Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Patient and Tumor Characteristics
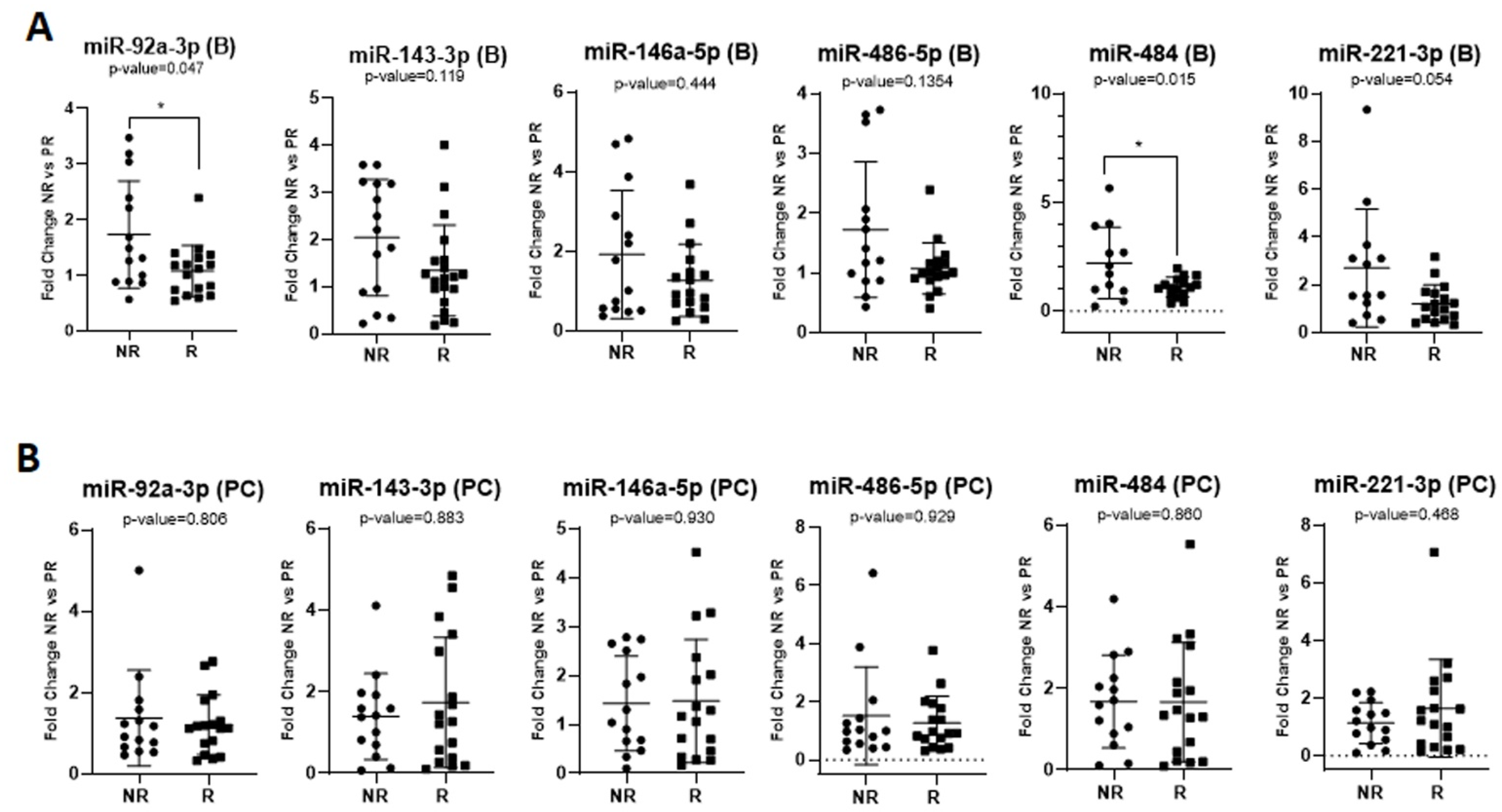
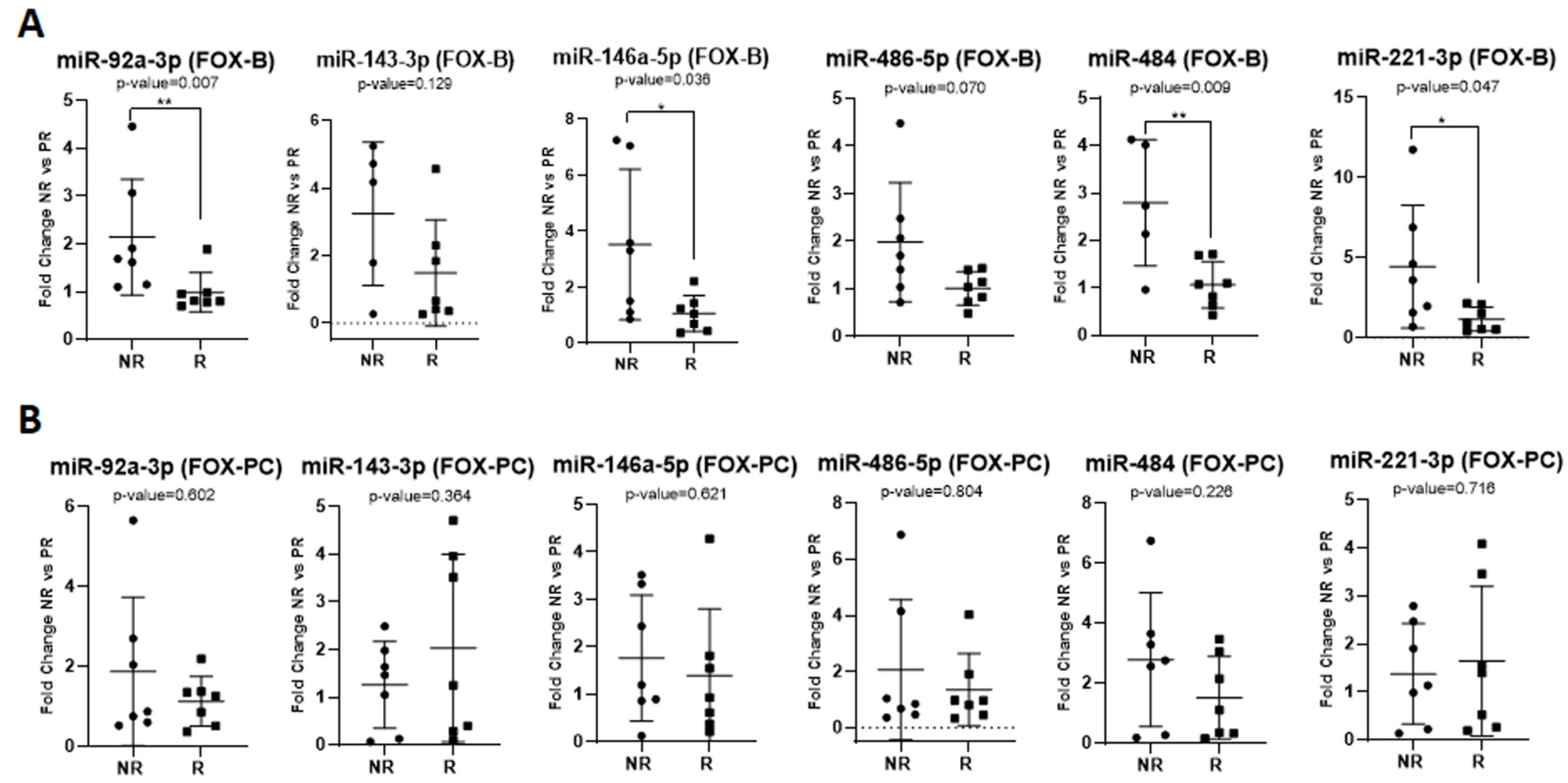
2.2. Investigating the Exosomal miRNAs of Interest
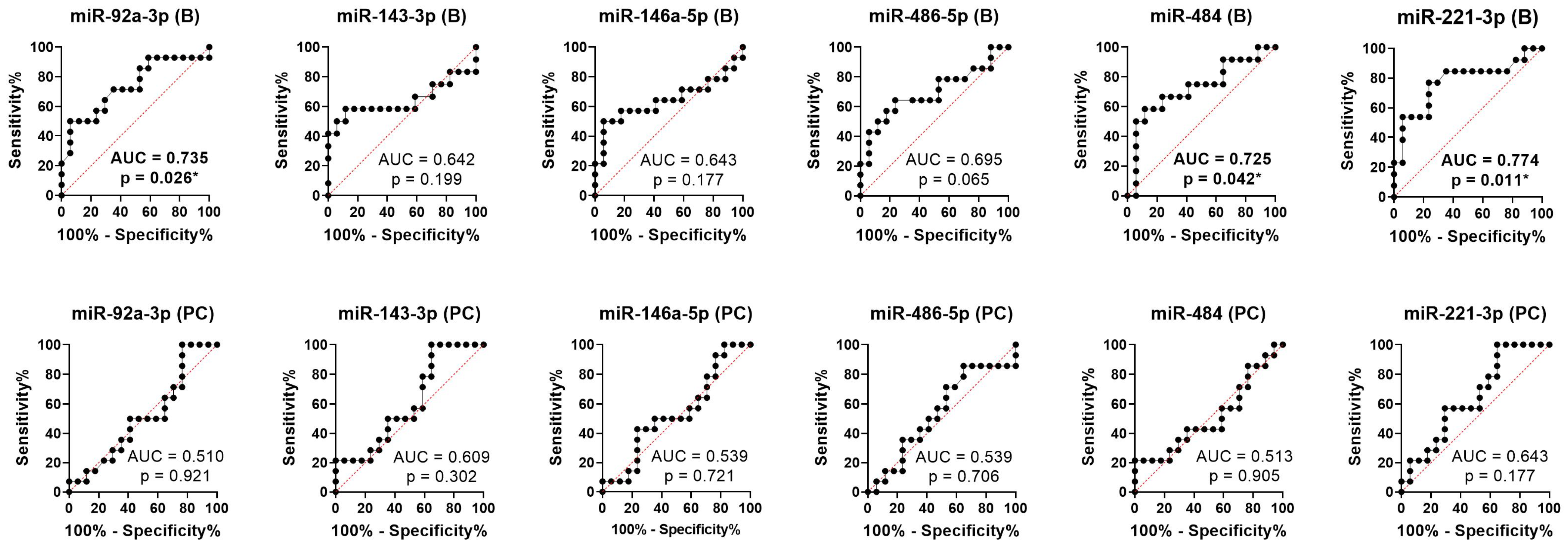
2.3. Predictive Role of Exosomal miRNAs for Chemotherapy Response to First-Line Chemotherapy in mCRC
2.4. Predictive Role of Exosomal miRNAs for Progression-Free Survival and Overall Survival in mCRC
3. Discussion
4. Materials and Methods
4.1. Study Design and Patients Included
4.2. Treatments Administered and Response Evaluation
4.3. Plasma Collection
4.4. Isolation of Plasma-Derived Exosomes and RNA Extraction from Them
4.5. Selection of miRNAs of Interest
4.6. Assessment of the Expression of miRNAs of Interest
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Hajjo, R.; Sabbah, D.A.; Al Bawab, A.Q. Unlocking the Potential of the Human Microbiome for Identifying Disease Diagnostic Biomarkers. Diagnostics 2022, 12, 1742. [Google Scholar] [CrossRef]
- Zhou, Z.; Ge, S.; Li, Y.; Ma, W.; Liu, Y.; Hu, S.; Zhang, R.; Ma, Y.; Du, K.; Syed, A.; et al. Human Gut Microbiome-Based Knowledgebase as a Biomarker Screening Tool to Improve the Predicted Probability for Colorectal Cancer. Front. Microbiol. 2020, 11, 596027. [Google Scholar] [CrossRef]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef]
- Gherman, A.; Balacescu, L.; Gheorghe-Cetean, S.; Vlad, C.; Balacescu, O.; Irimie, A.; Lisencu, C. Current and New Predictors for Treatment Response in Metastatic Colorectal Cancer. The Role of Circulating miRNAs as Biomarkers. Int. J. Mol. Sci. 2020, 21, 2089. [Google Scholar] [CrossRef]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. microRNAs in cancer management. Lancet Oncol. 2012, 13, e249–e258. [Google Scholar] [CrossRef]
- Okayama, H.; Schetter, A.J.; Harris, C.C. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig. Dis. 2012, 30 (Suppl. S2), 9–15. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, G.; Gallo, G.; Vescio, G.; Picciariello, A.; De Paola, G.; Trompetto, M.; Currò, G.; Ammendola, M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. J. Clin. Med. 2020, 9, 2852. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Vautrot, V.; Chanteloup, G.; Elmallah, M.; Cordonnier, M.; Aubin, F.; Garrido, C.; Gobbo, J. Exosomal miRNA: Small Molecules, Big Impact in Colorectal Cancer. J. Oncol. 2019, 2019, 8585276. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Balacescu, O.; Sur, D.; Cainap, C.; Visan, S.; Cruceriu, D.; Manzat-Saplacan, R.; Muresan, M.-S.; Balacescu, L.; Lisencu, C.; Irimie, A. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int. J. Mol. Sci. 2018, 19, 3711. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Poel, D.; Gootjes, E.C.; Bakkerus, L.; Trypsteen, W.; Dekker, H.; van der Vliet, H.J.; Van Grieken, N.C.T.; Verhoef, C.; Buffart, T.E.; Verheul, H.M.W. A specific microRNA profile as predictive biomarker for systemic treatment in patients with metastatic colorectal cancer. Cancer Med. 2020, 9, 7558–7571. [Google Scholar] [CrossRef] [PubMed]
- Conev, N.V.; Donev, I.S.; Konsoulova-Kirova, A.A.; Chervenkov, T.G.; Kashlov, J.K.; Ivanov, K.D. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci. Trends 2015, 9, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Ke, T.-W.; Wei, P.-L.; Yeh, K.-T.; Chen, W.T.-L.; Cheng, Y.-W. MiR-92a Promotes Cell Metastasis of Colorectal Cancer Through PTEN-Mediated PI3K/AKT Pathway. Ann. Surg. Oncol. 2015, 22, 2649–2655. [Google Scholar] [CrossRef]
- Sahami-Fard, M.H.; Kheirandish, S.; Sheikhha, M.H. Expression levels of miR-143-3p and -424-5p in colorectal cancer and their clinical significance. Cancer Biomark. 2019, 24, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lorca, A.; Novillo, A.; Gaibar, M.; Gilsanz, M.F.; Galán, M.; Beltrán, L.; Antón, B.; Malón, D.; Moreno, A.; Fernández-Santander, A. miR-7, miR-10a and miR-143 Expression May Predict Response to Bevacizumab Plus Chemotherapy in Patients with Metastatic Colorectal Cancer. Pharmgenomics Pers. Med. 2021, 14, 1263–1273. [Google Scholar] [CrossRef]
- Simmer, F.; Venderbosch, S.; Dijkstra, J.R.; Vink-Börger, E.M.; Faber, C.; Mekenkamp, L.J.; Koopman, M.; De Haan, A.F.; Punt, C.J.; Nagtegaal, I.D. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget 2015, 6, 22996–23007. [Google Scholar] [CrossRef]
- Lu, D.; Yao, Q.; Zhan, C.; Le-Meng, Z.; Liu, H.; Cai, Y.; Tu, C.; Li, X.; Zou, Y.; Zhang, S. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget 2017, 8, 22674–22684. [Google Scholar] [CrossRef]
- Cai, K.; Shen, F.; Cui, J.-H.; Yu, Y.; Pan, H.-Q. Expression of miR-221 in colon cancer correlates with prognosis. Int. J. Clin. Exp. Med. 2015, 8, 2794–2798. [Google Scholar]
- Sukocheva, O.A.; Liu, J.; Neganova, M.E.; Beeraka, N.M.; Aleksandrova, Y.R.; Manogaran, P.; Grigorevskikh, E.M.; Chubarev, V.N.; Fan, R. Perspectives of using microRNA-loaded nanocarriers for epigenetic reprogramming of drug resistant colorectal cancers. Semin. Cancer Biol. 2022, 86, 358–375. [Google Scholar] [CrossRef]
- Ulivi, P.; Canale, M.; Passardi, A.; Marisi, G.; Valgiusti, M.; Frassineti, G.L.; Calistri, D.; Amadori, D.; Scarpi, E. Circulating Plasma Levels of miR-20b, miR-29b and miR-155 as Predictors of Bevacizumab Efficacy in Patients with Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 307. [Google Scholar] [CrossRef]
- Dokhanchi, M.; Pakravan, K.; Zareian, S.; Hussen, B.M.; Farid, M.; Razmara, E.; Mossahebi-Mohammadi, M.; Cho, W.C.; Babashah, S. Colorectal cancer cell-derived extracellular vesicles transfer miR-221-3p to promote endothelial cell angiogenesis via targeting suppressor of cytokine signaling 3. Life Sci. 2021, 285, 119937. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Wang, P.; Lin, D.; Dai, J.; Liu, Q.; Guan, Y.; Zhan, Y.; Yang, Y.; Wang, W.; Wang, J.; et al. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021, 112, 3744–3755. [Google Scholar] [CrossRef] [PubMed]
- Kjersem, J.B.; Ikdahl, T.; Lingjaerde, O.C.; Guren, T.; Tveit, K.M.; Kure, E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014, 8, 59–67. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.; Wu, M.; Zhu, P.; Zhou, Z.; Gong, Y.; Gu, Y. LncRNA LINC01315 silencing modulates cancer stem cell properties and epithelial-to-mesenchymal transition in colorectal cancer via miR-484/DLK1 axis. Cell Cycle 2022, 21, 851–873. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, X.; Zeng, K.; Xu, M.; He, B.; Pan, Y.; Sun, H.; Pan, B.; Xu, X.; Xu, T.; et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Bjørnetrø, T.; Redalen, K.R.; Meltzer, S.; Thusyanthan, N.S.; Samiappan, R.; Jegerschöld, C.; Handeland, K.R.; Ree, A.H. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J. Extracell. Vesicles 2019, 8, 1567219. [Google Scholar] [CrossRef]
- Wang, X.; Kattan, M.W. Cohort Studies: Design, Analysis, and Reporting. Chest 2020, 158, S72–S78. [Google Scholar] [CrossRef]
- Baldasici, O.; Balacescu, L.; Cruceriu, D.; Roman, A.; Lisencu, C.; Fetica, B.; Visan, S.; Cismaru, A.; Jurj, A.; Barbu-Tudoran, L.; et al. Circulating Small EVs miRNAs as Predictors of Pathological Response to Neo-Adjuvant Therapy in Breast Cancer Patients. Int. J. Mol. Sci. 2022, 23, 12625. [Google Scholar] [CrossRef]
- Yan, S.; Cao, Y.; Mao, A. MicroRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Front. Biosci. 2015, 20, 1092–1103. [Google Scholar] [CrossRef]
- Stiegelbauer, V.; Perakis, S.; Deutsch, A.; Ling, H.; Gerger, A.; Pichler, M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J. Gastroenterol. 2014, 20, 11727–11735. [Google Scholar] [CrossRef]
- Yang, Y.; Meng, W.-J.; Wang, Z.-Q. MicroRNAs (miRNAs): Novel potential therapeutic targets in colorectal cancer. Front. Oncol. 2022, 12, 1054846. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.O.; Senda, T. Circulating microRNA-92a-3p in colorectal cancer: A review. Med. Mol. Morphol. 2021, 54, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Khan, A.Q.; Inchakalody, V.P.; Mestiri, S.; Yoosuf, Z.S.K.M.; Bedhiafi, T.; El-Ella, D.M.A.; Taib, N.; Hydrose, S.; Akbar, S.; et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 99. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]

| Variable | N = 31 | |
|---|---|---|
| Age | Mean | 59.35 |
| Median | 61 | |
| Gender | Male | 19 (61.29%) |
| Female | 12 (38.71%) | |
| Tumor location | Right colon | 10 (32.26%) |
| Left colon | 21 (67.74%) | |
| Tumor grade | G1 | 4 (12.90%) |
| G2 | 21 (67.74%) | |
| G3 | 6 (19.36%) | |
| Metastases type | Synchronous | 19 (61.29%) |
| Metachronous | 12 (38.71%) | |
| Metastatic sites | single organ | 16 (51.61%) |
| multiple organs | 15 (48.39%) | |
| Chemotherapy protocol in first-line treatment | FOLFOX 1 | 14 (45.16%) |
| FOLFIRI 2 | 17 (54.84%) | |
| No. | miARN Name/ Accession Number | miRNA Code | FOLFIRI | FOLFOX | ||
|---|---|---|---|---|---|---|
| CT (B) | CT (PC) | CT (B) | CT (PC) | |||
| 1 | cel-miR-39 (MIMAT0000069) | uagcagcacguaaauauuggcg | 18.29 | 18.07 | 18.67 | 18.27 |
| 2 | hsa-miR-16-5p (MIMAT0000069) | uagcagcacguaaauauuggcg | 25.88 | 26.02 | 26.15 | 26.76 |
| 3 | hsa-miR-92a-3p (MIMAT0000092) | uauugcacuugucccggccugu | 24.71 | 26.10 | 24.58 | 24.49 |
| 4 | hsa-miR-143-3p (MIMAT0000435) | ugagaugaagcacuguagcuc | 30.29 | 30.66 | 29.49 | 30.05 |
| 5 | hsa-miR-146a-5p (MIMAT0000449) | ugagaacugaauuccauggguu | 28.28 | 29.54 | 28.34 | 28.57 |
| 6 | hsa-miR-486-5p (MIMAT0002177) | uccuguacugagcugccccgag | 24.84 | 26.46 | 24.29 | 24.20 |
| 7 | hsa-miR-484 (MIMAT0002174) | ucaggcucaguccccucccgau | 29.11 | 29.28 | 28.05 | 29.09 |
| 8 | hsa-miR-221-3p (MIMAT0000278) | agcuacauugucugcuggguuuc | 26.46 | 27.18 | 25.94 | 26.06 |
| miR-92a-3p p-Value | mir-143-3p p-Value | miR-146a-5p p-Value | miR-486-5p p-Value | miR-484 p-Value | mir-221-3p p-Value | ||
|---|---|---|---|---|---|---|---|
| PFS1 | |||||||
| UA High vs. Low | B | 0.003 ** HR > 1 | 0.06 HR > 1 | 0.07 HR > 1 | 0.003 ** HR > 1 | 0.074 HR > 1 | 0.193 HR > 1 |
| PC | 0.718 HR > 1 | 0.699 HR < 1 | 0.32 HR > 1 | 0.887 HR < 1 | 0.975 HR < 1 | 0.4848 HR < 1 | |
| MA High vs. Low | B | 0.512 HR > 1 | - | - | 0.44 HR > 1 | - | - |
| PP PC CC | - | - | - | - | - | - | |
| OS | |||||||
| UA High vs. Low | B | 0.008 ** HR > 1 | 0.009 ** HR > 1 | 0.112 HR > 1 | 0.019 * HR > 1 | 0.161 HR > 1 | 0.016 * HR > 1 |
| PC | 0.097 HR > 1 | 0.428 HR > 1 | 0.452 HR > 1 | 0.181 HR > 1 | 0.19 HR > 1 | 0.601 HR > 1 | |
| MA High vs. Low | B | 0.768 HR > 1 | 0.096 HR > 1 | - | 0.765 HR < 1 | - | 0.91 HR < 1 |
| PC | - | - | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gherman, A.; Balacescu, L.; Popa, C.; Cainap, C.; Vlad, C.; Cainap, S.S.; Balacescu, O. Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 10622. https://doi.org/10.3390/ijms241310622
Gherman A, Balacescu L, Popa C, Cainap C, Vlad C, Cainap SS, Balacescu O. Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(13):10622. https://doi.org/10.3390/ijms241310622
Chicago/Turabian StyleGherman, Alexandra, Loredana Balacescu, Calin Popa, Calin Cainap, Catalin Vlad, Simona S. Cainap, and Ovidiu Balacescu. 2023. "Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer" International Journal of Molecular Sciences 24, no. 13: 10622. https://doi.org/10.3390/ijms241310622
APA StyleGherman, A., Balacescu, L., Popa, C., Cainap, C., Vlad, C., Cainap, S. S., & Balacescu, O. (2023). Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer. International Journal of Molecular Sciences, 24(13), 10622. https://doi.org/10.3390/ijms241310622







