Transcriptional and Histone Acetylation Changes Associated with CRE Elements Expose Key Factors Governing the Regulatory Circuit in the Early Stage of Huntington’s Disease Models
Abstract
1. Introduction
2. Results
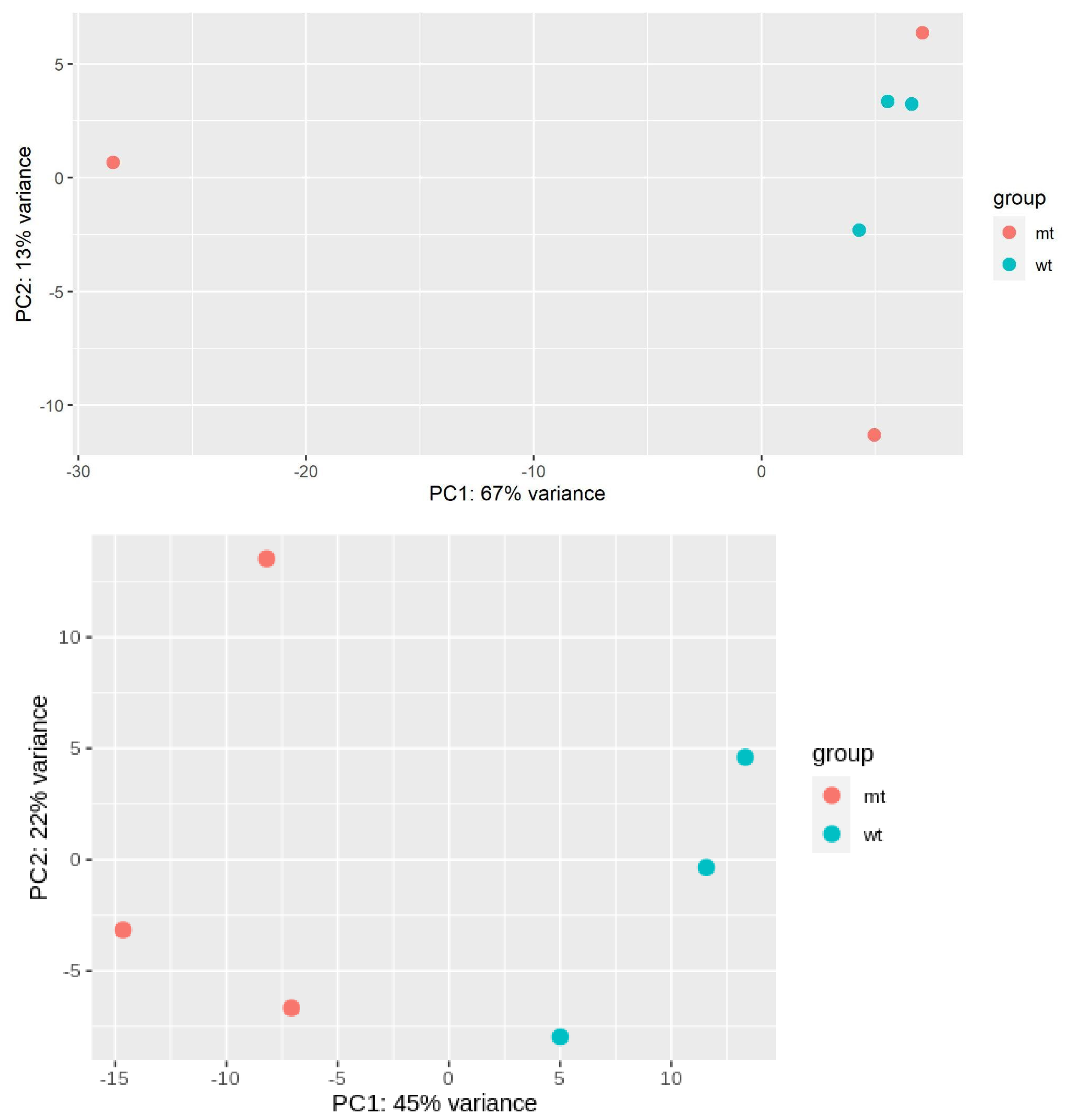
2.1. Protein Aggregation and Transcriptional Changes in Huntington’s Disease Are Detectable in Pre-Symptomatic Stages
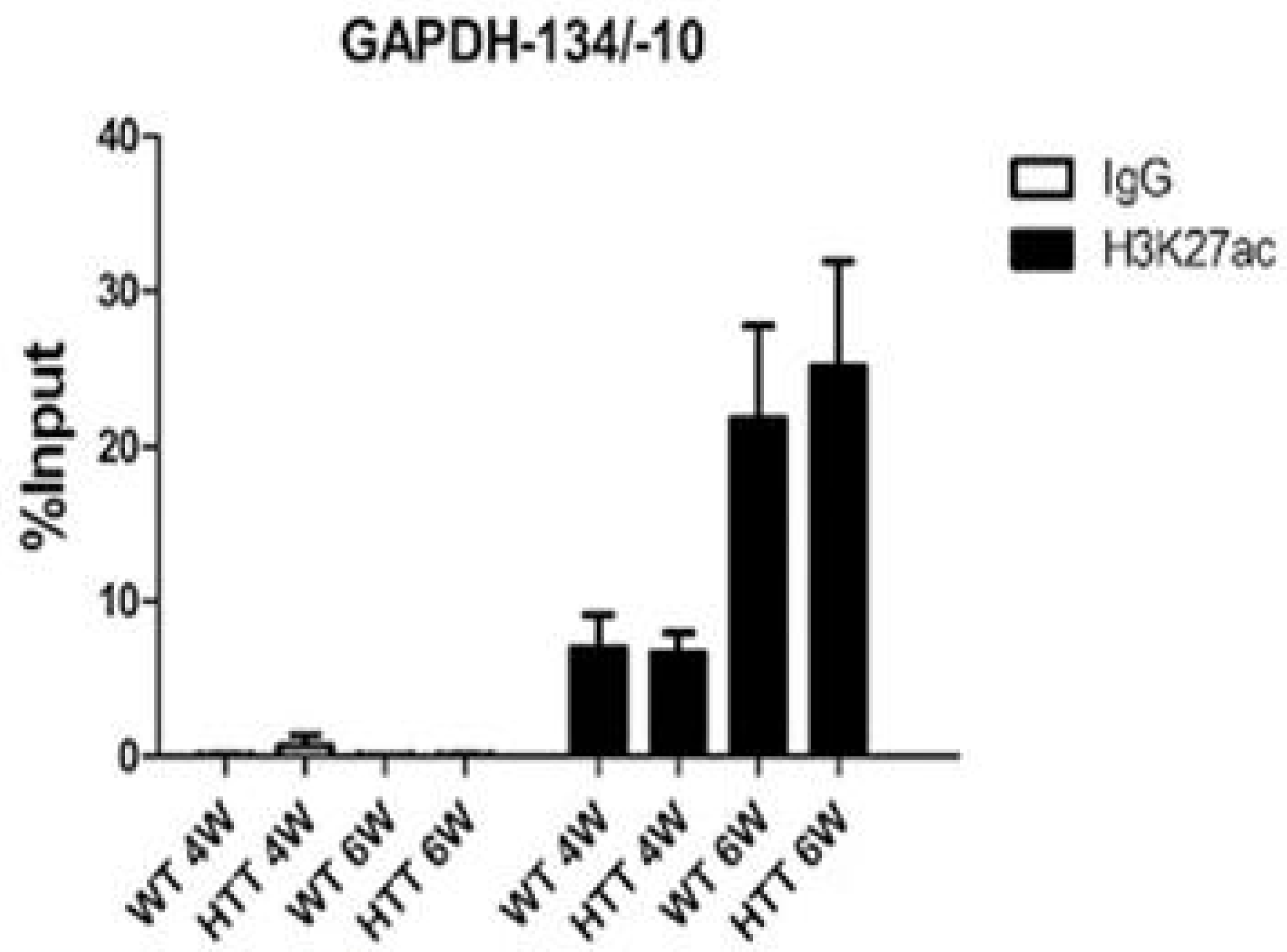
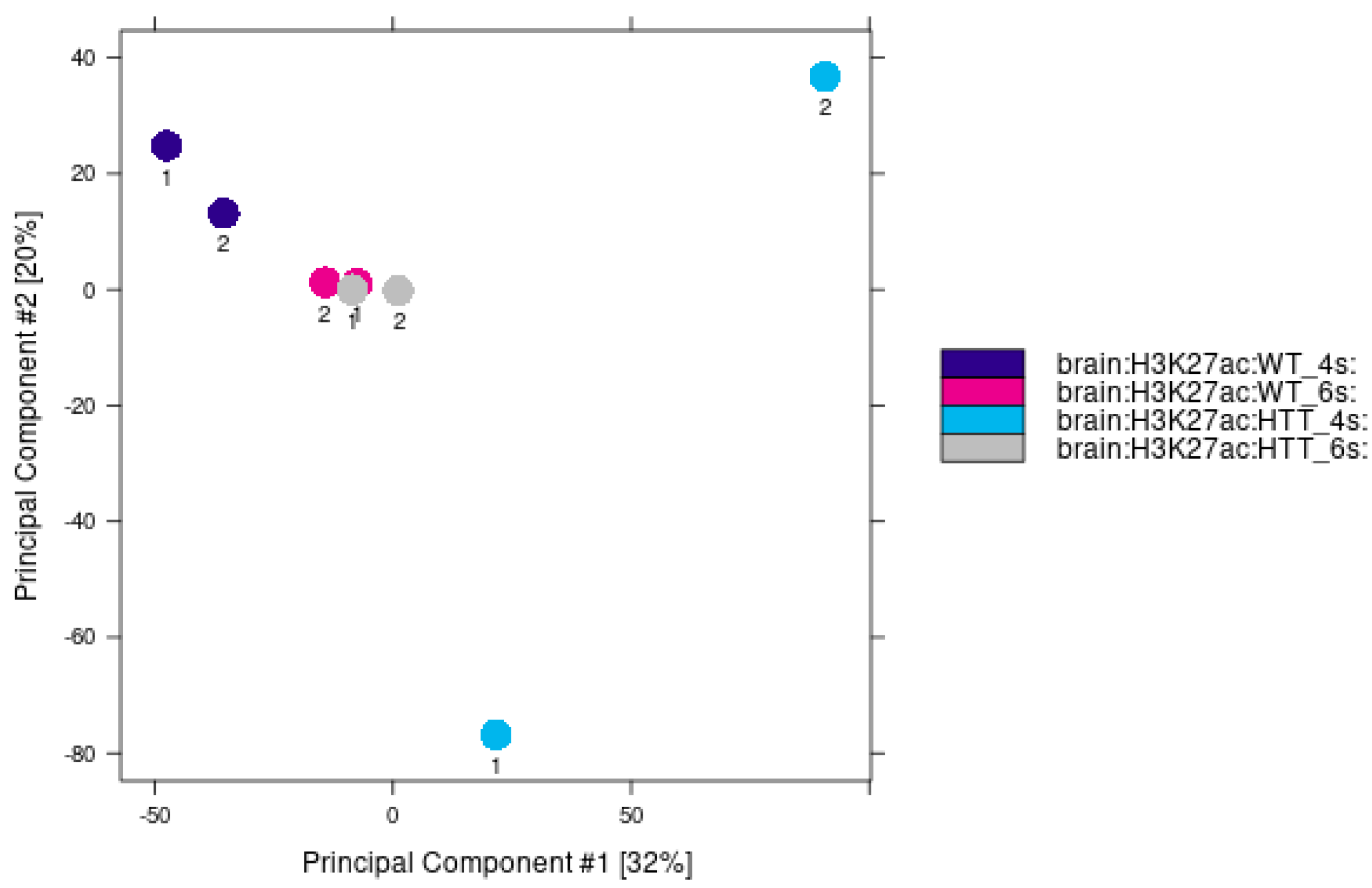
2.2. Decreased Levels of H3K27ac Are Detectable at Early Stages of Huntington’s Disease
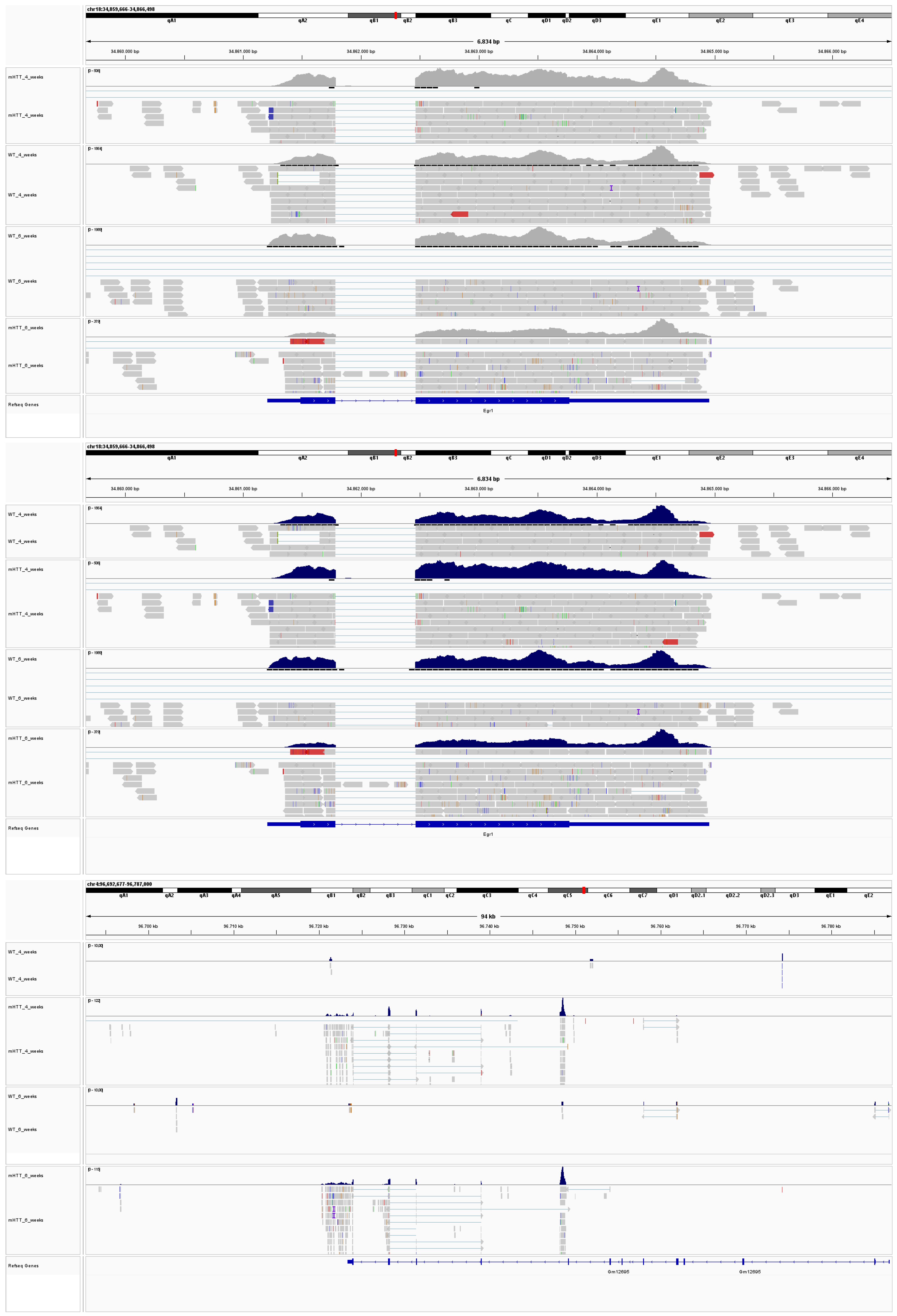
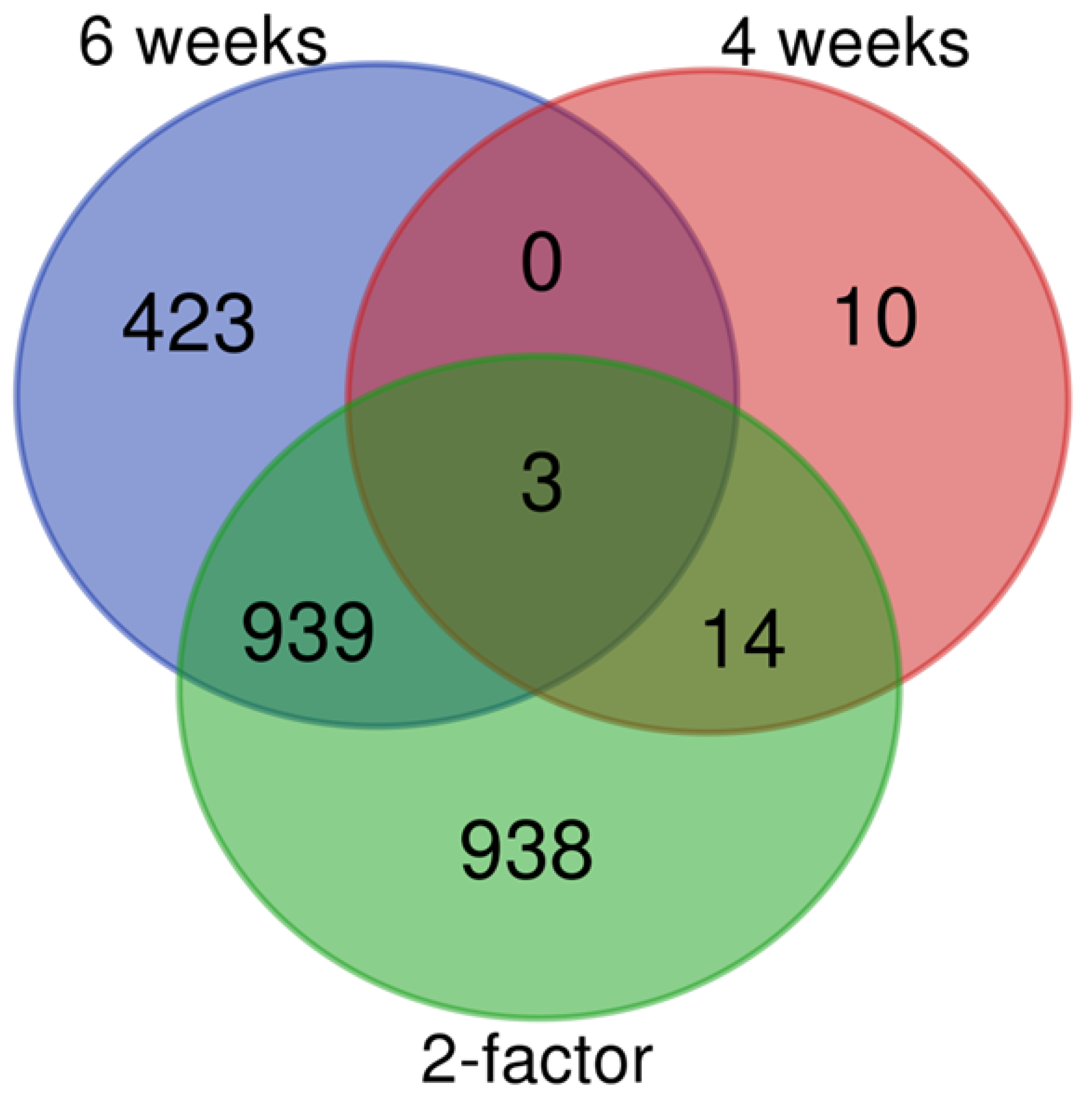
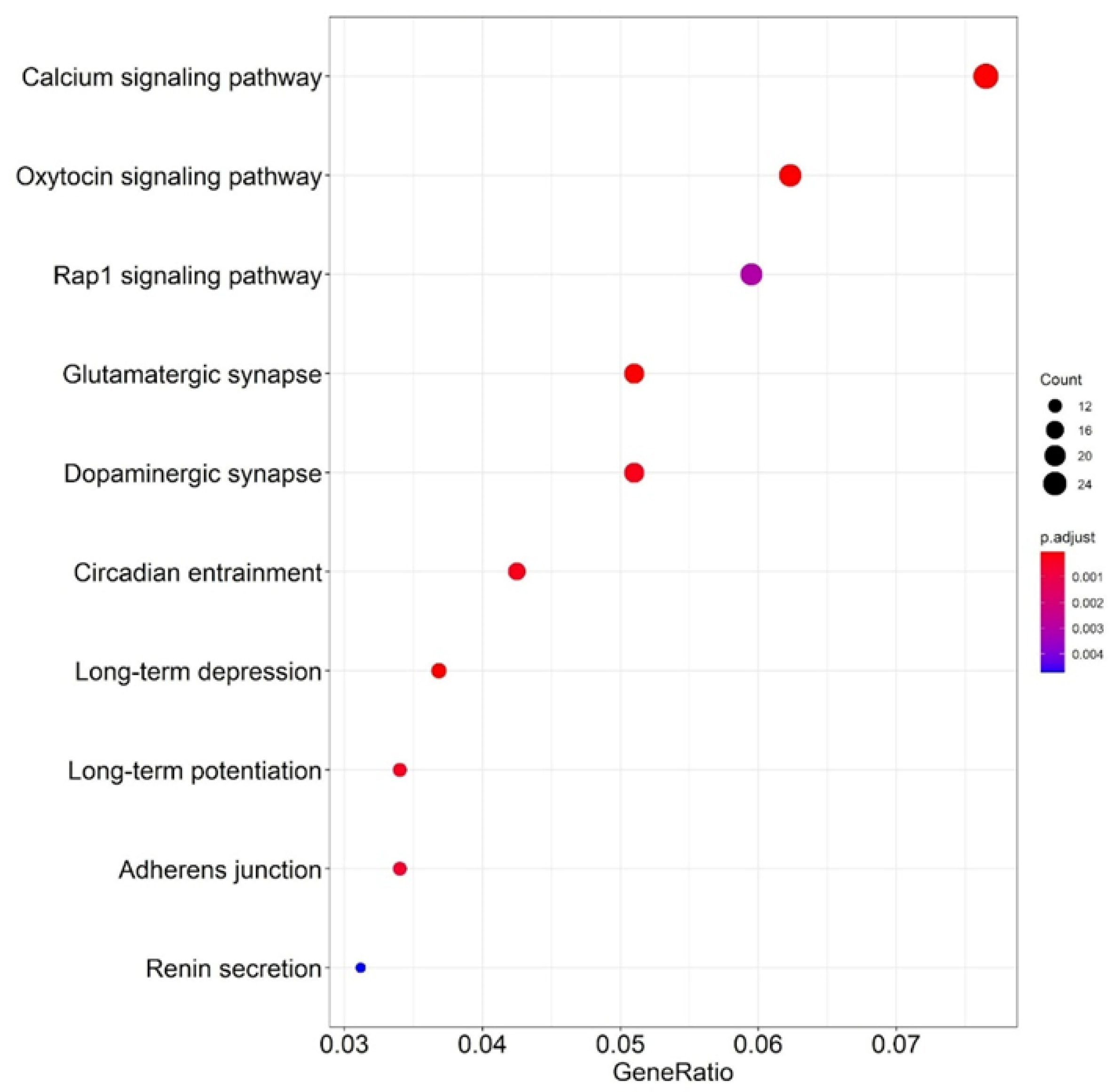
2.3. Altered Acetylation Levels Reshape the Transcriptional Cascades in Presymptomatic Stages of HD
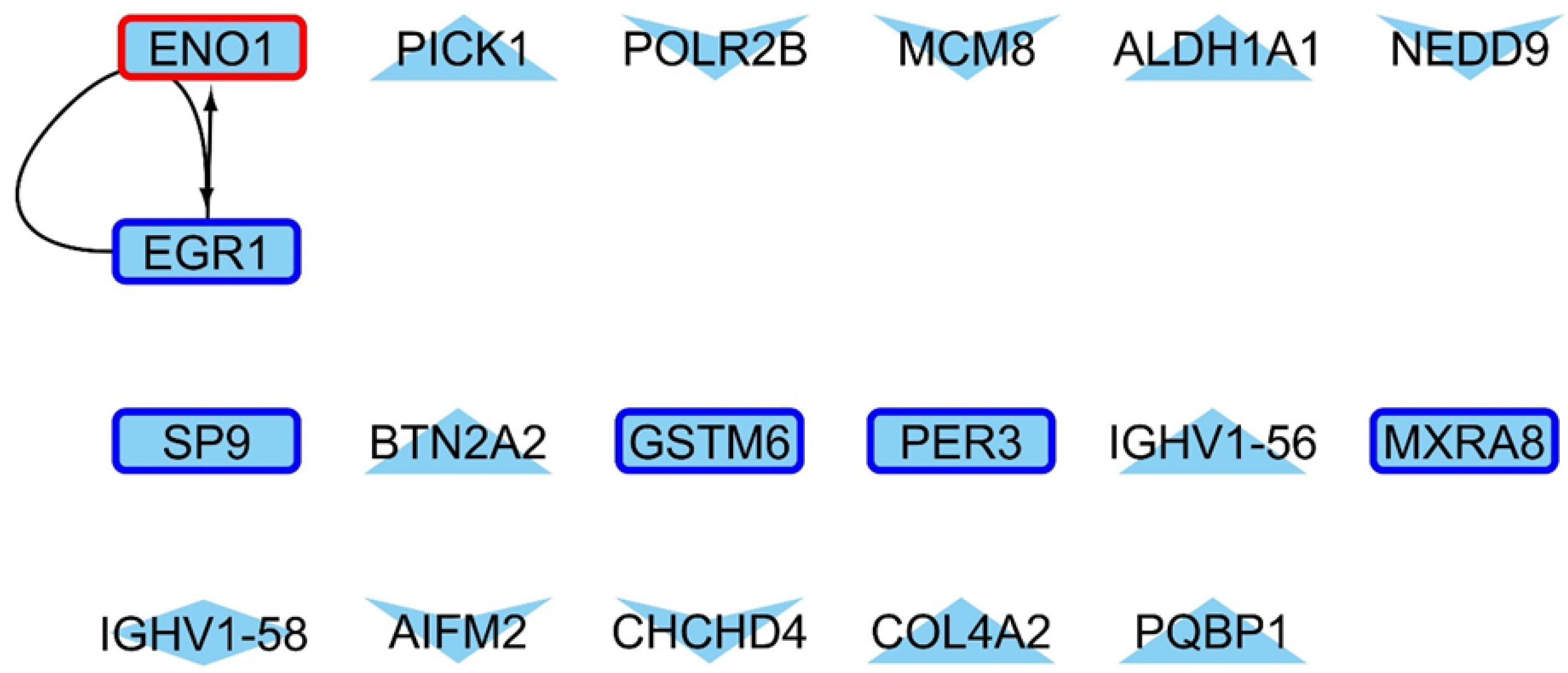
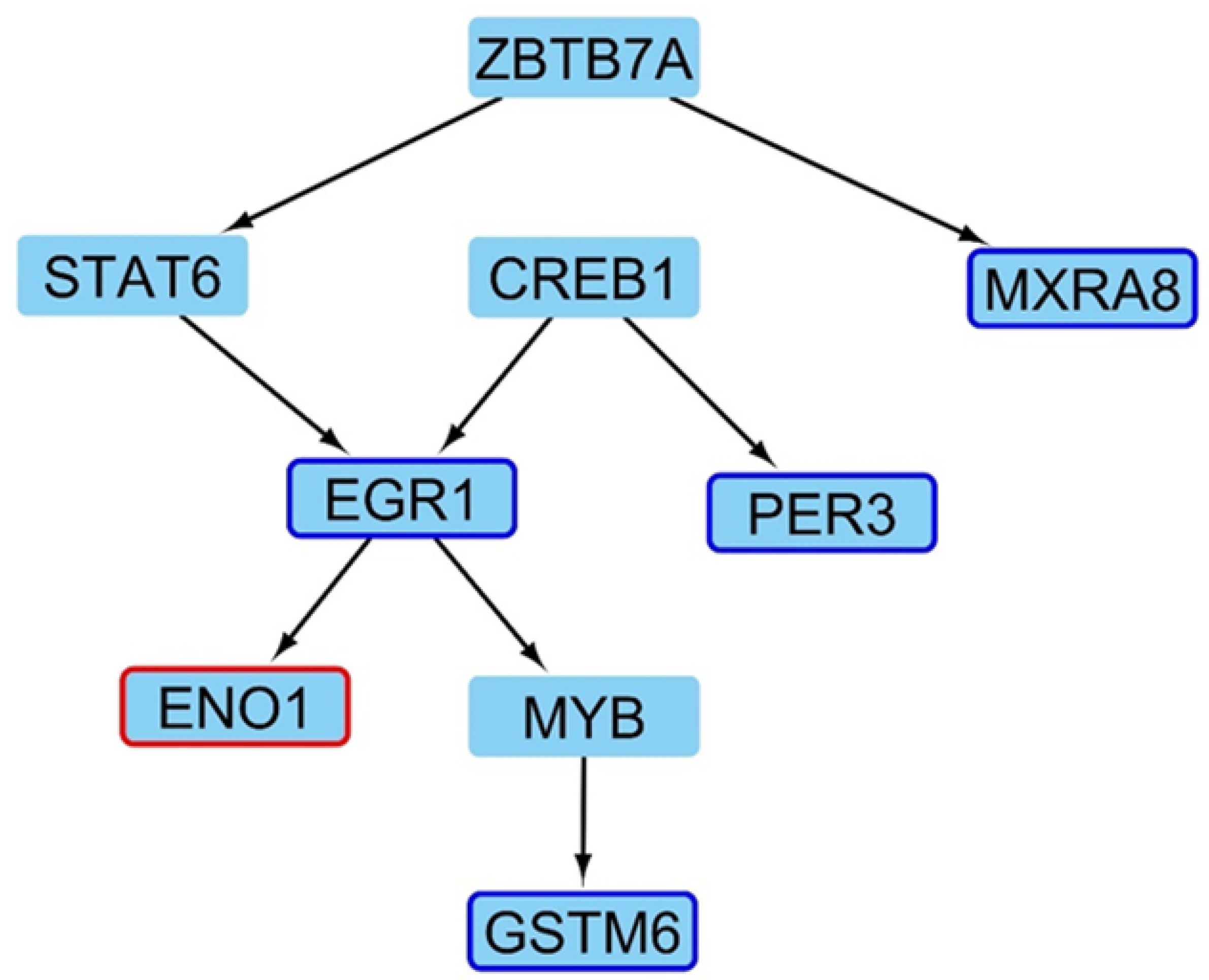
2.4. Master Regulators Guiding the Transcriptional Landscape in HD Presents Changes in Their Expression Profiles
3. Discussion
4. Materials and Methods
4.1. Mouse Strains
4.2. Transcriptomic Analysis of Striatal Tissues from R6/2 Mice
4.3. Identification of Differentially Expressed Genes in the R6/2 HD Mouse Model
4.4. ChIP Analysis in the R6/2 Model
4.5. Determination of mHtt Protein Levels in Brain Tissue
4.6. Soluble and Insoluble Huntingtin Species
4.7. Identification of H3K27ac Level in R6/2 Murine Models
4.8. Quantification of Differentially Acetylated Promoters
4.9. Determination of CRE Regions and the Genes Associated with These Regions in Mice
4.10. Generation of Gene Regulatory Networks in the R6/2 Murine Model of HD and Their Respective Control
4.11. Inference of Master Regulators
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HD | Huntington’s disease |
| polyQ | polyglutamine |
| mHTT | Mutant Huntingtin protein |
| CREB | cAMP response element-binding protein |
| CBP | CREB binding protein |
| TBP | TATA-binding protein |
| SP1 | specificity protein 1 |
| TFs | transcription factors |
| GRN | Gene Regulatory Network |
| MRs | Master Regulators |
| WT | Wild-Type |
| DE | differentially expressed |
| GO | gene ontology |
| ChIP | Chromatin Immunoprecipitation |
| MSN | medium spiny neuron |
Appendix A. Figures




References
- McCandless, S.E.; Cassidy, S.B. Nontraditional inheritance. Princ. Mol. Med. 2006, 39, 9–18. [Google Scholar]
- Paulson, H. Repeat expansion diseases. Handb. Clin. Neurol. 2018, 147, 105–123. [Google Scholar] [PubMed]
- Gonzalez-Alegre, P. Recent advances in molecular therapies for neurological disease: Triplet repeat disorders. Hum. Mol. Genet. 2019, 28, R80–R87. [Google Scholar] [CrossRef] [PubMed]
- Nopoulos, P.C. Huntington disease: A single-gene degenerative disorder of the striatum. Dialogues Clin. Neurosci. 2022, 18, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Sathasivam, K.; Neueder, A.; Gipson, T.A.; Landles, C.; Benjamin, A.C.; Bondulich, M.K.; Smith, D.L.; Faull, R.L.; Roos, R.A.; Howland, D.; et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. USA 2013, 110, 2366–2370. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Zuccato, C.; Valenza, M.; Cattaneo, E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010, 90, 905–981. [Google Scholar] [CrossRef]
- Gatto, E.M.; Rojas, N.G.; Persi, G.; Etcheverry, J.L.; Cesarini, M.E.; Perandones, C. Huntington disease: Advances in the understanding of its mechanisms. Clin. Park. Relat. Disord. 2020, 3, 100056. [Google Scholar] [CrossRef]
- Zheng, P.; Kozloski, J. Striatal network models of Huntington’s disease dysfunction phenotypes. Front. Comput. Neurosci. 2017, 11, 70. [Google Scholar] [CrossRef]
- Bhattacharyya, K.B. The story of George Huntington and his disease. Ann. Indian Acad. Neurol. 2016, 19, 25–28. [Google Scholar] [CrossRef]
- Cohen-Carmon, D.; Meshorer, E. Polyglutamine (polyQ) disorders: The chromatin connection. Nucleus 2012, 3, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Landles, C.; Bates, G.P. Huntingtin and the molecular pathogenesis of Huntington’s disease. EMBO Rep. 2004, 5, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Schilling, G.; Sharp, A.H.; Loev, S.J.; Wagster, M.V.; Li, S.H.; Stine, O.C.; Ross, C.A. Expression of the Huntington’s disease (IT15) protein product in HD patients. Hum. Mol. Genet. 1995, 4, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Ament, S.A.; Pearl, J.R.; Grindeland, A.; Claire, J.S.; Earls, J.C.; Kovalenko, M.; Gillis, T.; Mysore, J.; Gusella, J.F.; Lee, J.M.; et al. High resolution time-course mapping of early transcriptomic, molecular and cellular phenotypes in Huntington’s disease CAG knock-in mice across multiple genetic backgrounds. Hum. Mol. Genet. 2017, 26, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Bossy-Wetzel, E.; Petrilli, A.; Knott, A.B. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008, 31, 609–616. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Leggo, J.; Coles, R.; Almqvist, E.; Biancalana, V.; Cassiman, J.J.; Chotai, K.; Connarty, M.; Craufurd, D.; Curtis, A.; et al. Phenotypic Characterization of Individuals with 30–40 CAG Repeats in the Huntington Disease (HD) Gene Reveals HD Cases with 36 Repeats and Apparently Normal Elderly Individuals with 36–39 Repeats. Am. J. Hum. Genet. 1996, 59, 16. [Google Scholar]
- Shin, A.; Shin, B.; Shin, J.W.; Kim, K.H.; Atwal, R.S.; Hope, J.M.; Gillis, T.; Leszyk, J.D.; Shaffer, S.A.; Lee, R.; et al. Novel allele-specific quantification methods reveal no effects of adult onset CAG repeats on HTT mRNA and protein levels. Hum. Mol. Genet. 2017, 26, 1258–1267. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, M.; Licitra, F.; Underwood, B.R.; Rubinsztein, D.C. Huntington’s Disease: Mechanisms of Pathogenesis and Therapeutic Strategies. Cold Spring Harb. Perspect. Med. 2017, 7, a024240. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Lazarovici, P.; Quirion, R.; Zheng, W. cAMP Response Element-Binding Protein (CREB): A Possible Signaling Molecule Link in the Pathophysiology of Schizophrenia. Front. Mol. Neurosci. 2018, 11, 255. [Google Scholar] [CrossRef]
- Reinius, B.; Blunder, M.; Brett, F.M.; Eriksson, A.; Patra, K.; Jonsson, J.; Jazin, E.; Kullander, K. Conditional targeting of medium spiny neurons in the striatal matrix. Front. Behav. Neurosci. 2015, 9, 71. [Google Scholar] [CrossRef]
- Schulte, J.; Littleton, J.T. The biological function of the Huntingtin protein and its relevance to Huntington’s Disease pathology. Curr. Trends Neurol. 2011, 5, 65. [Google Scholar] [PubMed]
- Kim, S.H.; Thomas, C.A.; André, V.M.; Cummings, D.M.; Cepeda, C.; Levine, M.S.; Ehrlich, M.E. Forebrain striatal-specific expression of mutant huntingtin protein in vivo induces cell-autonomous age-dependent alterations in sensitivity to excitotoxicity and mitochondrial function. ASN Neuro 2011, 3, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Akimaru, H.; Tanaka, Y.; Hou, D.X.; Yasukawa, T.; Kanei-Ishii, C.; Takahashi, T.; Ishii, S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996, 10, 528–540. [Google Scholar] [CrossRef]
- Henry, R.A.; Kuo, Y.M.; Andrews, A.J. Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 2013, 52, 5746–5759. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mut, J.V.; Gräff, J. Epigenetic alterations in Alzheimer’s disease. Front. Behav. Neurosci. 2015, 9, 347. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Sasi, M.; Vignoli, B.; Canossa, M.; Blum, R. Neurobiology of local and intercellular BDNF signaling. PflüGers Arch. Eur. J. Physiol. 2017, 469, 593–610. [Google Scholar] [CrossRef]
- Miller, R.S.; Wolfe, A.; He, L.; Radovick, S.; Wondisford, F.E. CREB Binding Protein (CBP) Activation Is Required for Luteinizing Hormone Beta Expression and Normal Fertility in Mice. Mol. Cell. Biol. 2012, 32, 2349–2358. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef]
- McManus, K.J.; Hendzel, M.J. CBP, a transcriptional coactivator and acetyltransferase. Biochem. Cell Biol. 2001, 79, 253–266. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Valor, L.M.; Pulopulos, M.M.; Jimenez-Minchan, M.; Olivares, R.; Lutz, B.; Barco, A. Ablation of CBP in Forebrain Principal Neurons Causes Modest Memory and Transcriptional Defects and a Dramatic Reduction of Histone Acetylation But Does Not Affect Cell Viability. J. Neurosci. 2011, 31, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Nucifora, J.; Sasaki, M.; Peters, M.F.; Huang, H.; Cooper, J.K.; Yamada, M.; Takahashi, H.; Tsuji, S.; Troncoso, J.; Dawson, V.L.; et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 2001, 291, 2423–2428. [Google Scholar] [CrossRef]
- Steffan, J.S.; Kazantsev, A.; Spasic-Boskovic, O.; Greenwald, M.; Zhu, Y.Z.; Gohler, H.; Wanker, E.E.; Bates, G.P.; Housman, D.E.; Thompson, L.M. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 2000, 97, 6763–6768. [Google Scholar] [CrossRef]
- Paldino, E.; Cardinale, A.; D’Angelo, V.; Sauve, I.; Giampà, C.; Fusco, F.R. Selective Sparing of Striatal Interneurons after Poly (ADP-Ribose) Polymerase 1 Inhibition in the R6/2 Mouse Model of Huntington’s Disease. Front. Neuroanat. 2017, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Achour, M.; Gras, S.L.; Keime, C.; Parmentier, F.; Lejeune, F.X.; Boutillier, A.L.; Néri, C.; Davidson, I.; Merienne, K. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum. Mol. Genet. 2015, 24, 3481–3496. [Google Scholar] [CrossRef]
- Alcalá-Vida, R.; Seguin, J.; Lotz, C.; Molitor, A.M.; Irastorza-Azcarate, I.; Awada, A.; Karasu, N.; Bombardier, A.; Cosquer, B.; Skarmeta, J.L.G.; et al. Age-related and disease locus-specific mechanisms contribute to early remodelling of chromatin structure in Huntington’s disease mice. Nat. Commun. 2021, 12, 364. [Google Scholar] [CrossRef]
- Langfelder, P.; Cantle, J.P.; Chatzopoulou, D.; Wang, N.; Gao, F.; Al-Ramahi, I.; Lu, X.H.; Ramos, E.M.; El-Zein, K.; Zhao, Y.; et al. Integrated genomics and proteomics define huntingtin CAG length–dependent networks in mice. Nat. Neurosci. 2016, 19, 623–633. [Google Scholar] [CrossRef]
- Kohl, Z.; Regensburger, M.; Aigner, R.; Kandasamy, M.; Winner, B.; Aigner, L.; Winkler, J. Impaired adult olfactory bulb neurogenesis in the R6/2 mouse model of Huntington’s disease. BMC Neurosci. 2010, 11, 114. [Google Scholar] [CrossRef]
- Ragauskas, S.; Leinonen, H.; Puranen, J.; Rönkkö, S.; Nymark, S.; Gurevicius, K.; Lipponen, A.; Kontkanen, O.; Puoliväli, J.; Tanila, H.; et al. Early Retinal Function Deficit without Prominent Morphological Changes in the R6/2 Mouse Model of Huntington’s Disease. PLoS ONE 2014, 9, e113317. [Google Scholar] [CrossRef]
- Stack, E.C.; Dedeoglu, A.; Smith, K.M.; Cormier, K.; Kubilus, J.K.; Bogdanov, M.; Matson, W.R.; Yang, L.; Jenkins, B.G.; Luthi-Carter, R.; et al. Neuroprotective Effects of Synaptic Modulation in Huntington’s Disease R6/2 Mice. J. Neurosci. 2007, 27, 12908–12915. [Google Scholar] [CrossRef]
- Yildirim, F.; Ng, C.W.; Kappes, V.; Ehrenberger, T.; Rigby, S.K.; Stivanello, V.; Gipson, T.A.; Soltis, A.R.; Vanhoutte, P.; Caboche, J.; et al. Early epigenomic and transcriptional changes reveal Elk-1 transcription factor as a therapeutic target in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2019, 116, 24840–24851. [Google Scholar] [CrossRef] [PubMed]
- Rikani, A.A.; Choudhry, Z.; Choudhry, A.M.; Rizvi, N.; Ikram, H.; Mobassarah, N.J.; Tulli, S. The mechanism of degeneration of striatal neuronal subtypes in Huntington disease. Ann. Neurosci. 2014, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Blumenstock, S.; Dudanova, I. Cortical and Striatal Circuits in Huntington’s Disease. Front. Neurosci. 2020, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Duclot, F.; Kabbaj, M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 2017, 11, 35. [Google Scholar] [CrossRef]
- Crapser, J.D.; Ochaba, J.; Soni, N.; Reidling, J.C.; Thompson, L.M.; Green, K.N. Microglial depletion prevents extracellular matrix changes and striatal volume reduction in a model of Huntington’s disease. Brain 2020, 143, 266–288. [Google Scholar] [CrossRef]
- Miyazaki, H.; Oyama, F.; Inoue, R.; Aosaki, T.; Abe, T.; Kiyonari, H.; Kino, Y.; Kurosawa, M.; Shimizu, J.; Ogiwara, I.; et al. Singular localization of sodium channel β4 subunit in unmyelinated fibres and its role in the striatum. Nat. Commun. 2014, 5, 5525. [Google Scholar] [CrossRef] [PubMed]
- Oyama, F.; Miyazaki, H.; Sakamoto, N.; Becquet, C.; Machida, Y.; Kaneko, K.; Uchikawa, C.; Suzuki, T.; Kurosawa, M.; Ikeda, T.; et al. Sodium channel β4 subunit: Down-regulation and possible involvement in neuritic degeneration in Huntington’s disease transgenic mice. J. Neurochem. 2006, 98, 518–529. [Google Scholar] [CrossRef]
- Bergonzoni, G.; Döring, J.; Biagioli, M. D1R- and D2R-Medium-Sized Spiny Neurons Diversity: Insights Into Striatal Vulnerability to Huntington’s Disease Mutation. Front. Cell. Neurosci. 2021, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Alkass, K.; Bernard, S.; Salehpour, M.; Perl, S.; Tisdale, J.; Possnert, G.; Druid, H.; Frisén, J. Neurogenesis in the striatum of the adult human brain. Cell 2014, 156, 1072–1083. [Google Scholar] [CrossRef]
- Fedele, V.; Roybon, L.; Nordström, U.; Li, J.Y.; Brundin, P. Neurogenesis in the R6/2 mouse model of Huntington’s disease is impaired at the level of NeuroD1. Neuroscience 2011, 173, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Gil-Mohapel, J.; Simpson, J.M.; Ghilan, M.; Christie, B.R. Neurogenesis in Huntington’s disease: Can studying adult neurogenesis lead to the development of new therapeutic strategies? Brain Res. 2011, 1406, 84–105. [Google Scholar] [CrossRef]
- Malla, B.; Guo, X.; Senger, G.; Chasapopoulou, Z.; Yildirim, F. A Systematic Review of Transcriptional Dysregulation in Huntington’s Disease Studied by RNA Sequencing. Front. Genet. 2021, 12, 1898. [Google Scholar] [CrossRef]
- Chen, J.Y.; Wang, E.A.; Cepeda, C.; Levine, M.S. Dopamine imbalance in Huntington’s disease: A mechanism for the lack of behavioral flexibility. Front. Neurosci. 2013, 7, 114. [Google Scholar] [CrossRef]
- Cepeda, C.; Murphy, K.P.; Parent, M.; Levine, M.S. The role of dopamine in huntington’s disease. Prog. Brain Res. 2014, 211, 235–254. [Google Scholar]
- Kolobkova, Y.A.; Vigont, V.A.; Shalygin, A.V.; Kaznacheyeva, E.V. Huntington’s Disease: Calcium Dyshomeostasis and Pathology Models. Acta Naturae 2017, 9, 34. [Google Scholar] [CrossRef]
- Davis, T.L.; Rebay, I. Master regulators in development: Views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev. Biol. 2017, 421, 93–107. [Google Scholar] [CrossRef]
- Wei, L.L.; Wu, X.J.; Gong, C.C.; Pei, D.S. Egr-1 suppresses breast cancer cells proliferation by arresting cell cycle progression via down-regulating CyclinDs. Int. J. Clin. Exp. Pathol. 2017, 10, 10212. [Google Scholar]
- Yuan, C.; Wang, L.; Zhou, L.; Fu, Z. The function of FOXO1 in the late phases of the cell cycle is suppressed by PLK1-mediated phosphorylation. Cell Cycle 2014, 13, 807–819. [Google Scholar] [CrossRef]
- Bromberg, J.; Darnell, J.E. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000, 19, 2468–2473. [Google Scholar] [CrossRef]
- Fu, Q.F.; Liu, Y.; Fan, Y.; Hua, S.N.; Qu, H.Y.; Dong, S.W.; Li, R.L.; Zhao, M.Y.; Zhen, Y.; Yu, X.L.; et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J. Hematol. Oncol. 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.D.; Mitton, B.; Lacayo, N.J.; Sakamoto, K.M. Replication factor C3 is a CREB target gene that regulates cell cycle progression through the modulation of chromatin loading of PCNA. Leukemia 2014, 29, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Lin, A.; Chen, J.; Zhang, X.; Liu, H.; Li, H.; Hu, Y.; Zhang, X.; Zhang, J.; Qiu, L.; et al. CREB1 directly activates the transcription of ribonucleotide reductase small subunit M2 and promotes the aggressiveness of human colorectal cancer. Oncotarget 2016, 7, 78055–78068. [Google Scholar] [CrossRef] [PubMed]
- Dickey, A.S.; Pineda, V.V.; Tsunemi, T.; Liu, P.P.; Miranda, H.C.; Gilmore-Hall, S.K.; Lomas, N.; Sampat, K.R.; Buttgereit, A.; Torres, M.J.M.; et al. PPAR-δ is repressed in Huntington’s disease, is required for normal neuronal function and can be targeted therapeutically. Nat. Med. 2015, 22, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Lemprière, S. Huntington disease alters early neurodevelopment. Nat. Rev. Neurol. 2020, 16, 459. [Google Scholar] [CrossRef]
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dötsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. p63, a p53 Homolog at 3q27–29, Encodes Multiple Products with Transactivating, Death-Inducing, and Dominant-Negative Activities. Mol. Cell 1998, 2, 305–316. [Google Scholar] [CrossRef]
- Kurita, T.; Medina, R.T.; Mills, A.A.; Cunha, G.R. Role of p63 and basal cells in the prostate. Development 2004, 131, 4955–4964. [Google Scholar] [CrossRef]
- Zhao, X.; Strong, R.; Zhang, J.; Sun, G.; Tsien, J.Z.; Cui, Z.; Grotta, J.C.; Aronowski, J. Neuronal PPARγ Deficiency Increases Susceptibility to Brain Damage after Cerebral Ischemia. J. Neurosci. 2009, 29, 6186–6195. [Google Scholar] [CrossRef]
- Haubst, N.; Berger, J.; Radjendirane, V.; Graw, J.; Favor, J.; Saunders, G.F.; Stoykova, A.; Götz, M. Molecular dissection of Pax6 function: The specific roles of the paired domain and homeodomain in brain development. Development 2004, 131, 6131–6140. [Google Scholar] [CrossRef]
- Block, R.C.; Dorsey, E.R.; Beck, C.A.; Brenna, J.T.; Shoulson, I. Altered cholesterol and fatty acid metabolism in Huntington disease. J. Clin. Lipidol. 2010, 4, 17–23. [Google Scholar] [CrossRef]
- Dong, X.; Tsuji, J.; Labadorf, A.; Roussos, P.; Chen, J.F.; Myers, R.H.; Akbarian, S.; Weng, Z. The Role of H3K4me3 in Transcriptional Regulation Is Altered in Huntington’s Disease. PLoS ONE 2015, 10, e0144398. [Google Scholar] [CrossRef] [PubMed]
- Vashishtha, M.; Ng, C.W.; Yildirim, F.; Gipson, T.A.; Kratter, I.H.; Bodai, L.; Song, W.; Lau, A.; Labadorf, A.; Vogel-Ciernia, A.; et al. Targeting H3K4 trimethylation in Huntington disease. Proc. Natl. Acad. Sci. USA 2013, 110, E3027–E3036. [Google Scholar] [CrossRef] [PubMed]
- Valor, L.M.; Guiretti, D.; Lopez-Atalaya, J.P.; Barco, A. Genomic Landscape of Transcriptional and Epigenetic Dysregulation in Early Onset Polyglutamine Disease. J. Neurosci. 2013, 33, 10471–10482. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nussbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; Lee, R.V.D.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Wu, C.; Miao, H.; Wu, H. RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015, 2015, bav095. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Martin, A.J.; Dominguez, C.; Contreras-Riquelme, S.; Holmes, D.S.; Perez-Acle, T. Graphlet Based Metrics for the Comparison of Gene Regulatory Networks. PLoS ONE 2016, 11, e0163497. [Google Scholar] [CrossRef]
- Santander, N.; Lizama, C.; Murgas, L.; Contreras, S.; Martin, A.J.; Molina, P.; Quiroz, A.; Rivera, K.; Salas-Pérez, F.; Godoy, A.; et al. Transcriptional profiling of embryos lacking the lipoprotein receptor SR-B1 reveals a regulatory circuit governing a neurodevelopmental or metabolic decision during neural tube closure. BMC Genom. 2018, 19, 731. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
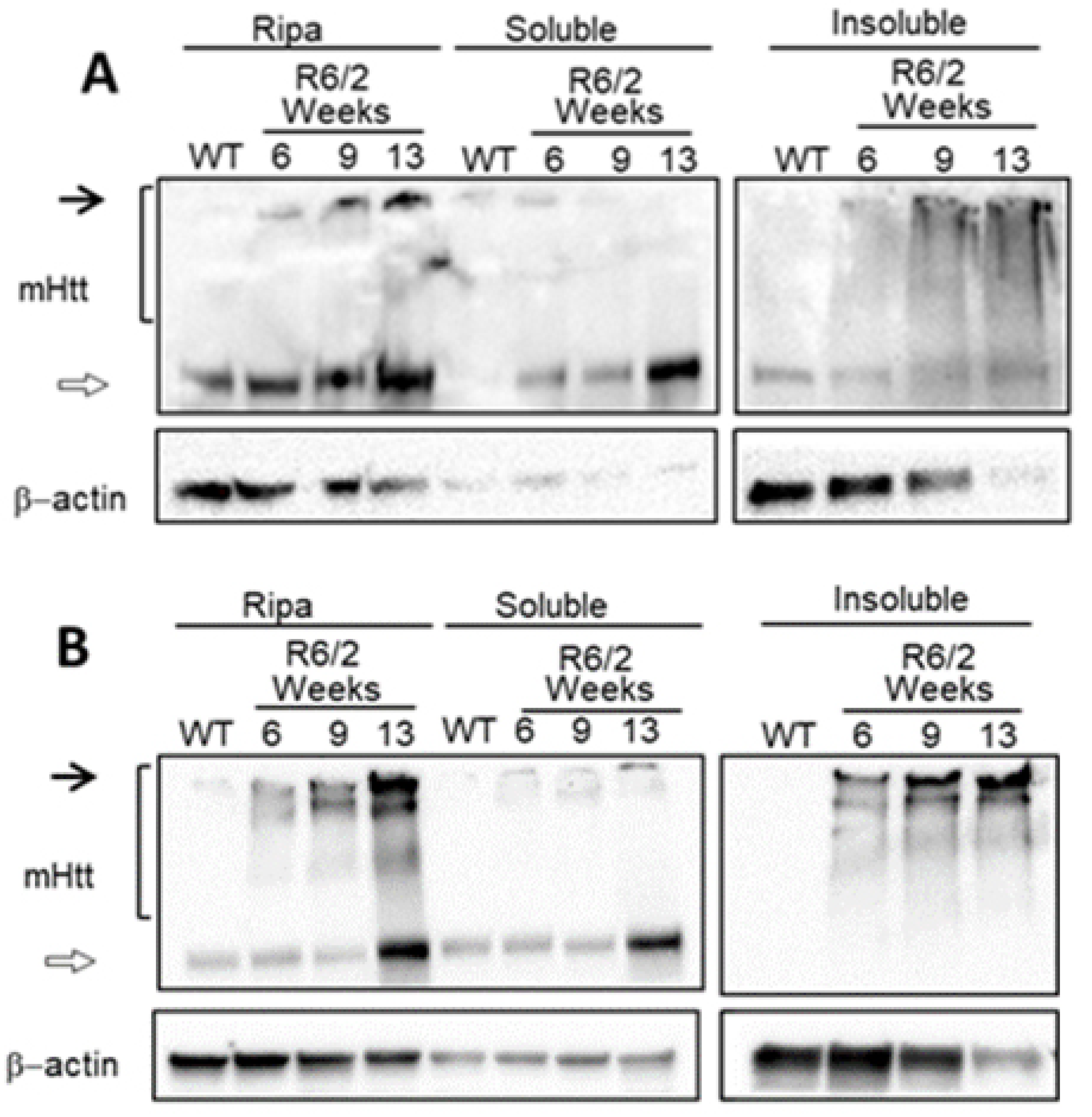
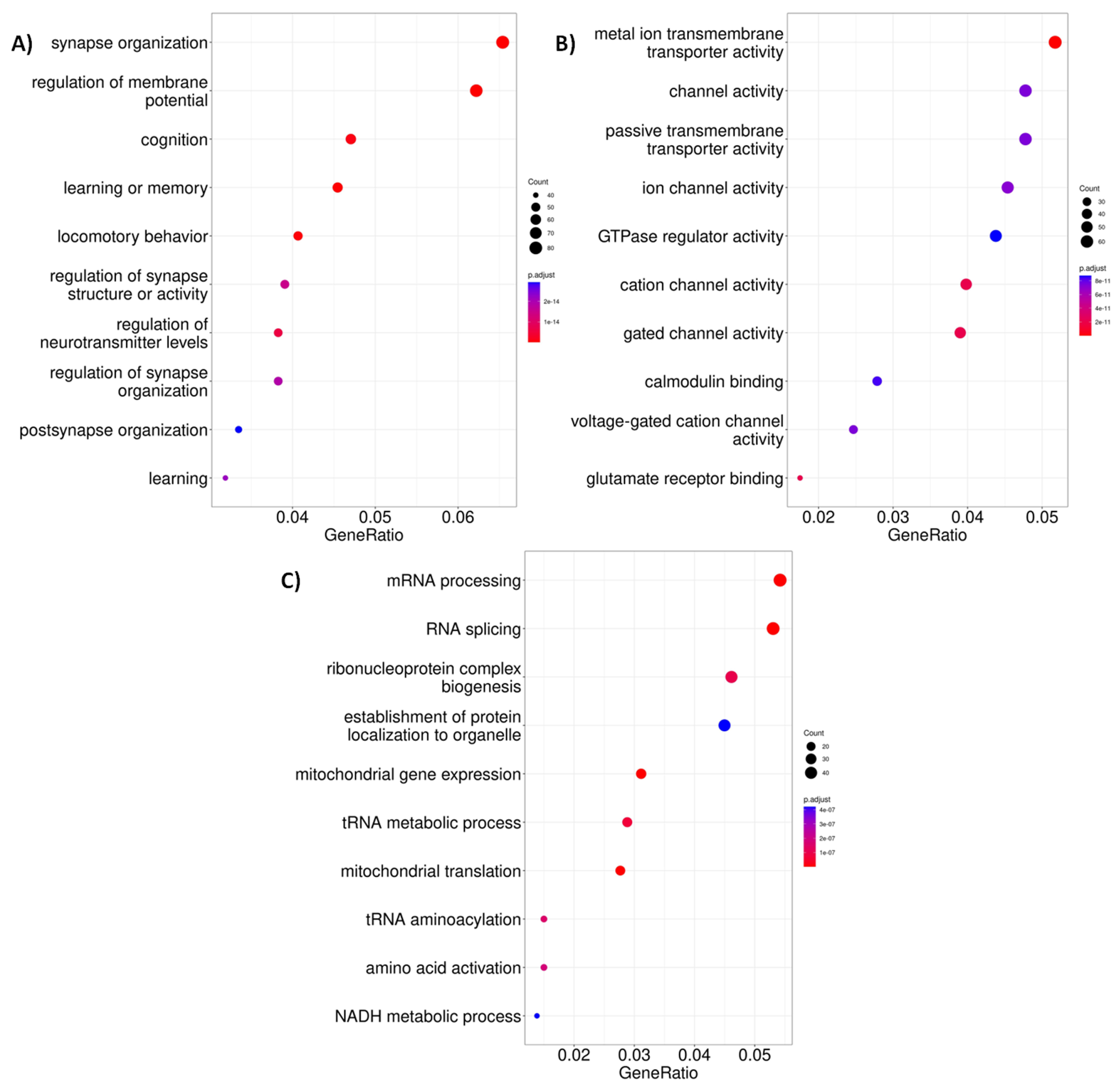
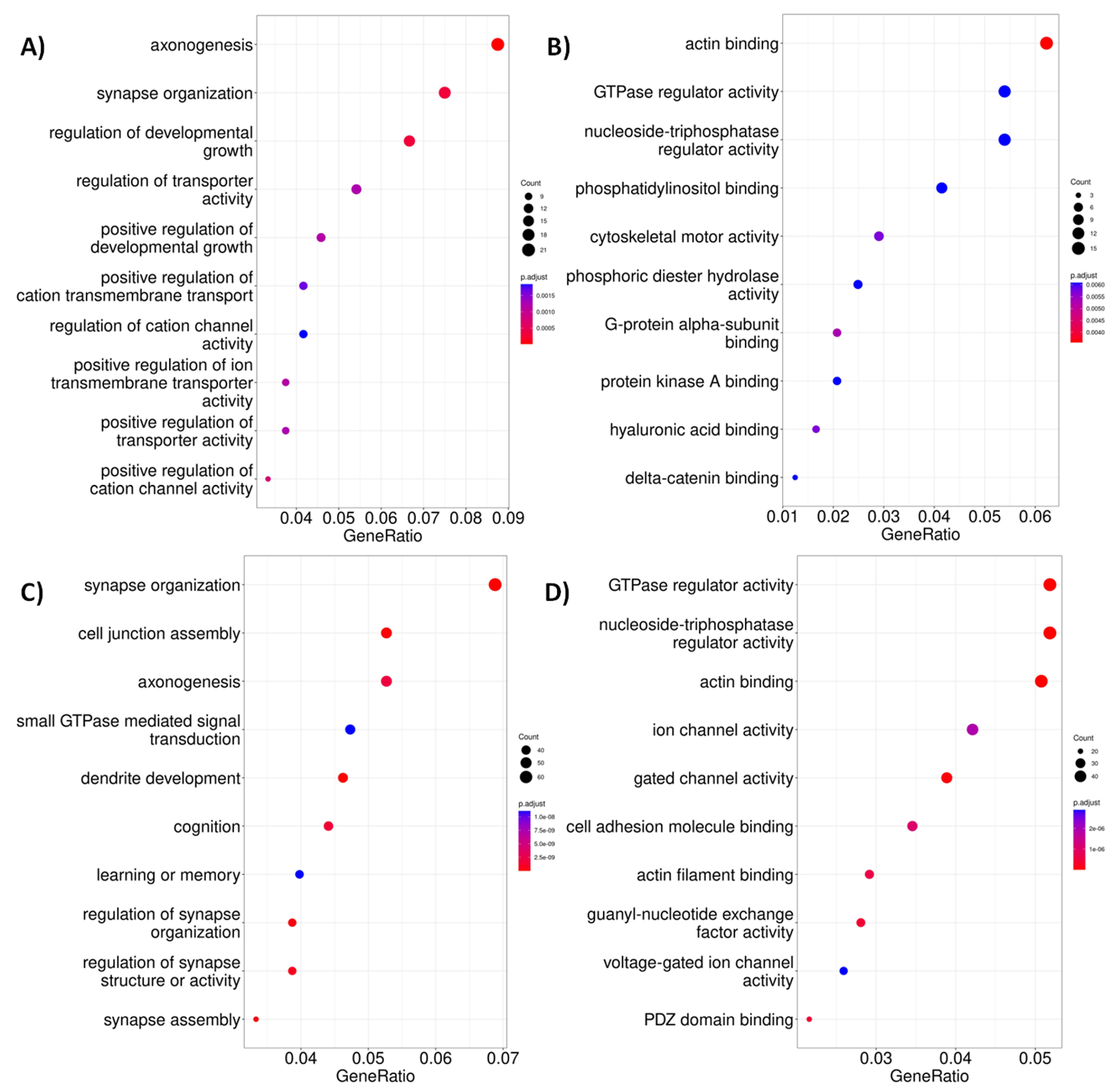
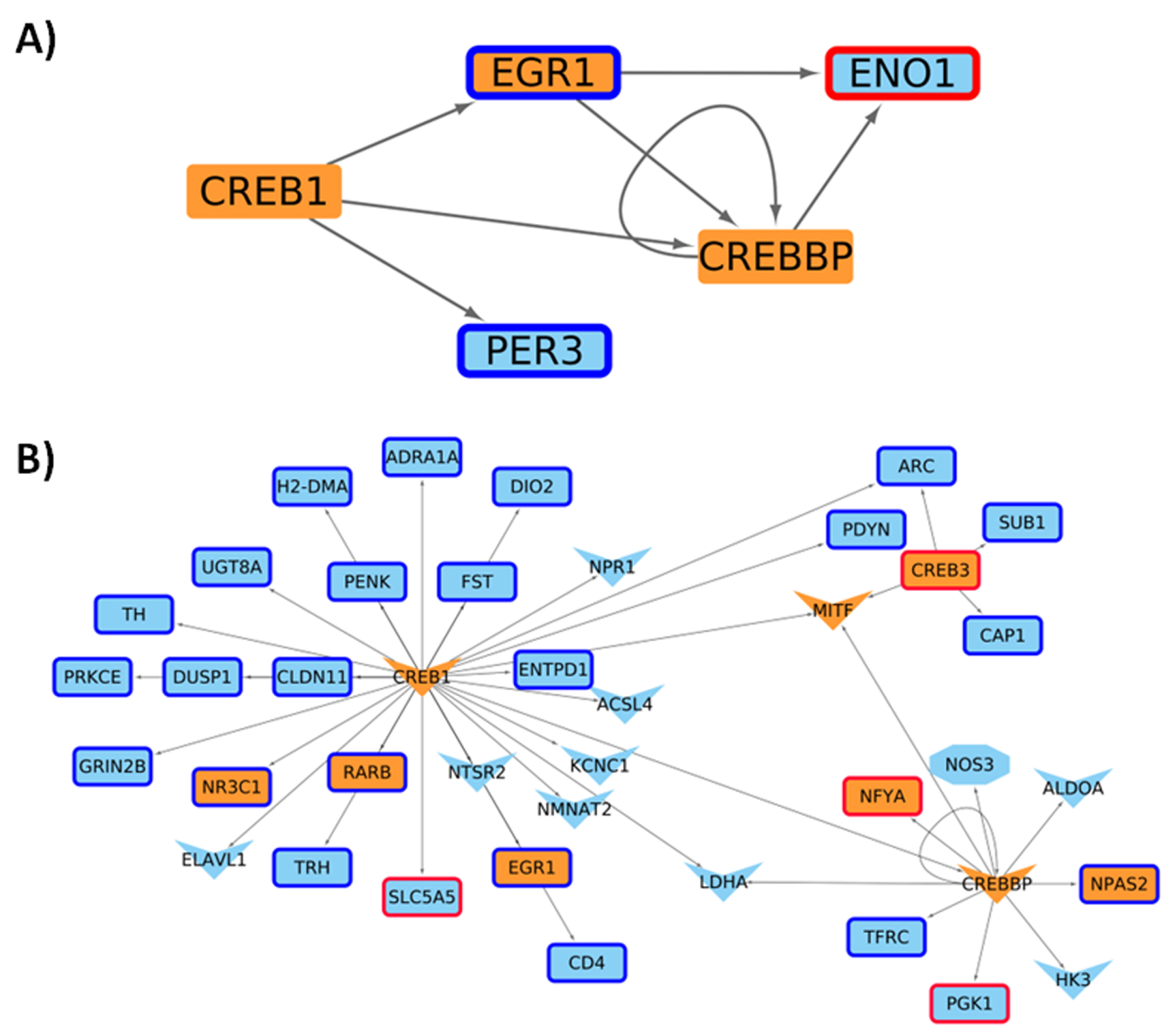
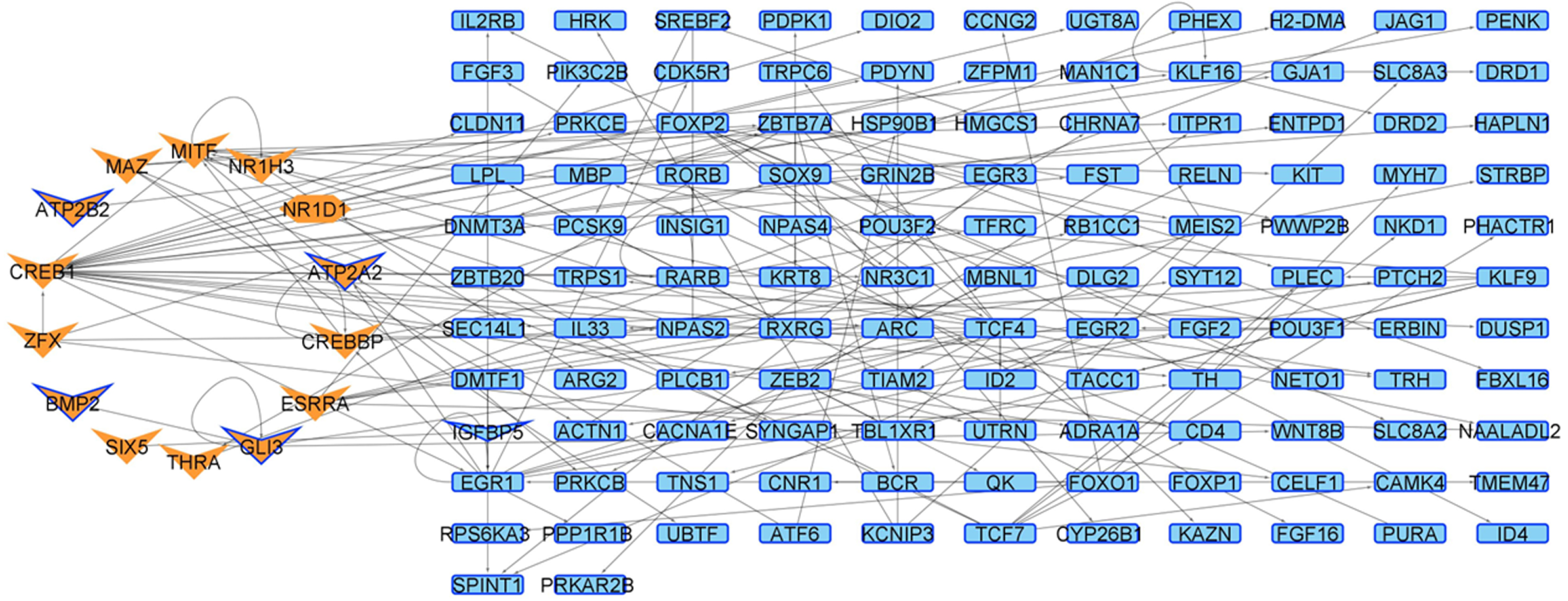
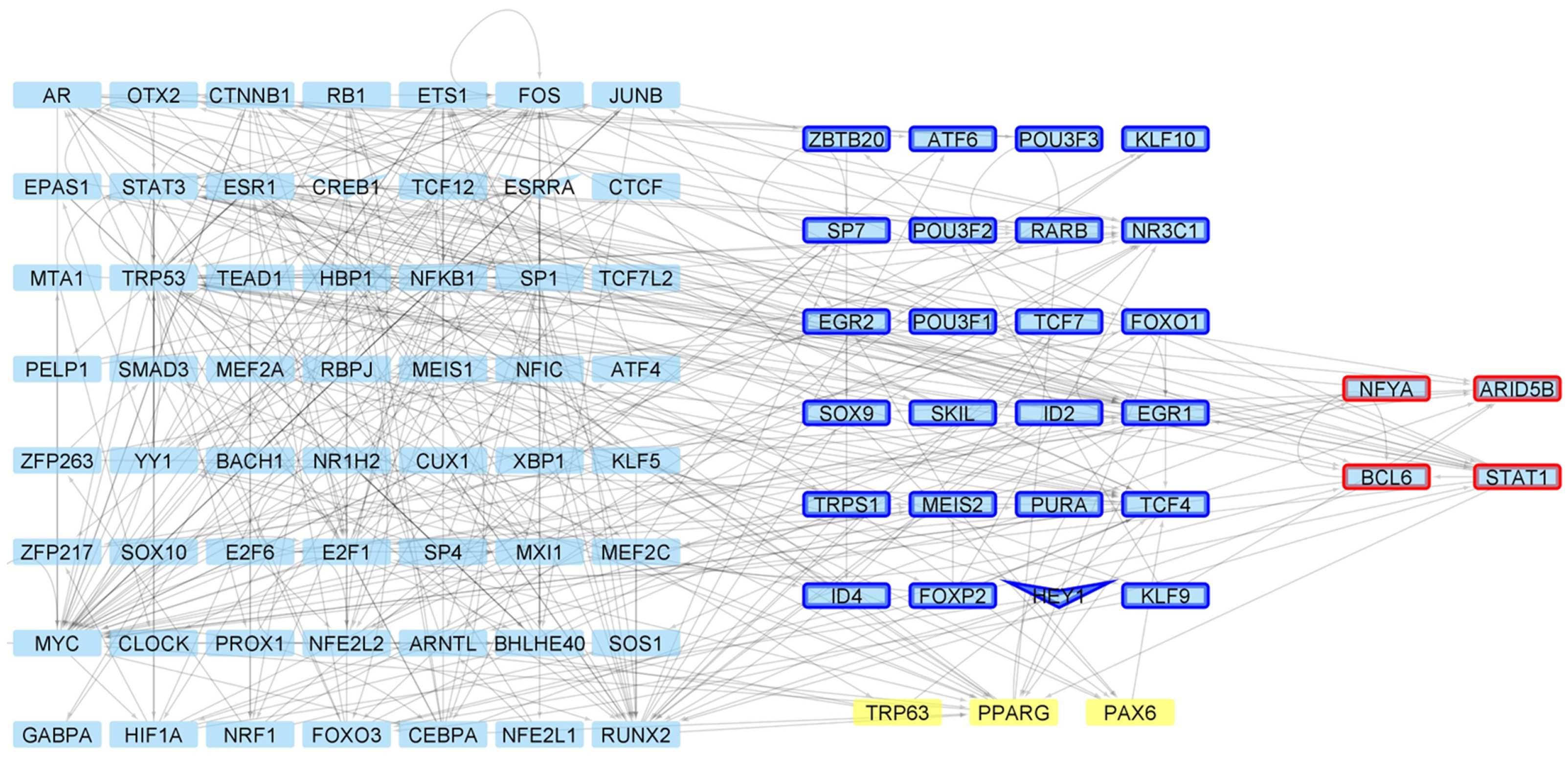
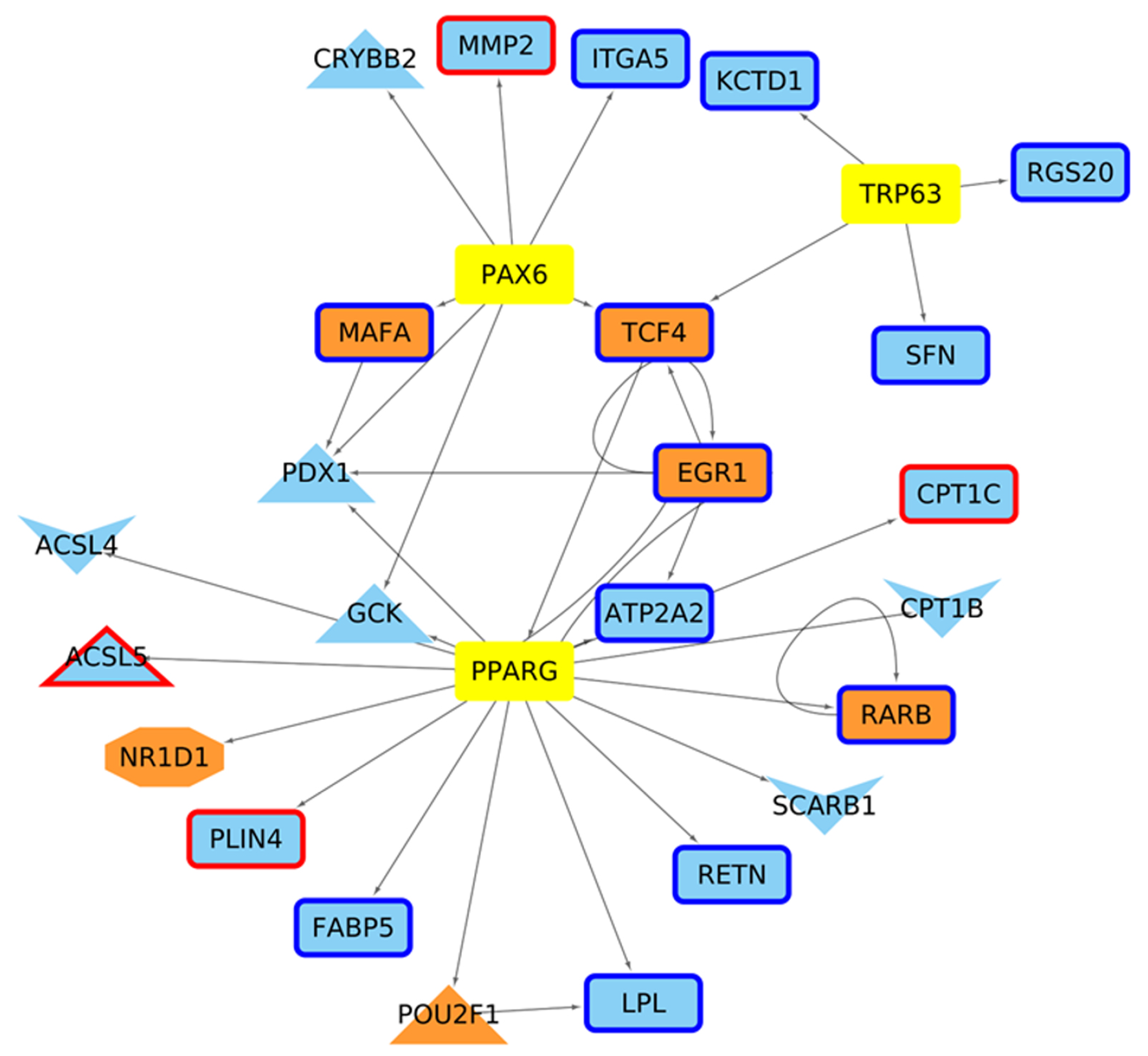
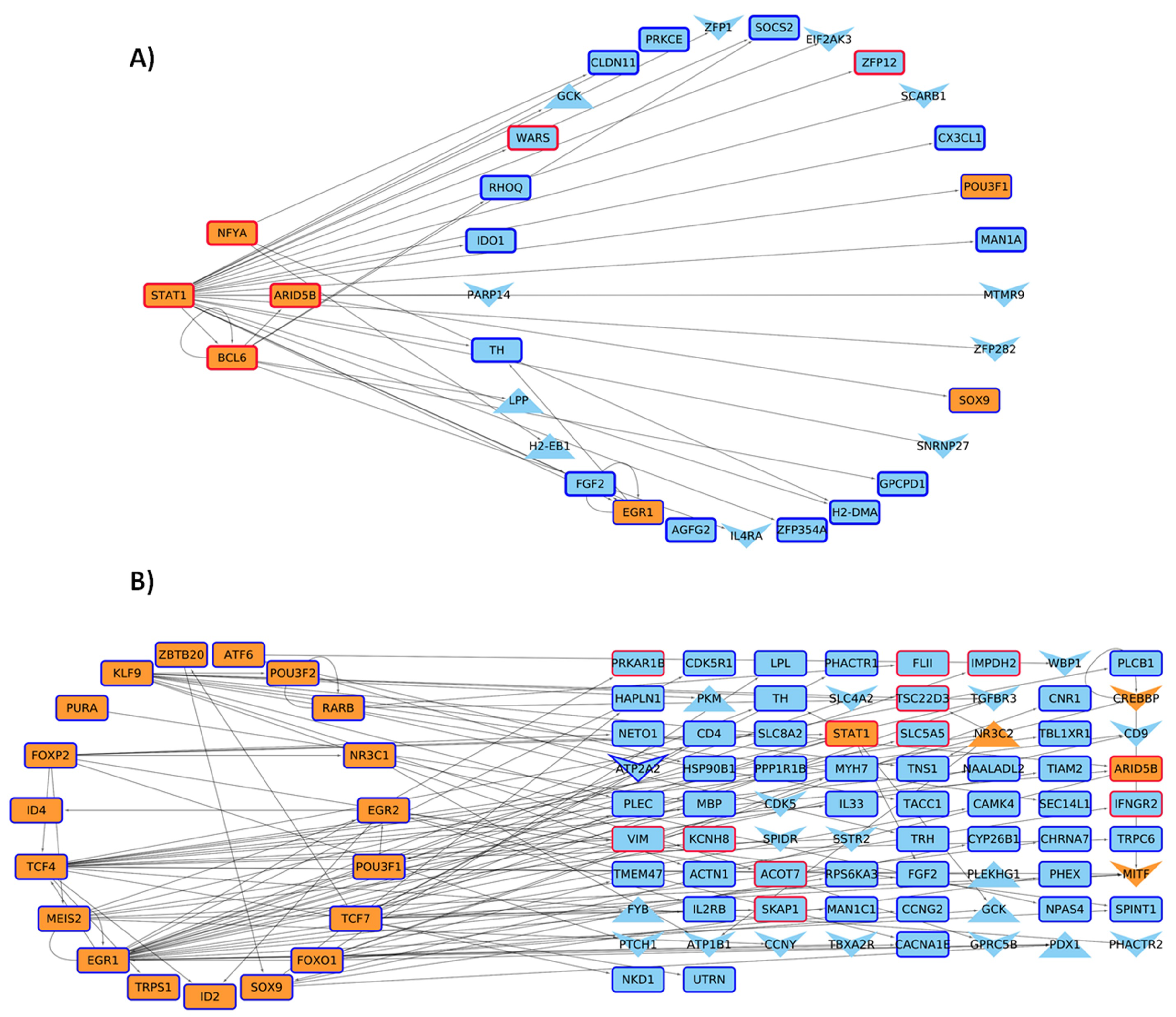
| Network | Total Nodes (TFs) | Unique Nodes (TFs) | Total Connections | Total Connections |
|---|---|---|---|---|
| 4 weeks Control | 5876 (643) | 0 | 16,635 | 0 |
| 4 weeks Mutant | 5972 (680) | 96 (9) | 17,292 | 657 |
| 6 weeks Control | 5841 (641) | 1 (1) | 16,412 | 22 |
| 6 weeks Mutant | 5925 (663) | 84 (8) | 16,990 | 600 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arancibia-Opazo, S.; Contreras-Riquelme, J.S.; Sánchez, M.; Cisternas-Olmedo, M.; Vidal, R.L.; Martin, A.J.M.; Sáez, M.A. Transcriptional and Histone Acetylation Changes Associated with CRE Elements Expose Key Factors Governing the Regulatory Circuit in the Early Stage of Huntington’s Disease Models. Int. J. Mol. Sci. 2023, 24, 10848. https://doi.org/10.3390/ijms241310848
Arancibia-Opazo S, Contreras-Riquelme JS, Sánchez M, Cisternas-Olmedo M, Vidal RL, Martin AJM, Sáez MA. Transcriptional and Histone Acetylation Changes Associated with CRE Elements Expose Key Factors Governing the Regulatory Circuit in the Early Stage of Huntington’s Disease Models. International Journal of Molecular Sciences. 2023; 24(13):10848. https://doi.org/10.3390/ijms241310848
Chicago/Turabian StyleArancibia-Opazo, Sandra, J. Sebastián Contreras-Riquelme, Mario Sánchez, Marisol Cisternas-Olmedo, René L. Vidal, Alberto J. M. Martin, and Mauricio A. Sáez. 2023. "Transcriptional and Histone Acetylation Changes Associated with CRE Elements Expose Key Factors Governing the Regulatory Circuit in the Early Stage of Huntington’s Disease Models" International Journal of Molecular Sciences 24, no. 13: 10848. https://doi.org/10.3390/ijms241310848
APA StyleArancibia-Opazo, S., Contreras-Riquelme, J. S., Sánchez, M., Cisternas-Olmedo, M., Vidal, R. L., Martin, A. J. M., & Sáez, M. A. (2023). Transcriptional and Histone Acetylation Changes Associated with CRE Elements Expose Key Factors Governing the Regulatory Circuit in the Early Stage of Huntington’s Disease Models. International Journal of Molecular Sciences, 24(13), 10848. https://doi.org/10.3390/ijms241310848






