | gmx00561 | 10 | 158 | 0.008016 | 0.043641 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, S.; Patel, J.; Silliman, K.; Hall, N.; Bowen, K.; Koebernick, J. Comparative Transcriptome Profiling Unfolds a Complex Defense and Secondary Metabolite Networks Imparting Corynespora cassiicola Resistance in Soybean (Glycine max (L.) Merrill). Int. J. Mol. Sci. 2023, 24, 10563. https://doi.org/10.3390/ijms241310563
Patel S, Patel J, Silliman K, Hall N, Bowen K, Koebernick J. Comparative Transcriptome Profiling Unfolds a Complex Defense and Secondary Metabolite Networks Imparting Corynespora cassiicola Resistance in Soybean (Glycine max (L.) Merrill). International Journal of Molecular Sciences. 2023; 24(13):10563. https://doi.org/10.3390/ijms241310563
Chicago/Turabian StylePatel, Sejal, Jinesh Patel, Katherine Silliman, Nathan Hall, Kira Bowen, and Jenny Koebernick. 2023. "Comparative Transcriptome Profiling Unfolds a Complex Defense and Secondary Metabolite Networks Imparting Corynespora cassiicola Resistance in Soybean (Glycine max (L.) Merrill)" International Journal of Molecular Sciences 24, no. 13: 10563. https://doi.org/10.3390/ijms241310563
APA StylePatel, S., Patel, J., Silliman, K., Hall, N., Bowen, K., & Koebernick, J. (2023). Comparative Transcriptome Profiling Unfolds a Complex Defense and Secondary Metabolite Networks Imparting Corynespora cassiicola Resistance in Soybean (Glycine max (L.) Merrill). International Journal of Molecular Sciences, 24(13), 10563. https://doi.org/10.3390/ijms241310563








