Technological Development and Application of Plant Genetic Transformation
Abstract
:1. Introduction
2. Techniques of Plant Genetic Transformation
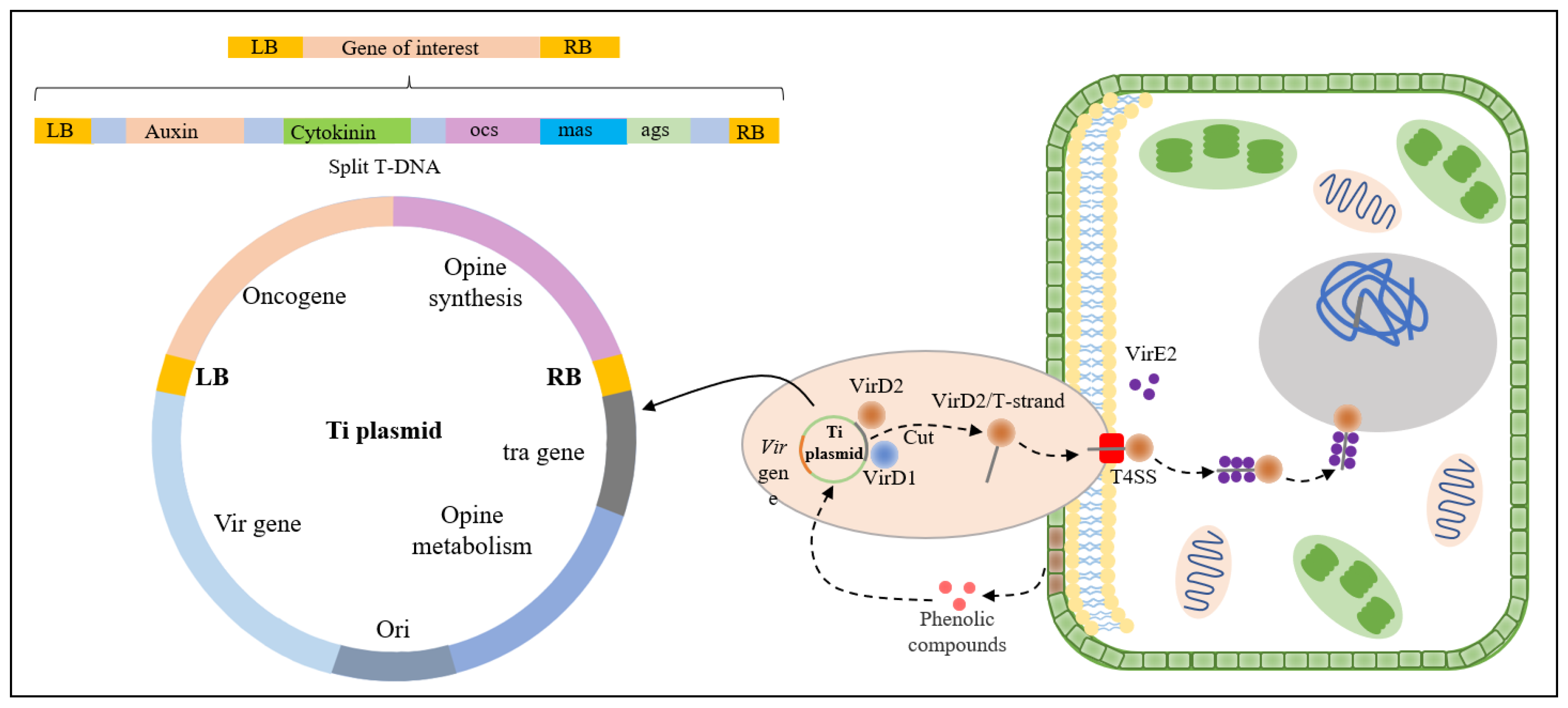
2.1. Indirect Genetic Transformation
2.2. Direct Genetic Transformation
2.2.1. Particle-Bombardment-Mediated Transformation
2.2.2. Electroporation-Mediated Plant Transformation
2.2.3. Liposome-Mediated Plant Genetic Transformation
2.2.4. Silicon-Carbide-Whisker-Mediated Transformation
2.2.5. Microinjection-Mediated Plant Genetic Transformation
2.2.6. Pollen-Tube-Pathway-Mediated Transformation
2.3. Key Factors Affecting Plant Genetic Transformation
3. Strategies to Overcome the Limitations of Traditional Gene Transfer Methods
3.1. Nanoparticle-Mediated Gene Delivery
3.1.1. Magnetic-Nanoparticle-Mediated Gene Delivery
3.1.2. Peptide-Mediated Gene Delivery
3.1.3. Layered-Double-Hydroxide-Mediated Gene Delivery
3.1.4. DNA-Nanostructure-Mediated Gene Delivery
3.1.5. Carbon-Nanotube-Mediated Gene Delivery
3.2. CRISPR/Cas/ZFN-Mediated Targeted DNA Insertion
3.2.1. CRISPR/Cas-Mediated Targeted DNA Insertion
3.2.2. ZFN-Mediated Targeted DNA Insertion
4. Ways to Overcome the Difficulty of Transformed Plant Regeneration
4.1. Baby Boom and Wuschel2
4.2. GRF
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, Y.; Zhu, X.; Yu, Y.; Li, C.; Zhang, Z.; Wang, F. Nanotechnology Strategies for Plant Genetic Engineering. Adv. Mater. 2022, 34, 2106945. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, T.R.; Lenka, S.K.; Arya, S.S.; Bansal, K.C. A Short History and Perspectives on Plant Genetic Transformation. In Biolistic DNA Delivery in Plants; Rustgi, S., Luo, H., Eds.; Humana: New York, NY, USA, 2020; pp. 39–68. [Google Scholar]
- Xu, N.; Kang, M.; Zobrist, J.D.; Wang, K.; Fei, S. Genetic Transformation of Recalcitrant Upland Switchgrass Using Morphogenic Genes. Front. Plant Sci. 2022, 12, 781565. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.M. Particle bombardment: An established weapon in the arsenal of plant biotechnologists. In Plant Transformation Technologies; Stewart, C.N., Touraev, A., Citovsky, V., Tzfira, T., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2011; pp. 53–71. [Google Scholar]
- Rao, A.Q.; Bakhsh, A.; Kiani, S.; Shahzad, K.; Shahid, A.A.; Husnain, T.; Riazuddin, S. The Myth of Plant Transformation. Biotechnol. Adv. 2009, 27, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Anami, S.; Njuguna, E.; Coussens, G.; Aesaert, S.; Lijsebettens, M.V. Higher Plant Transformation: Principles and Molecular Tools. Int. J. Dev. Biol. 2013, 57, 483–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demirer, G.S.; Zhang, H.; Matos, J.L.; Goh, N.S.; Cunningham, F.J.; Sung, Y.; Chang, R.; Aditham, A.J.; Chio, L.; Cho, M.J.; et al. High Aspect Ratio Nanomaterials Enable Delivery of Functional Genetic Material without DNA Integration in Mature Plants. Nat. Nanotechnol. 2019, 14, 456–464. [Google Scholar] [CrossRef]
- Cunningham, F.J.; Goh, N.S.; Demirer, G.S.; Matos, J.L.; Landry, M.P. Nanoparticle-Mediated Delivery towards Advancing Plant Genetic Engineering. Trends Biotechnol. 2018, 36, 882–897. [Google Scholar] [CrossRef]
- Lv, Z.; Jiang, R.; Chen, J.; Chen, W. Nanoparticle-mediated Gene Transformation Strategies for Plant Genetic Engineering. Plant J. 2020, 104, 880–891. [Google Scholar] [CrossRef]
- Lee, K.; Wang, K. Strategies for Genotype-Flexible Plant Transformation. Curr. Opin. Biotechnol. 2023, 79, 102848. [Google Scholar] [CrossRef]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid Genotype “Independent” Zea Mays L. (Maize) Transformation via Direct Somatic Embryogenesis. Vitr. Cell. Dev. Biol.-Plant 2018, 54, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Ryan, L.; Sardesai, N.; Wu, E.; Lenderts, B.; Lowe, K.; Che, P.; Anand, A.; Worden, A.; van Dyk, D.; et al. Leaf Transformation for Efficient Random Integration and Targeted Genome Modification in Maize and Sorghum. Nat. Plants 2023, 9, 255–270. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-Transcriptional Control of GRF Transcription Factors by microRNA miR396 and GIF Co-Activator Affects Leaf Size and Longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF Chimeric Protein Improves the Regeneration Efficiency of Transgenic Plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 Controls Grain Size and Yield in Rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef]
- Qiu, F.; Xing, S.; Xue, C.; Liu, J.; Chen, K.; Chai, T.; Gao, C. Transient Expression of a TaGRF4-TaGIF1 Complex Stimulates Wheat Regeneration and Improves Genome Editing. Sci. China Life Sci. 2022, 65, 731–738. [Google Scholar] [CrossRef]
- Keshavareddy, G.; Kumar, A.R.V.; Ramu, V.S. Methods of Plant Transformation—A Review. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 2656–2668. [Google Scholar] [CrossRef]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the Plant Genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef]
- Ramachandra, S.R.; Ravishankar, G.A. Plant Cell Cultures: Chemical Factories of Secondary Metabolites. Biotechnol. Adv. 2002, 20, 101–153. [Google Scholar] [CrossRef] [PubMed]
- Flores, H.E.; Vivanco, J.M.; Loyola-Vargas, V.M. ‘Radicle’ biochemistry: The biology of root-specific metabolism. Trends Plant Sci. 1999, 4, 220–226. [Google Scholar] [CrossRef]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut–dip–budding Delivery System Enables Genetic Modifications in Plants without Tissue Culture. Innovation 2023, 4, 100345. [Google Scholar] [CrossRef]
- Mirzaee, M.; Osmani, Z.; Frébortová, J.; Frébort, I. Recent Advances in Molecular Farming Using Monocot Plants. Biotechnol. Adv. 2022, 58, 107913. [Google Scholar] [CrossRef]
- Kausch, A.P.; Nelson-Vasilchik, K.; Hague, J.; Mookkan, M.; Quemada, H.; Dellaporta, S.; Fragoso, C.; Zhang, Z.J. Edit at Will: Genotype Independent Plant Transformation in the Era of Advanced Genomics and Genome Editing. Plant Sci. 2019, 281, 186–205. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Bass, S.H.; Wu, E.; Wang, N.; McBride, K.E.; Annaluru, N.; Miller, M.; Hua, M.; Jones, T.J. An Improved Ternary Vector System for Agrobacterium-Mediated Rapid Maize Transformation. Plant Mol. Biol. 2018, 97, 187–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayta, S.; Smedley, M.A.; Demir, S.U.; Blundell, R.; Hinchliffe, A.; Atkinson, N.; Harwood, W.A. An Efficient and Reproducible Agrobacterium-Mediated Transformation Method for Hexaploid Wheat (Triticum aestivum L.). Plant Methods 2019, 15, 121. [Google Scholar] [CrossRef] [PubMed]
- Hensel, G.; Marthe, C.; Kumlehn, J. Agrobacterium-Mediated Transformation of Wheat Using Immature Embryos. In Wheat (Triticum aestivum L.) Transformation Using Immature Embryos; Bhalla, P.L., Singh, M.B., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1679, pp. 129–139. [Google Scholar]
- Kumar, R.; Mamrutha, H.M.; Kaur, A.; Venkatesh, K.; Sharma, D.; Singh, G.P. Optimization of Agrobacterium-Mediated Transformation in Spring Bread Wheat Using Mature and Immature Embryos. Mol. Biol. Rep. 2019, 46, 1845–1853. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Du, L.; Ye, X. Generation of Marker-Free Transgenic Hexaploid Wheat via an Agrobacterium -Mediated Co-Transformation Strategy in Commercial Chinese Wheat Varieties. Plant Biotechnol. J. 2017, 15, 614–623. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Tsuda, K.; Nosaka-Takahashi, M.; Suzuki, T.; Ono, S.; Ta, K.N.; Yoshida, Y.; Nonomura, K.-I.; Sato, Y. Agrobacterium-Mediated Genetic Transformation of Wild Oryza Species Using Immature Embryos. Rice 2020, 13, 33. [Google Scholar] [CrossRef]
- Samara Shekar Reddy, S.; Singh, B.; John Peter, A.; Venkateswar Rao, T. Genetic Transformation of Indica Rice Varieties Involving Am-SOD Gene for Improved Abiotic Stress Tolerance. Saudi J. Biol. Sci. 2019, 26, 294–300. [Google Scholar] [CrossRef]
- Liu, S.; Shi, Y.; Liu, F.; Guo, Y.; Lu, M. LaCl3 Treatment Improves Agrobacterium-Mediated Immature Embryo Genetic Transformation Frequency of Maize. Plant Cell Rep. 2022, 41, 1439–1448. [Google Scholar] [CrossRef]
- Zalewski, W.; Orczyk, W.; Gasparis, S.; Nadolska-Orczyk, A. HvCKX2 Gene Silencing by Biolistic or Agrobacterium-Mediated Transformation in Barley Leads to Different Phenotypes. BMC Plant Biol. 2012, 12, 206. [Google Scholar] [CrossRef] [Green Version]
- Harwood, W.A. A Protocol for High-Throughput Agrobacterium-Mediated Barley Transformation. In Cereal Genomics; Henry, R.J., Furtado, A., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1099, pp. 251–260. [Google Scholar]
- Wang, X.; Chen, S.; Zhang, H.; Luo, P.; Zhou, F.; Zeng, B.; Xu, J.; Fan, C. Agrobacterium-Mediated Genetic Transformation of the Most Widely Cultivated Superior Clone Eucalyptus urophylla × Eucalyptus grandis DH32-29 in Southern China. Front. Plant Sci. 2022, 13, 1011245. [Google Scholar] [CrossRef]
- Song, C.; Lu, L.; Guo, Y.; Xu, H.; Li, R. Efficient Agrobacterium-Mediated Transformation of the Commercial Hybrid Poplar Populus alba × Populus glandulosa Uyeki. IJMS 2019, 20, 2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, S.S.; Ge, X.L.; Wang, R.; Yang, H.F.; Bai, Y.E.; Guo, Y.H.; Zhang, J.; Lu, M.Z.; Zhao, S.T.; Wang, L.Q. An Efficient Agrobacterium-Mediated Transformation Method for Hybrid Poplar 84K (Populus alba × P. glandulosa) Using Calli as Explants. IJMS 2022, 23, 2216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Ji, J.J.; Jiang, F.; Tian, X.R.; Li, J.K.; Gao, J.P. Establishment of a Genetic Transformation System for Codonopsis pilosula Callus. Plant Biotechnol. 2022, 39, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Chen, C.; Xu, Y.; Hu, S.; Wang, L.; Sun, K.; Chen, X.; Li, X. Optimization of Agrobacterium tumefaciens -Mediated Transformation Systems in Tea Plant (Camellia sinensis). Hortic. Plant J. 2017, 3, 105–109. [Google Scholar] [CrossRef]
- Li, M.; Wang, D.; Long, X.; Hao, Z.; Lu, Y.; Zhou, Y.; Peng, Y.; Cheng, T.; Shi, J.; Chen, J. Agrobacterium-Mediated Genetic Transformation of Embryogenic Callus in a Liriodendron Hybrid (L. chinense × L. tulipifera). Front. Plant Sci. 2022, 13, 802128. [Google Scholar] [CrossRef]
- Karmakar, S.; Molla, K.A.; Gayen, D.; Karmakar, A.; Das, K.; Sarkar, S.N.; Datta, K.; Datta, S.K. Development of a Rapid and Highly Efficient Agrobacterium -Mediated Transformation System for Pigeon Pea [ Cajanus cajan (L.) Millsp]. GM Crops Food 2019, 10, 115–138. [Google Scholar] [CrossRef] [Green Version]
- Ning, W.; Zhai, H.; Yu, J.; Liang, S.; Yang, X.; Xing, X.; Huo, J.; Pang, T.; Yang, Y.; Bai, X. Overexpression of Glycine Soja WRKY20 Enhances Drought Tolerance and Improves Plant Yields under Drought Stress in Transgenic Soybean. Mol. Breed. 2017, 37, 19. [Google Scholar] [CrossRef]
- Ge, X.; Xu, J.; Yang, Z.; Yang, X.; Wang, Y.; Chen, Y.; Wang, P.; Li, F. Efficient Genotype-independent Cotton Genetic Transformation and Genome Editing. JIPB 2023, 65, 907–917. [Google Scholar] [CrossRef]
- Sanford, J.C. The Development of the Biolistics Process. Vitr. Cell. Dev. Biol. -Plant 2000, 36, 303–308. [Google Scholar] [CrossRef]
- Paszkowski, J.; Shillito, R.D.; Saul, M.; Mandák, V.; Hohn, T.; Hohn, B.; Potrykus, I. Direct Gene Transfer to Plants. EMBO J. 1984, 3, 2717–2722. [Google Scholar] [CrossRef]
- Altpeter, F.; Baisakh, N.; Beachy, R.; Bock, R.; Capell, T.; Christou, P.; Daniell, H.; Datta, K.; Datta, S.; Dix, P.J.; et al. Particle Bombardment and the Genetic Enhancement of Crops: Myths and Realities. Mol. Breed. 2005, 15, 305–327. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Targeted DNA Insertion in Plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.B.; Gray, B.N.; January, E.; Gordon, G.C.; He, Y.; Liu, H.; Wu, X.; Brutnell, T.P.; Mockler, T.C.; Oufattole, M. Precise Insertion and Guided Editing of Higher Plant Genomes Using Cpf1 CRISPR Nucleases. Sci. Rep. 2017, 7, 11606. [Google Scholar] [CrossRef] [Green Version]
- Bonawitz, N.D.; Ainley, W.M.; Itaya, A.; Chennareddy, S.R.; Cicak, T.; Effinger, K.; Jiang, K.; Mall, T.K.; Marri, P.R.; Samuel, J.P.; et al. Zinc Finger Nuclease-mediated Targeting of Multiple Transgenes to an Endogenous Soybean Genomic Locus via Non-homologous End Joining. Plant Biotechnol. J. 2019, 17, 750–761. [Google Scholar] [CrossRef]
- Saeed, T.; Shahzad, A. Basic Principles Behind Genetic Transformation in Plants. In Biotechnological Strategies for the Conservation of Medicinal and Ornamental Climbers; Shahzad, A., Sharma, S., Siddiqui, S., Eds.; Springer: Cham, Switzerland, 2016; pp. 327–350. [Google Scholar]
- Ozyigit, I.I. Gene Transfer to Plants by Electroporation: Methods and Applications. Mol. Biol. Rep. 2020, 47, 3195–3210. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, T.; Rems, L.; Tarek, M.; Miklavčič, D. Membrane Electroporation and Electropermeabilization: Mechanisms and Models. Annu. Rev. Biophys. 2019, 48, 63–91. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Loganathan, M.; Dey, K.; Shinde, P.; Chang, H.-Y.; Nagai, M.; Santra, T.S. Single-Cell Electroporation: Current Trends, Applications and Future Prospects. J. Micromech. Microeng. 2018, 28, 123002. [Google Scholar] [CrossRef]
- Bates, G.W. Plant Transformation via Protoplast Electroporation. In Plant Cell Culture Protocols; Hall, R.D., Ed.; Humana Press: New York, NY, USA, 1999; pp. 359–366. [Google Scholar]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, Preparation, and Applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A. Liposomes in Drug Delivery: Progress and Limitations. Int. J. Pharm. 1997, 154, 123–140. [Google Scholar] [CrossRef]
- Gad, A.E.; Rosenberg, N.; Altman, A. Liposome-Mediated Gene Delivery into Plant Cells. Physiol. Plant 1990, 79, 177–183. [Google Scholar] [CrossRef]
- van Wordragen, M.; Shakya, R.; Verkerk, R.; Peytavis, R.; van Kammen, A.; Zabel, P. Liposome-Mediated Transfer of YAC DNA to Tobacco Cells. Plant Mol. Biol. Rep. 1997, 15, 170–178. [Google Scholar] [CrossRef]
- Hassan, M.; Akram, Z.; Ali, S.; Ali, G.M.; Zafar, Y.; Shah, Z.H.; Alghabari, F. Whisker-Mediated Transformation of Peanut with Chitinase Gene Enhances Resistance to Leaf Spot Disease. Crop Breed. Appl. Biotechnol. 2016, 16, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Mutsuddy, B.C. Electrokinetic Behavior of Aqueous Silicon Carbide Whisker Suspensions. J. Am. Ceram. Soc. 1990, 73, 2747–2749. [Google Scholar] [CrossRef]
- Kaeppler, H.F.; Gu, W.; Somers, D.A.; Rines, H.W.; Cockburn, A.F. Silicon Carbide Fiber-Mediated DNA Delivery into Plant Cells. Plant Cell Rep. 1990, 9, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, H.F.; Somers, D.A.; Rines, H.W.; Cockburn, A.F. Silicon Carbide Fiber-Mediated Stable Transformation of Plant Cells. Theoret. Appl. Genet. 1992, 84, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Takahashi, W.; Ohyama, T.; Shimada, T.; Tanaka, O. Improvement of the Aluminum Borate Whisker-Mediated Method of DNA Delivery into Rice Callus. Plant Prod. Sci. 2004, 7, 45–49. [Google Scholar] [CrossRef]
- Crossway, A.; Oakes, J.V.; Irvine, J.M.; Ward, B.; Knauf, V.C.; Shewmaker, C.K. Integration of Foreign DNA Following Microinjection of Tobacco Mesophyll Protoplasts. Mol. Gen. Genet. 1986, 202, 179–185. [Google Scholar] [CrossRef]
- Morikawa, H.; Yamada, Y. Capillary Microinjection into Protoplasts and Intranuclear Localization of Injected Materials. Plant Cell Physiol. 1985, 26, 229–236. [Google Scholar]
- Lorz, H.; Paszkowski, J.; Dierks-Ventling, C.; Potrykus, I. Isolation and Characterization of Cytoplasts and Miniprotoplasts Derived from Protoplasts of Cultured Cells. Physiol. Plant 1981, 53, 385–391. [Google Scholar] [CrossRef]
- Matsumoto, T.K.; Gonsalves, D. Biolistic and Other Non-Agrobacterium Technologies of Plant Transformation. In Plant Biotechnology and Agriculture; Altman, A., Hasegawa, P.W., Eds.; Academic Press: Cambridge, MA, USA, 2012; pp. 117–129. [Google Scholar]
- Luo, Z.; Wu, R. A Simple Method for the Transformation of Rice via the Pollen-Tube Pathway. Plant Mol. Biol. Report. 1989, 7, 69–77. [Google Scholar] [CrossRef]
- Ali, A.; Bang, S.W.; Chung, S.-M.; Staub, J.E. Plant Transformation via Pollen Tube-Mediated Gene Transfer. Plant Mol. Biol. Rep. 2015, 33, 742–747. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, X.; Yang, A.; Li, G.; Zhang, J. Stability of Inheritance of Transgenes in Maize (Zea mays L.) Lines Produced Using Different Transformation Methods. Euphytica 2005, 144, 11–22. [Google Scholar] [CrossRef]
- Hu, C.Y.; Wang, L. In Planta Soybean Transformation Technologies Developed in China: Procedure, Confirmation and Field Performance. Vitr. Cell. Dev. Biol. -Plant 1999, 35, 417–420. [Google Scholar] [CrossRef]
- Zhang, L.; Gu, L.; Ringler, P.; Smith, S.; Rushton, P.J.; Shen, Q.J. Three WRKY Transcription Factors Additively Repress Abscisic Acid and Gibberellin Signaling in Aleurone Cells. Plant Sci. 2015, 236, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla Hemoglobin Increases Waterlogging Tolerance in Arabidopsis and Maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef] [Green Version]
- Belide, S.; Vanhercke, T.; Petrie, J.R.; Singh, S.P. Robust Genetic Transformation of Sorghum (Sorghum bicolor L.) Using Differentiating Embryogenic Callus Induced from Immature Embryos. Plant Methods 2017, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Yang, Y.; Wang, C.; Guo, J.; Zhou, D.; Wu, Q.; Su, Y.; Xu, L.; Que, Y. Transgenic Sugarcane with a Cry1Ac Gene Exhibited Better Phenotypic Traits and Enhanced Resistance against Sugarcane Borer. PLoS ONE 2016, 11, e0153929. [Google Scholar] [CrossRef] [Green Version]
- Miroshnichenko, D.; Ashin, D.; Pushin, A.; Dolgov, S. Genetic Transformation of Einkorn (Triticum monococcum L. ssp. Monococcum L.), a Diploid Cultivated Wheat Species. BMC Biotechnol. 2018, 18, 68. [Google Scholar]
- Duan, X.; Hou, Q.; Liu, G.; Pang, X.; Niu, Z.; Wang, X.; Zhang, Y.; Li, B.; Liang, R. Expression of Pinellia Pedatisecta Lectin Gene in Transgenic Wheat Enhances Resistance to Wheat Aphids. Molecules 2018, 23, 748. [Google Scholar] [CrossRef] [Green Version]
- Allam, M.A.; Saker, M.M. Microprojectile Bombardment Transformation of Date Palm Using the Insecticidal Cholesterol Oxidase (ChoA) Gene. In Date Palm Biotechnology Protocols Volume I.; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1637, pp. 281–293. [Google Scholar]
- Das, D.K. Expression of a Bacterial Chitinase (ChiB) Gene Enhances Resistance against Erysiphae polygoni Induced Powdery Mildew Disease in the Transgenic Black Gram (Vigna mungo L.) (Cv. T9). AJPS 2018, 09, 1759–1770. [Google Scholar] [CrossRef] [Green Version]
- Grazziotin, M.A.G.D.; Cabral, G.B.; Ibrahim, A.B.; Machado, R.B.; Aragão, F.J.L. Expression of the Arcelin 1 gene from Phaseolus vulgaris L. in cowpea seeds (Vigna unguiculata L.) confers bruchid resistant. Ann. Appl. Biol. 2020, 176, 268–274. [Google Scholar] [CrossRef]
- Wu, H.; Acanda, Y.; Jia, H.; Wang, N.; Zale, J. Biolistic Transformation of Carrizo Citrange (Citrus sinensis Osb. × Poncirus trifoliata L. Raf.). Plant Cell Rep. 2016, 35, 1955–1962. [Google Scholar] [CrossRef] [PubMed]
- Kawai, J.; Kanazawa, M.; Suzuki, R.; Kikuchi, N.; Hayakawa, Y.; Sekimoto, H. Highly Efficient Transformation of the Model Zygnematophycean Alga Closterium Peracerosum-strigosum-littorale Complex by Square-pulse Electroporation. New Phytol. 2022, 233, 569–578. [Google Scholar] [CrossRef]
- Sorokin, A.P.; Ke, X.; Chen, D.; Elliott, M.C. Production of Fertile Transgenic Wheat Plants via Tissue Electroporation. Plant Sci. 2000, 156, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Karny, A.; Zinger, A.; Kajal, A.; Shainsky-Roitman, J.; Schroeder, A. Therapeutic Nanoparticles Penetrate Leaves and Deliver Nutrients to Agricultural Crops. Sci. Rep. 2018, 8, 7589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petolino, J.F.; Hopkins, N.L.; Kosegi, B.D.; Skokut, M. Whisker-Mediated Transformation of Embryogenic Callus of Maize. Plant Cell Rep. 2000, 19, 781–786. [Google Scholar] [CrossRef]
- Asad, S.; Mukhtar, Z.; Nazir, F.; Hashmi, J.A.; Mansoor, S.; Zafar, Y.; Arshad, M. Silicon Carbide Whisker-Mediated Embryogenic Callus Transformation of Cotton (Gossypium hirsutum L.) and Regeneration of Salt Tolerant Plants. Mol. Biotechnol. 2008, 40, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Asad, S. Embryogenic Calli Explants and Silicon Carbide Whisker-Mediated Transformation of Cotton (Gossypium hirsutum L.). In Transgenic Cotton, Zhang, B., Eds.; Springer: New York, NY, USA, 2019; pp. 75–91. [Google Scholar]
- Holm, P.B.; Olsen, O.; Schnorf, M.; Brinch-Pedersen, H. Transformation of Barley by Microinjection into Isolated Zygote Protoplasts. Transgenic Res. 2000, 9, 21–32. [Google Scholar] [CrossRef]
- Masani, M.Y.A.; Noll, G.A.; Parveez, G.K.A.; Sambanthamurthi, R.; Prüfer, D. Efficient Transformation of Oil Palm Protoplasts by PEG-Mediated Transfection and DNA Microinjection. PLoS ONE 2014, 9, e96831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.; Su, Q.; An, L. Ovary-Drip Transformation: A Simple Method for Directly Generating Vector- and Marker-Free Transgenic Maize (Zea mays L.) with a Linear GFP Cassette Transformation. Planta 2009, 229, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, R.; Zhang, B.; Wang, Q. Pollen Tube Pathway-Mediated Cotton Transformation. In Transgenic Cotton; Zhang, B., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1902, pp. 67–73. [Google Scholar]
- Li, X.; Liu, L.; Liu, X.; Hou, Y.; Xu, B.; Luan, F.; Wang, X. Construction of Expression Vectors of the Melon Resistance Gene Fom-2 and Genetic Transformation. Pak. J. Bot 2019, 51, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Luo, J.; Xiao, D.; Wang, A.; He, L.; Zhan, J. An Efficient Method for the Production of Transgenic Peanut Plants by Pollen Tube Transformation Mediated by Agrobacterium tumefaciens. Plant Cell Tiss. Organ. Cult. 2023, 152, 207–214. [Google Scholar] [CrossRef]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, S.; Samad, A.A.; Rahmat, Z. Agrobacterium-Mediated Transformation of Rice: Constraints and Possible Solutions. Rice Sci. 2019, 26, 133–146. [Google Scholar] [CrossRef]
- Shahzad, A.; Yadav, V.; Ahmad, Z. Plant Tissue Culture: Profile of Pioneers. In Biotechnological Strategies for the Conservation of Medicinal and Ornamental Climbers; Shahzad, A., Sharma, S., Siddiqui, S., Eds.; Springer: Cham, Switzerland, 2016; pp. 141–162. [Google Scholar]
- Kotnik, T.; Frey, W.; Sack, M.; Meglič, S.H.; Peterka, M.; Miklavčič, D. Electroporation-Based Applications in Biotechnology. Trends Biotechnol. 2015, 33, 480–488. [Google Scholar] [CrossRef]
- Zhao, X.; Meng, Z.; Wang, Y.; Chen, W.; Sun, C.; Cui, B.; Cui, J.; Yu, M.; Zeng, Z.; Guo, S.; et al. Pollen Magnetofection for Genetic Modification with Magnetic Nanoparticles as Gene Carriers. Nat. Plants 2017, 3, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M. Peptides, Polypeptides and Peptide-Polymer Hybrids as Nucleic Acid Carriers. Biomater. Sci. 2017, 5, 2188–2211. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.W.; Hu, J.W.; Liu, B.R.; Lee, C.Y.; Li, J.F.; Chou, J.C.; Lee, H.J. Arginine-Rich Intracellular Delivery Peptides Synchronously Deliver Covalently and Noncovalently Linked Proteins into Plant Cells. J. Agric. Food Chem. 2010, 58, 2288–2294. [Google Scholar] [CrossRef]
- Rosenbluh, J.; Singh, S.K.; Gafni, Y.; Graessmann, A.; Loyter, A. Non-Endocytic Penetration of Core Histones into Petunia Protoplasts and Cultured Cells: A Novel Mechanism for the Introduction of Macromolecules into Plant Cells. Biochim. Et. Biophys. Acta (BBA)—Biomembr. 2004, 1664, 230–240. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Chou, J.C.; Lee, H.J. Cellular Internalization of Fluorescent Proteins via Arginine-Rich Intracellular Delivery Peptide in Plant Cells. Plant Cell Physiol. 2005, 46, 482–488. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Chou, J.; Chen, C.; Liu, B.R.; Lee, H. Noncovalent Protein Transduction in Plant Cells by Macropinocytosis. New Phytol. 2007, 174, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Chuah, J.-A.; Numata, K. Stimulus-Responsive Peptide for Effective Delivery and Release of DNA in Plants. Biomacromolecules 2018, 19, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, T.; Oikawa, K.; Chuah, J.A.; Kodama, Y.; Numata, K. Selective Gene Delivery for Integrating Exogenous DNA into Plastid and Mitochondrial Genomes Using Peptide-DNA Complexes. Biomacromolecules 2018, 19, 1582–1591. [Google Scholar] [CrossRef] [Green Version]
- Thagun, C.; Chuah, J.; Numata, K. Targeted Gene Delivery into Various Plastids Mediated by Clustered Cell-Penetrating and Chloroplast-Targeting Peptides. Adv. Sci. 2019, 6, 1902064. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.P.; Zeng, Q.H.; Lu, G.Q.; Yu, A.B. Inorganic Nanoparticles as Carriers for Efficient Cellular Delivery. Chem. Eng. Sci. 2006, 61, 1027–1040. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, H.; Lu, S. Preparation and Application of Layered Double Hydroxide Nanosheets. RSC Adv. 2021, 11, 24254–24281. [Google Scholar] [CrossRef]
- Bao, W.; Wang, J.; Wang, Q.; O’Hare, D.; Wan, Y. Layered Double Hydroxide Nanotransporter for Molecule Delivery to Intact Plant Cells. Sci. Rep. 2016, 6, 26738. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Vaghasiya, K.; Verma, R.K.; Yadav, A.B. DNA Nanostructures: Chemistry, Self-Assembly, and Applications. In Emerging Applications of Nanoparticles and Architecture Nanostructures; Barhoum, A., Makhlouf, A.S.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 71–94. [Google Scholar]
- Wang, Y.; Benson, E.; Fördős, F.; Lolaico, M.; Baars, I.; Fang, T.; Teixeira, A.; Högberg, B. DNA Origami Penetration in Cell Spheroid Tissue Models Is Enhanced by Wireframe Design. Adv. Mater. 2021, 33, e2008457. [Google Scholar] [CrossRef]
- Mohanta, D.; Patnaik, S.; Sood, S.; Das, N. Carbon Nanotubes: Evaluation of Toxicity at Biointerfaces. J. Pharm. Anal. 2019, 9, 293–300. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, B.; Wang, Q.; Shi, X.; Xiao, Z.; Lin, J.; Fang, X. Carbon Nanotubes as Molecular Transporters for Walled Plant Cells. Nano Lett. 2009, 9, 1007–1010. [Google Scholar] [CrossRef]
- Singh, S.; Chaudhary, R.; Deshmukh, R.; Tiwari, S. Opportunities and Challenges with CRISPR-Cas Mediated Homologous Recombination Based Precise Editing in Plants and Animals. Plant Mol. Biol. 2023, 111, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Collonnier, C.; Guyon-Debast, A.; Maclot, F.; Mara, K.; Charlot, F.; Nogué, F. Towards Mastering CRISPR-Induced Gene Knock-in in Plants: Survey of Key Features and Focus on the Model Physcomitrella patens. Methods 2017, 121, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency Gene Targeting in Hexaploid Wheat Using DNA Replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Wang, M.; Chen, M.; Dong, J.; Zhang, T.; Li, F.; Lei, M.; et al. Targeted, Efficient Sequence Insertion and Replacement in Rice. Nat. Biotechnol. 2020, 38, 1402–1407. [Google Scholar] [CrossRef]
- Schiml, S.; Fauser, F.; Puchta, H. The CRISPR/Cas System Can Be Used as Nuclease for in Planta Gene Targeting and as Paired Nickases for Directed Mutagenesis in Arabidopsis Resulting in Heritable Progeny. Plant J. 2014, 80, 1139–1150. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-Frequency, Precise Modification of the Tomato Genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [Green Version]
- Carroll, D. Genome Engineering With Zinc-Finger Nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Wright, D.A.; Townsend, J.A.; Winfrey, R.J.; Irwin, P.A.; Rajagopal, J.; Lonosky, P.M.; Hall, B.D.; Jondle, M.D.; Voytas, D.F. High-Frequency Homologous Recombination in Plants Mediated by Zinc-Finger Nucleases: Recombination and Zinc-Finger Nucleases. Plant J. 2005, 44, 693–705. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise Genome Modification in the Crop Species Zea Mays Using Zinc-Finger Nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Kumar, S.; AlAbed, D.; Worden, A.; Novak, S.; Wu, H.; Ausmus, C.; Beck, M.; Robinson, H.; Minnicks, T.; Hemingway, D.; et al. A Modular Gene Targeting System for Sequential Transgene Stacking in Plants. J. Biotechnol. 2015, 207, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Z.; Zheng, D.; Zhang, T.; Li, X.; Zhang, C.; Yu, R.; Wei, J.; Wu, Z. Efficient and Genotype Independent Maize Transformation Using Pollen Transfected by DNA-coated Magnetic Nanoparticles. JIPB 2022, 64, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Martin-Ortigosa, S.; Peterson, D.J.; Valenstein, J.S.; Lin, V.S.Y.; Trewyn, B.G.; Lyznik, L.A.; Wang, K. Mesoporous Silica Nanoparticle-Mediated Intracellular Cre Protein Delivery for Maize Genome Editing via LoxP Site Excision. Plant Physiol. 2014, 164, 537–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, N.; Ather, L.; Shafiq, M.; Nawaz-ul-Rehman, M.S.; Haseeb, M.; Anjum, T.; Abbas, Q.; Hussain, M.; Ali, N.; Asad Abbas, S.A.A.; et al. Magnetofection Approach for the Transformation of Okra Using Green Iron Nanoparticles. Sci. Rep. 2022, 12, 16568. [Google Scholar] [CrossRef]
- Odahara, M.; Horii, Y.; Itami, J.; Watanabe, K.; Numata, K. Functional Peptide-Mediated Plastid Transformation in Tobacco, Rice, and Kenaf. Front. Plant Sci. 2022, 13, 989310. [Google Scholar] [CrossRef]
- Yong, J.; Zhang, R.; Bi, S.; Li, P.; Sun, L.; Mitter, N.; Carroll, B.J.; Xu, Z.P. Sheet-like Clay Nanoparticles Deliver RNA into Developing Pollen to Efficiently Silence a Target Gene. Plant Physiol. 2021, 187, 886–899. [Google Scholar] [CrossRef]
- Zhang, H.; Demirer, G.S.; Zhang, H.; Ye, T.; Goh, N.S.; Aditham, A.J.; Cunningham, F.J.; Fan, C.; Landry, M.P. DNA Nanostructures Coordinate Gene Silencing in Mature Plants. Proc. Natl. Acad. Sci. USA 2019, 116, 7543–7548. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, T.; Tsakirpaloglou, N.; Septiningsih, E.M.; Thomson, M.J. Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds. Int. J. Mol. Sci. 2022, 23, 4081. [Google Scholar] [CrossRef]
- Demirer, G.S.; Zhang, H.; Goh, N.S.; Pinals, R.L.; Chang, R.; Landry, M.P. Carbon Nanocarriers Deliver SiRNA to Intact Plant Cells for Efficient Gene Knockdown. Sci. Adv. 2020, 6, eaaz0495. [Google Scholar] [CrossRef]
- Law, S.S.Y.; Liou, G.; Nagai, Y.; Giménez-Dejoz, J.; Tateishi, A.; Tsuchiya, K.; Kodama, Y.; Fujigaya, T.; Numata, K. Polymer-Coated Carbon Nanotube Hybrids with Functional Peptides for Gene Delivery into Plant Mitochondria. Nat. Commun. 2022, 13, 2417. [Google Scholar] [CrossRef]
- Su, Y.H.; Tang, L.P.; Zhao, X.Y.; Zhang, X.S. Plant Cell Totipotency: Insights into Cellular Reprogramming. J. Integr. Plant Biol. 2021, 63, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; Van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of Grain Size and Rice Yield by GL2-Mediated Brassinosteroid Responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef]
- Kumar, S.; Liu, Z.-B.; Sanyour-Doyel, N.; Lenderts, B.; Worden, A.; Anand, A.; Cho, H.-J.; Bolar, J.; Harris, C.; Huang, L.; et al. Efficient Gene Targeting in Soybean Using Ochrobactrum Haywardense -Mediated Delivery of a Marker-Free Donor Template. Plant Physiol. 2022, 189, 585–594. [Google Scholar] [CrossRef]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An Efficient DNA- and Selectable-Marker-Free Genome-Editing System Using Zygotes in Rice. Nat. Plants 2019, 5, 363–368. [Google Scholar] [CrossRef]
- Lu, L.; Wu, X.; Yin, X.; Morrand, J.; Chen, X.; Folk, W.R.; Zhang, Z.J. Development of Marker-Free Transgenic Sorghum [Sorghum bicolor (L.) Moench] Using Standard Binary Vectors with Bar as a Selectable Marker. Plant Cell Tiss. Organ Cult. 2009, 99, 97–108. [Google Scholar] [CrossRef]
- Puchta, H. Marker-Free Transgenic Plants. Plant Cell Tiss. Organ. Cult. 2003, 74, 123–134. [Google Scholar] [CrossRef]
- Wang, P.; Lombi, E.; Zhao, F.J.; Kopittke, P.M. Nanotechnology: A New Opportunity in Plant Sciences. Trends Plant Sci. 2016, 21, 699–712. [Google Scholar] [CrossRef]
- Maren, N.A.; Duan, H.; Da, K.; Yencho, G.C.; Ranney, T.G.; Liu, W. Genotype-Independent Plant Transformation. Hortic. Res. 2022, 9, uhac047. [Google Scholar] [CrossRef]


| Species | Explants | Genotype | Target Gene | Agrobacterium Strain | Vector | Selectable Marker | Efficiency (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Monocots | ||||||||
| Wheat | Immature embryos | Bobwhite SH98 26 | sGfp | AGL1 | pGH215 | Hpt | Up to 15% | [26] |
| Immature embryos | Fielder | gus | AGL1 | pGoldenGreenGate | Hpt | Up to 25% | [25] | |
| Immature embryos | CB037, Fielder, Jimai 22 Kenong 199, Shi 4185 | gus | C58C1 | pRK2013 | Bar | 2.8–53% | [28] | |
| Mature and immature embryos | DBW 88, DBW 90, DBW 93, DPW 621-50, HD 3086 and WH 1105 | gus | EHA105 | pCAMBIA3301 | Bar | 9.8–14.9% | [27] | |
| Rice | Immature embryos | a broad range of species | gfp | LAB4404 and EHA105 | pPUG1-1 | Hpt | - | [29] |
| Embryogenic calli | Sambha mahsuri | AmSOD1 | LAB4404 and EHA105 | pSFSOD1 | Hpt | - | [30] | |
| Maize | Immature embryos | ND101 and ND88 | DsRed | EHA105 | Bar | Up to 17.6% | [31] | |
| Immature embryos | HC69 and PH2RT | YFP | LBA4404THY- | pVIR | PAT/PMI | - | [24] | |
| Barley | Immature embryos | Scarlett and Golden Promise | HvCKX2 | AGL1 | pMCG161 | Bar | 3.47% | [32] |
| Immature embryos | Golden Promise | gus | AGL1 | pBRACT | Hpt | 25% | [33] | |
| Dicots | ||||||||
| Eucalyptus | Leaves | E. urophylla × E. grandis clone DH32- 29 | gus | GV3101, LBA4404, EHA105, and AGL1 | pBI121 | Npt II | 1.9% | [34] |
| Poplar | Leaves | Populus Alba×Populusglandulosa Uyeki | gus | GV3101 | 35S:GUS vector | - | - | [35] |
| Callus | 84K | gus | GV3101 | pCAM BIA1301 | Hygromycin B | greater than 50% | [36] | |
| Codonopsis pilosula | Stems | (Franch.) Nannf. | gus | GV3101 | pCAMBIA1381 | Hpt | 91.07% | [37] |
| Tea | Callus | Camellia sinensis | gus | EHA105 | PS1aG-3 | - | 3.6% | [38] |
| Liriodendron hybrid | Callus | 52053 | gus | EHA105 | pBI121 | Geneticin | 60.7% | [39] |
| Pigeon pea | Cotyledons | ICPL 85063 | gus/gfp | LBA4404 | pCAMBIA1301 | Hpt | 83% | [40] |
| Soybean | Cotyledonary node | Jack | GsWRKY20 | EHA101 | myc-pBA | Bar | - | [41] |
| Ailanthus altissima (Mill) Swingle | Shoots | - | gfp | K599 | pCAMBIA1300 | - | - | [21] |
| Cotton | Shoot apical meristem | Gossypium hirsutum, Gossypium barbadense and Gossypium arboreum | GFP and RUBY | - | pCAMBIA 2300 | AADA | Up to 8.01% | [42] |
| Species | Explants | Genes/Molecules | Gene Delivery System | Efficiency (%) | Reference |
|---|---|---|---|---|---|
| Barley | Seeds | OsWRKY70, OsWRKY53, and gus | Particle bombardment | - | [71] |
| Maize | Calli | VHb | Particle bombardment | - | [72] |
| Sorghum | Immature embryos | NptII | Particle bombardment | 46.6% | [73] |
| Sugarcane | Embryonic calli | cry1Ac and bar | Particle bombardment | - | [74] |
| Rice | Calli | Cpf1, crRNA, and repair templates | Particle bombardment | 8% | [47] |
| Wheat | Immature embryo | gfp and bar | Particle bombardment | - | [75] |
| Wheat | Callus | Ppa | Particle bombardment | - | [76] |
| Palm | Callus | ChoA | Particle bombardment | - | [77] |
| Blackgram | Embryonic axis | ChiB | Particle bombardment | 13% | [78] |
| Cowpea | Embryonic axis | Arc1 | Particle bombardment | - | [79] |
| Carrizo citrange | Immature epicotyl | nptII and gfp | Particle bombardment | 18.4% | [80] |
| Soybean | Embryogenic cells | Hpt, ZFN expression constructs, and HDR donor | Particle bombardment | About 2.84% | [48] |
| Zygnematophycean algae | Cells | GFP | Electroporation | - | [81] |
| Wheat | Immature embryos | bar and uidA | Electroporation | 0.4% | [82] |
| Tomato | Leaves | Fe and Mg | Liposomes | 33% | [83] |
| Maize | Embryogenic callus | gus and bar | Silicon carbide whisker | - | [84] |
| Cotton | Embryogenic callus | Gus and AVP1 | Silicon carbide whisker | Up to 94% | [85] |
| Cotton | Embryogenic callus | GUS, AVP1, and nptII | Silicon carbide whisker | - | [86] |
| Peanut | Callus | chitinase and hygromcin | Silicon carbide whisker | 6.88% | [58] |
| Barley | Protoplasts | Act1, gus, and nos | Microinjection | - | [87] |
| Oil palm | Protoplasts | GFP | Microinjection | 10–74.6% | [88] |
| Maize | Pollen | GFP | Pollen tube pathway | 0.86% | [89] |
| Cotton | Pollen | nptII | Pollen tube pathway | - | [90] |
| Melon | Pollen | Fom-2 | Pollen tube pathway | 3.28% and 4.26% | [91] |
| Peanut | Pollen | AhBI-1 | Pollen tube pathway | 50% | [92] |
| Transformation Methods | Tissue Type | Species | Delivery Type | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Agrobacterium | Cells, tissues, and whole plants | Monocot and dicot | DNA | It has high transformation efficiency and stability | It is species and genotype restricted, and random integration may result in gene destruction |
| Particle bombardment | Any intact tissue or explant | Monocot and dicot | DNA, siRNA, miRNA, and RNP | It is not limited by tissues or cell types | The transferred DNA is not protected, the transformation efficiency is lower than with Agrobacterium-mediated transformation, and the equipment used is costly. High copy numbers and extensive rearrangements of foreign DNA, as well as the integration of multiple copies of the same gene in the genome, often lead to gene silencing |
| Electroporation | Leaf blade, protoplast, meristem, and pollen grain | Green algae, monocot and dicot | DNA, siRNA, miRNA, and protein | It is possible to transform whole cells and tissues. The transformation efficiency depends on the plant’s material | It requires cell wall removal and is limited to an in vitro suspension system. It will cause tissue damage without specificity, and transformed cells have a 50% chance of survival |
| Liposome | Protoplast, callus, and pollen | Dicot | DNA, RNA, and protein | The wrapped nucleic acid can be protected from degradation by nucleases; specific cells as well as various cell types can be targeted. | Its transformation efficiency is low |
| Silicon carbide whisker | Callus and mature embryos | Monocot | DNA | Its operation is simple, and its cost is low | It has a low transformation efficiency, and silicon carbide whiskers are toxic |
| Microinjection | Protoplasts, immature embryos, and pollen | Monocot and dicot | DNA | The method, which is technically simple, may facilitate the transfer of genes to grains that are not easily regenerated from cultured cells | Its transformation efficiency and frequency are low, it takes a long time to complete, it is costly, and it requires trained and certified workers to conduct experiments |
| Pollen tube | Pollen tube | Monocot and dicot | DNA | It does not involve tissue culture or in vitro regeneration | Its transformation efficiency is low, and the transfer of exogenous genes is limited by natural flowering period |
| Species | Explants | Molecules | Nanoparticles | Efficiency (%) | Reference |
|---|---|---|---|---|---|
| Maize | Pollen | RFP, GUS, and EGFP | Magnetic nanoparticles | 32–55% (DNA entry) | [124] |
| Maize | Immature embryos | Cre recombinase Protein | Magnetic nanoparticles | 20% (bombarded embryos produced calli with the recombined loxP sites) | [125] |
| Okra | Embryo | mgfp | Magnetic nanoparticles | - | [126] |
| Cotton | Pollen | BTΔα-CPTI, and GUS | Magnetic nanoparticles | About 1% | [97] |
| Rice | Calli | aadA and gfp | Peptide nanoparticles | About 2.77% | [127] |
| Kenaf | Cotyledon or calli | aadA and gfp | Peptide nanoparticles | About 0.037% | [127] |
| Tomato | Pollen | dsRNA | Layered double hydroxides | - | [128] |
| Nicotiana benthamiana | Leaves | siRNAs | DNA nanostructures | - | [129] |
| Rice | Leaves and excised embryo | GFP, YFP, and GUS | Carbon nanotubes | - | [130] |
| Wheat | Leaves | sGFP | Carbon nanotubes | - | [7] |
| Nicotiana benthamiana | Leaves | sGFP | Carbon nanotubes | - | [7] |
| Cotton | Leaves | sGFP | Carbon nanotubes | - | [7] |
| Nicotiana Benthamiana | Leaves | siRNA | Carbon nanotubes | 95% (gene silencing rate within 24 h) | [131] |
| Arabidopsis thaliana | Seedlings | GFP and RLuc | Carbon nanotubes | - | [132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, W.; Xu, M.; Radani, Y.; Yang, L. Technological Development and Application of Plant Genetic Transformation. Int. J. Mol. Sci. 2023, 24, 10646. https://doi.org/10.3390/ijms241310646
Su W, Xu M, Radani Y, Yang L. Technological Development and Application of Plant Genetic Transformation. International Journal of Molecular Sciences. 2023; 24(13):10646. https://doi.org/10.3390/ijms241310646
Chicago/Turabian StyleSu, Wenbin, Mingyue Xu, Yasmina Radani, and Liming Yang. 2023. "Technological Development and Application of Plant Genetic Transformation" International Journal of Molecular Sciences 24, no. 13: 10646. https://doi.org/10.3390/ijms241310646
APA StyleSu, W., Xu, M., Radani, Y., & Yang, L. (2023). Technological Development and Application of Plant Genetic Transformation. International Journal of Molecular Sciences, 24(13), 10646. https://doi.org/10.3390/ijms241310646




