Promoting Photosynthetic Production of Dammarenediol-II in Chlamydomonas reinhardtii via Gene Loading and Culture Optimization
Abstract
:1. Introduction
2. Results
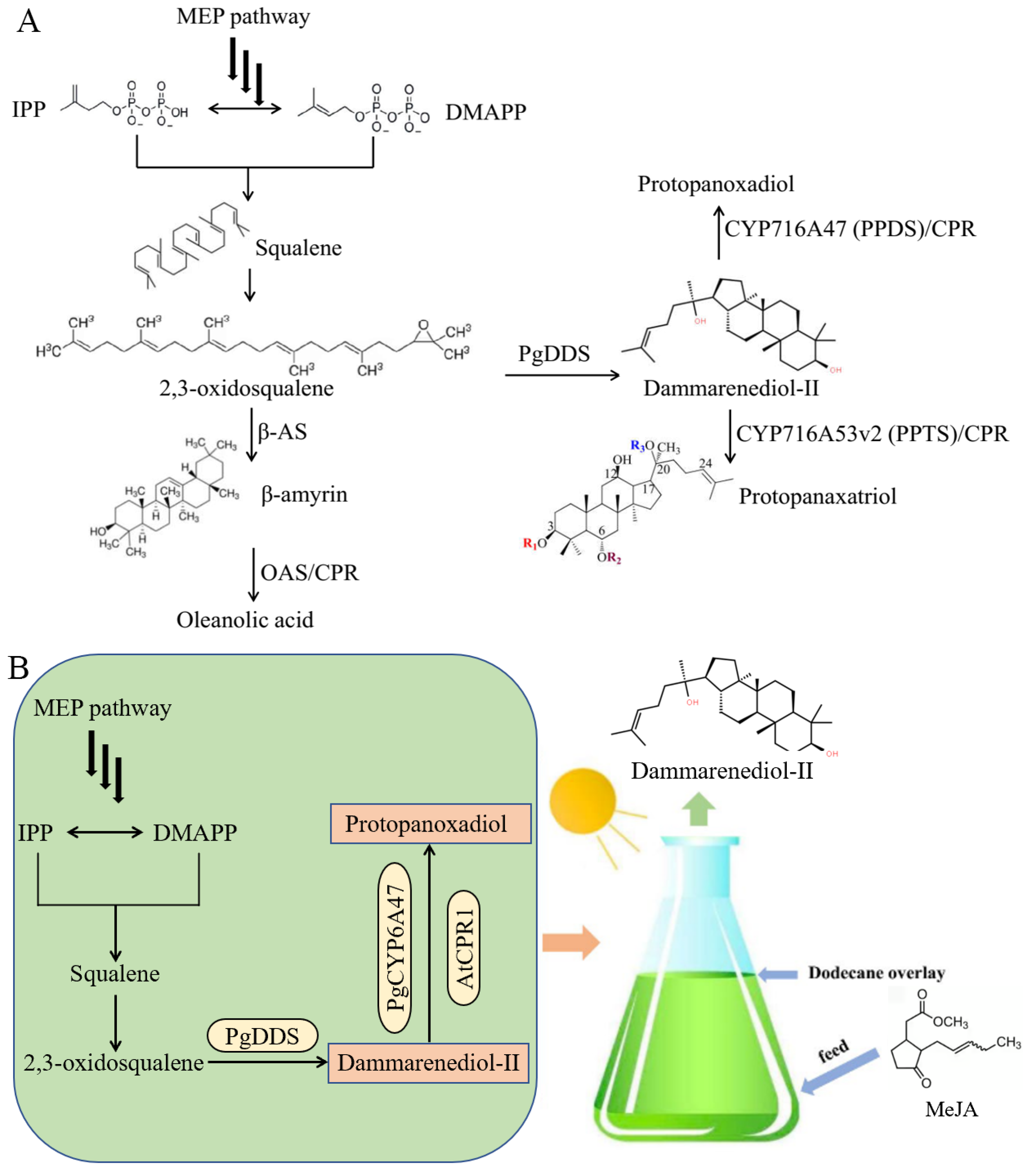
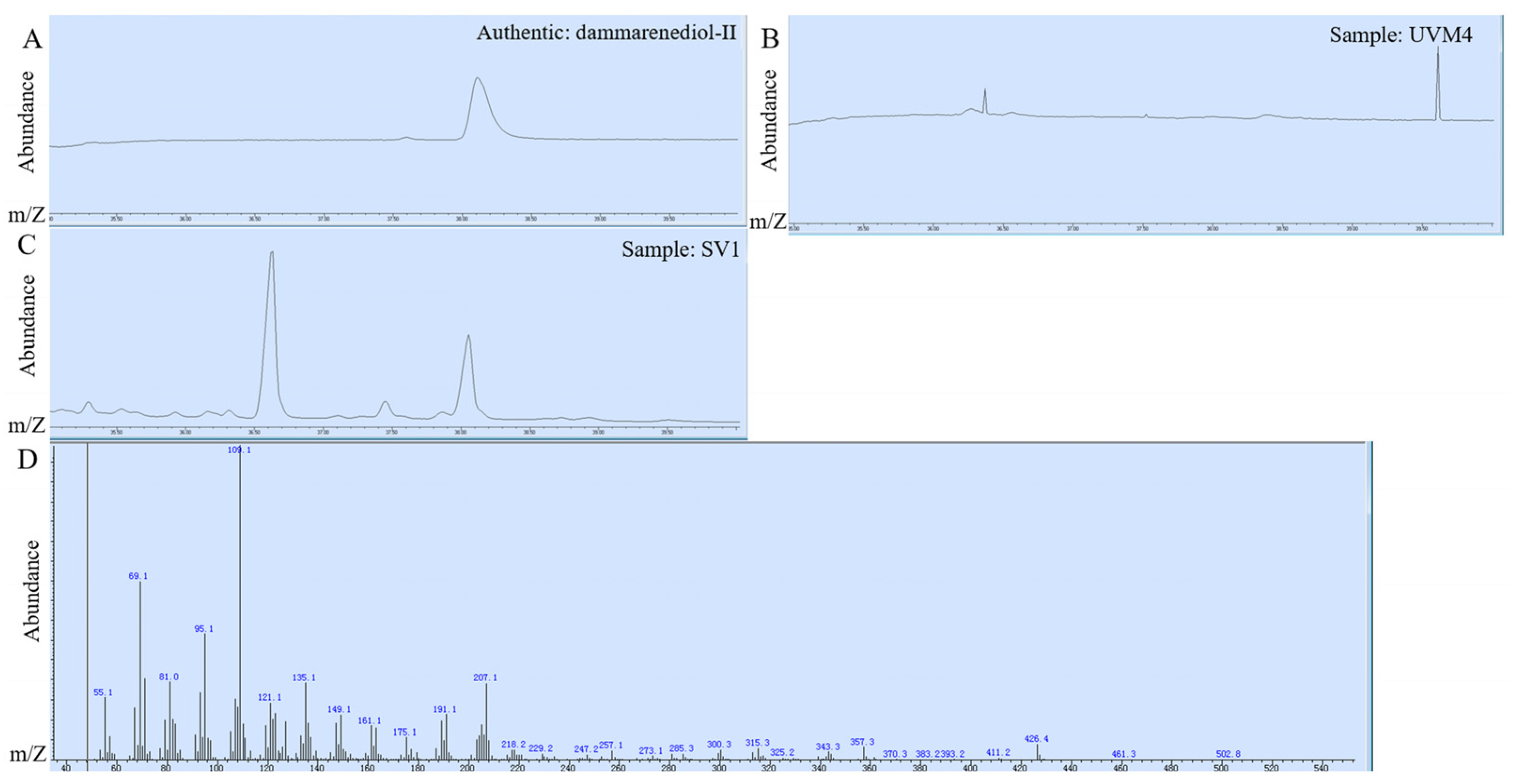
2.1. Construction of Dammarane-Type Ginsenosides Synthetic Pathway in C. reinhardtii
2.2. Dammarenediol-II Production Optimization
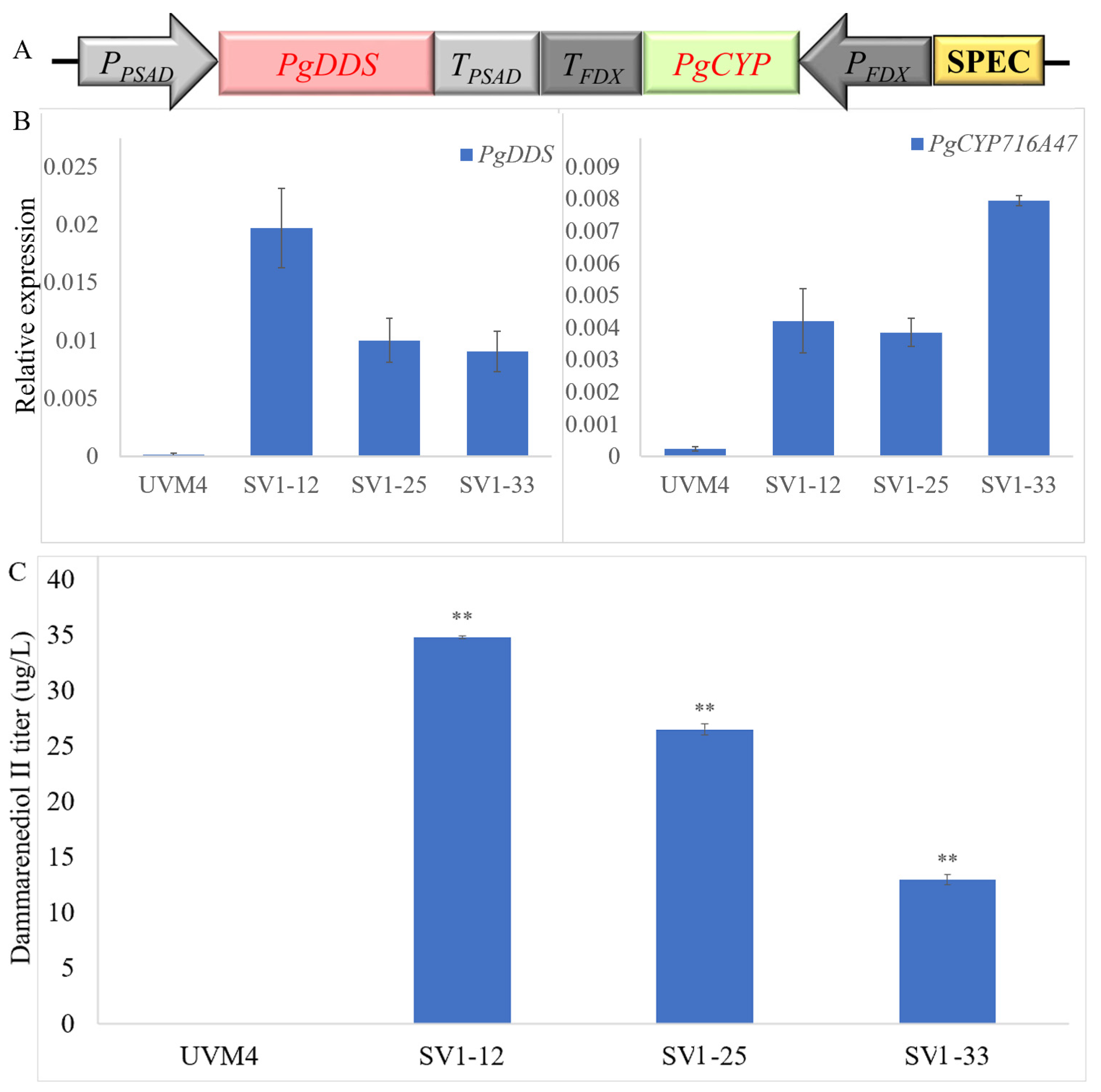
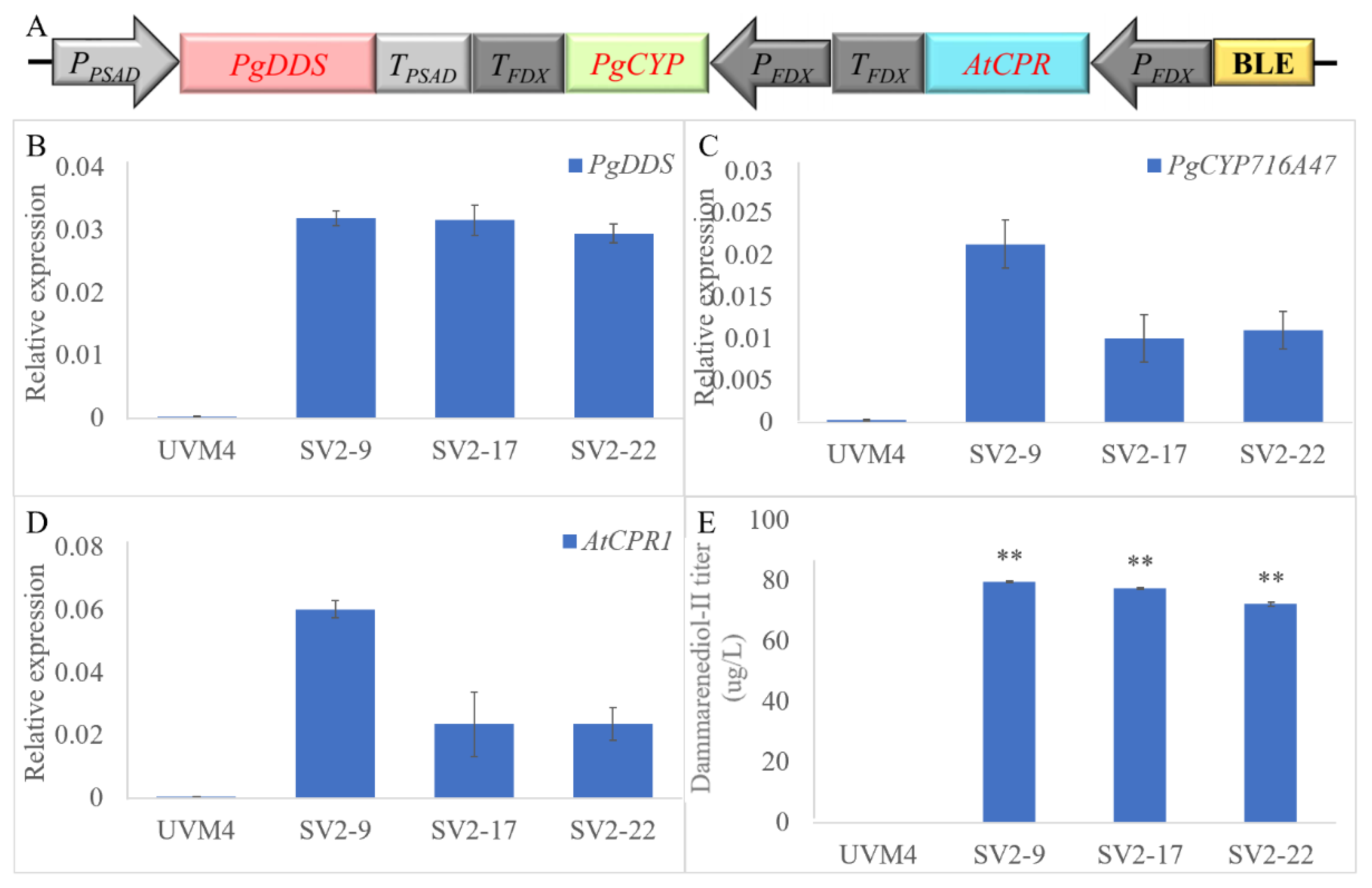
2.2.1. Employed Gene Loading Strategy to Increase Dammarane-Type Ginsenosides Synthesis
2.2.2. Culture Medium Optimization
2.2.3. Light Regime Optimization
2.2.4. Extraction Solvent Optimization
2.2.5. Employed MeJA to Increase Dammarane-Type Ginsenosides Synthesis
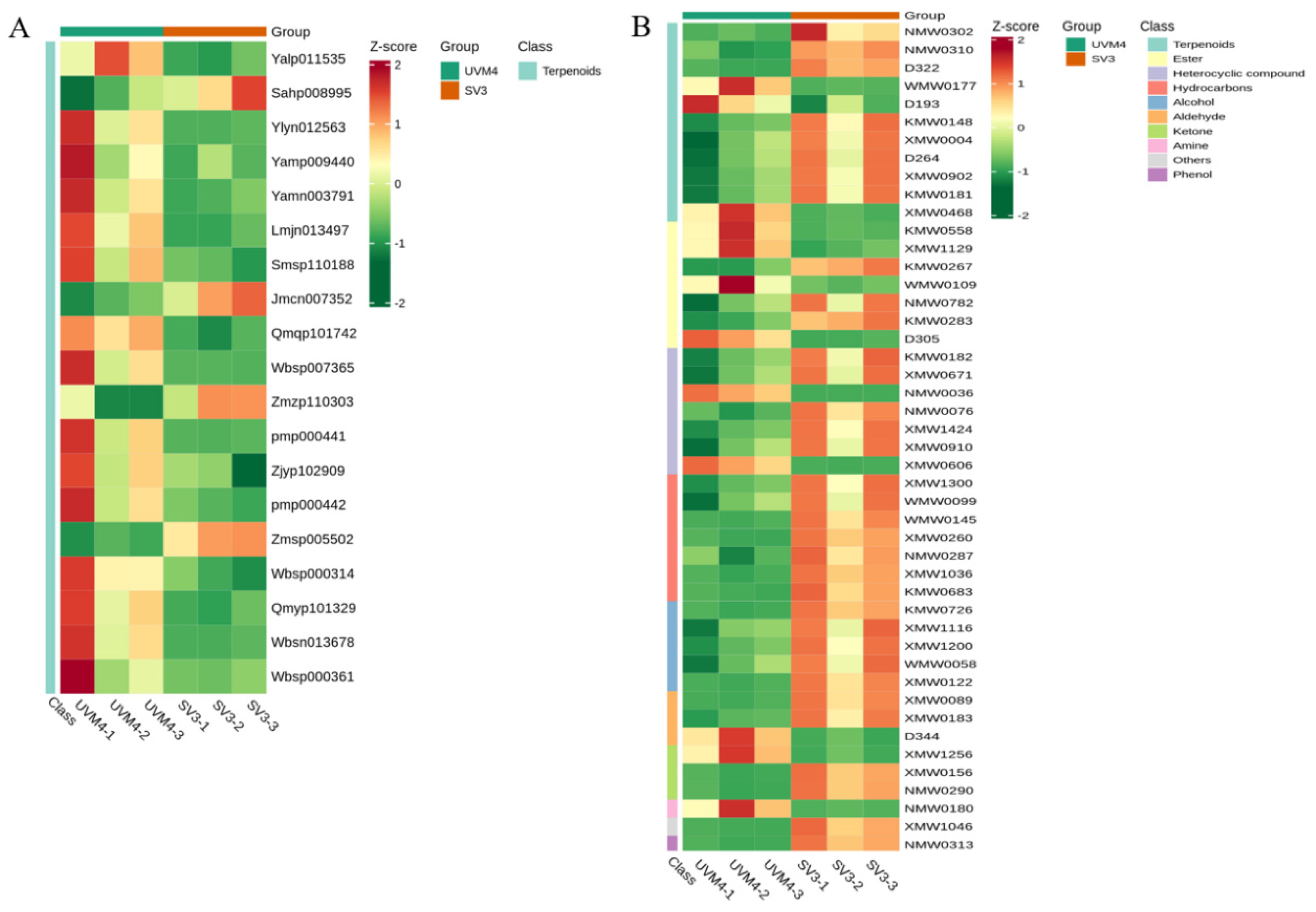
2.3. Metabolomic Profiling of Transgenic Algae
3. Discussion
4. Materials and Methods
4.1. C. reinhardtii Strain and Cultivation Conditions
4.2. Plasmid Construction
4.3. Strain Construction
4.4. Two-Phase Extractive Fermentation of Positive Clone Strain
4.5. Optimization of the Fermentation and Dammarenediol-II Extraction Conditions
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cox-Georgian, D.; Ramadoss, N.; Dona, C.; Basu, C. Therapeutic and Medicinal Uses of Terpenes. In Medicinal Plants; Joshee, N., Dhekney, S.A., Prahlad, P., Eds.; Springer: Cham, Switzerland, 2019; pp. 333–359. [Google Scholar]
- Lim, S.H.; Baek, J.I.; Jeon, B.M.; Seo, J.W.; Kim, M.S.; Byun, J.Y.; Park, S.H.; Kim, S.J.; Lee, J.Y.; Lee, J.H.; et al. CRISPRi-Guided Metabolic Flux Engineering for Enhanced Protopanaxadiol Production in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2021, 22, 11836. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhang, Z.Y.; Liao, F.L.; Jiang, T.L.; Tu, Y.Y. The birth of artemisinin. Pharmacol. Ther. 2020, 216, 107658. [Google Scholar] [CrossRef] [PubMed]
- Nah, S.-Y.; Kim, D.-H.; Rhim, H. Ginsenosides: Are Any of them Candidates for Drugs Acting on the Central Nervous System? CNS Drug Rev. 2007, 13, 381–404. [Google Scholar] [CrossRef]
- Gantait, S.; Mitra, M.; Chen, J.T. Biotechnological Interventions for Ginsenosides Production. Biomolecules 2020, 10, 538. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Liu, Y.; Zhang, X.; Shi, M.; Wang, B.; Wang, D.; Huang, L.; Zhang, X. Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab. Eng. 2013, 20, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Orsi, E.; Beekwilder, J.; van Gelder, D.; van Houwelingen, A.; Eggink, G.; Kengen, S.W.M.; Weusthuis, R.A. Functional replacement of isoprenoid pathways in Rhodobacter sphaeroides. Microb. Biotechnol. 2020, 13, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Qiu, S.; Blank, L.M. Recent Advances in Yeast Recombinant Biosynthesis of the Triterpenoid Protopanaxadiol and Glycosylated Derivatives Thereof. J. Agric. Food Chem. 2023, 71, 2197–2210. [Google Scholar] [CrossRef]
- Kim, J.E.; Jang, I.S.; Sung, B.H.; Kim, S.C.; Lee, J.Y. Rerouting of NADPH synthetic pathways for increased protopanaxadiol production in Saccharomyces cerevisiae. Sci. Rep. 2018, 8, 15820. [Google Scholar] [CrossRef] [Green Version]
- Hou, M.; Wang, R.; Zhao, S.; Wang, Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B 2021, 11, 1813–1834. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Nguyen, H.T.; Win, N.N.; Wong, C.P.; Huynh, K.L.V.; Hoang, N.N.; Do, K.M.; Nguyen, H.T.; Hoai, N.T.; Nguyen, M.D.; et al. Antimelanogenic Activity of Ocotillol-Type Saponins from Panax vietnamensis. Chem. Biodivers. 2020, 17, e2000037. [Google Scholar] [CrossRef]
- Chopra, P.; Chhillar, H.; Kim, Y.J.; Jo, I.H.; Kim, S.T.; Gupta, R. Phytochemistry of ginsenosides: Recent advancements and emerging roles. Crit. Rev. Food Sci. Nutr. 2023, 63, 613–640. [Google Scholar] [CrossRef]
- Tran, N.T.; Kaldenhoff, R. Metabolic engineering of ketocarotenoids biosynthetic pathway in Chlamydomonas reinhardtii strain CC-4102. Sci. Rep. 2020, 10, 10688. [Google Scholar] [CrossRef]
- Fayyaz, M.; Chew, K.W.; Show, P.L.; Ling, T.C.; Ng, I.-S.; Chang, J.-S. Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol. Adv. 2020, 43, 107554. [Google Scholar] [CrossRef]
- Perozeni, F.; Cazzaniga, S.; Baier, T.; Zanoni, F.; Zoccatelli, G.; Lauersen, K.J.; Wobbe, L.; Ballottari, M. Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii. Plant Biotechnol. J. 2020, 18, 2053–2067. [Google Scholar] [CrossRef] [Green Version]
- Li, X.Y.; Lan, C.X.; Li, X.Y.; Hu, Z.L.; Jia, B. A review on design-build-test-learn cycle to potentiate progress in isoprenoid engineering of photosynthetic microalgae. Bioresour. Technol. 2022, 363, 127981. [Google Scholar] [CrossRef]
- Lauersen, K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta 2019, 249, 155–180. [Google Scholar] [CrossRef]
- Lohr, M.; Schwender, J.; Polle, J.E. Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci. 2012, 185–186, 9–22. [Google Scholar] [CrossRef]
- Zhao, M.L.; Cai, W.S.; Zheng, S.Q.; Zhao, J.L.; Zhang, J.L.; Huang, Y.; Hu, Z.L.; Jia, B. Metabolic Engineering of the Isopentenol Utilization Pathway Enhanced the Production of Terpenoids in Chlamydomonas reinhardtii. Mar. Drugs 2022, 20, 577. [Google Scholar] [CrossRef]
- Lopez-Paz, C.; Liu, D.; Geng, S.; Umen, J.G. Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression. Plant J. 2017, 92, 1232–1244. [Google Scholar] [CrossRef] [Green Version]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Perez-Perez, M.E.; Auroy, P.; Rovira, A.G.; Sauret-Gueto, S.; et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [Green Version]
- Commault, A.S.; Fabris, M.; Kuzhiumparambil, U.; Adriaans, J.; Pernice, M.; Ralph, P.J. Methyl jasmonate treatment affects the regulation of the 2-C-methyl-D-erythritol 4-phosphate pathway and early steps of the triterpenoid biosynthesis in Chlamydomonas reinhardtii. Algal Res. 2019, 39, 101462. [Google Scholar] [CrossRef]
- Song, I.; Kim, J.; Baek, K.; Choi, Y.; Shin, B.; Jin, E. The generation of metabolic changes for the production of high-purity zeaxanthin mediated by CRISPR-Cas9 in Chlamydomonas reinhardtii. Microb. Cell Fact. 2020, 19, 220. [Google Scholar] [CrossRef]
- Overmans, S.; Lauersen, K.J. Biocompatible fluorocarbon liquid underlays for in situ extraction of isoprenoids from microbial cultures. RSC Adv. 2022, 12, 16632–16639. [Google Scholar] [CrossRef]
- Commault, A.S.; Kuzhiumparambil, U.; Herdean, A.; Fabris, M.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Ralph, P.J.; Pernice, M. Methyl Jasmonate and Methyl-beta-Cyclodextrin Individually Boost Triterpenoid Biosynthesis in Chlamydomonas Reinhardtii UVM4. Pharmaceuticals 2021, 14, 125. [Google Scholar] [CrossRef]
- Hu, Z.; Lin, L.; Li, H.; Li, P.; Weng, Y.; Zhang, C.; Yu, A.; Xiao, D. Engineering Saccharomyces cerevisiae for production of the valuable monoterpene d-limonene during Chinese baijiu fermentation. J. Ind. Microbiol. Biotechnol. 2020, 47, 511–523. [Google Scholar] [CrossRef]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef]
- Aw, R.; Polizzi, K.M. Can too many copies spoil the broth? Microb. Cell Fact. 2013, 12, 128. [Google Scholar] [CrossRef] [Green Version]
- Lauersen, K.J.; Baier, T.; Wichmann, J.; Wördenweber, R.; Mussgnug, J.H.; Hübner, W.; Huser, T.; Kruse, O. Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng. 2016, 38, 331–343. [Google Scholar] [CrossRef]
- Wichmann, J.; Baier, T.; Wentnagel, E.; Lauersen, K.; Kruse, O. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018, 45, 211–222. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Huber, I.; Wichmann, J.; Baier, T.; Leiter, A.; Gaukel, V.; Kartushin, V.; Rattenholl, A.; Steinweg, C.; von Riesen, L.; et al. Investigating the dynamics of recombinant protein secretion from a microalgal host. J. Biotechnol. 2015, 215, 62–71. [Google Scholar] [CrossRef]
- Jia, B.; Lan, C.; Li, X. Analysis of the initial response pattern of Chlamydomonas reinhardtii to methyl jasmonate (MeJA) treatment. J. Shenzhen Univ. Sci. Eng. 2022, 39, 237–244. [Google Scholar] [CrossRef]
- Liu, T.; Luo, T.; Guo, X.; Zou, X.; Zhou, D.; Afrin, S.; Li, G.; Zhang, Y.; Zhang, R.; Luo, Z. PgMYB2, a MeJA-Responsive Transcription Factor, Positively Regulates the Dammarenediol Synthase Gene Expression in Panax Ginseng. Int. J. Mol. Sci. 2019, 20, 2219. [Google Scholar] [CrossRef] [Green Version]
- Han, J.Y.; Wang, H.Y.; Choi, Y.E. Production of dammarenediol-II triterpene in a cell suspension culture of transgenic tobacco. Plant Cell Rep. 2014, 33, 225–233. [Google Scholar] [CrossRef]
- Huang, Z.; Lin, J.; Cheng, Z.; Xu, M.; Huang, X.; Yang, Z.; Zheng, J. Production of dammarane-type sapogenins in rice by expressing the dammarenediol-II synthase gene from Panax ginseng C.A. Mey. Plant Sci. 2015, 239, 106–114. [Google Scholar] [CrossRef]
- Liu, X.B.; Liu, M.; Tao, X.Y.; Zhang, Z.X.; Wang, F.Q.; Wei, D.Z. Metabolic engineering of pichia pastoris for the production of dammarenediol-II. J. Biotechnol. 2015, 216, 47–55. [Google Scholar] [CrossRef]
- Fuhrmann, M.; Oertel, W.; Hegemann, P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999, 19, 353–361. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Kruse, O.; Mussgnug, J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015, 99, 3491–3503. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef]
- Han, J.Y.; Kim, H.J.; Kwon, Y.S.; Choi, Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011, 52, 2062–2073. [Google Scholar] [CrossRef] [Green Version]


| Strain | Dammarenediol-II Titer | Cultivation Setup | Reference |
|---|---|---|---|
| Transgenic tobacco | 5.2 mg/L | Cell suspension culture | [34] |
| Transgenic rice | 0.44 mg/g | - | [35] |
| Transgenic Pichia pastoris | 13.233 mg/L | Shake flask fermentation | [36] |
| Transgenic C. reinhardtii | 3.3 mg/L | Shake flask fermentation with 1.5 mM MeJA | This study |
| Name | Description | Source | Antibiotic Resistance |
|---|---|---|---|
| pUC57 | Cloning vector with multiple cloning sites | GenScript | Ampicillin |
| PgDDS | Cloning PgDDS gene into pUC57 | This study | Ampicillin |
| PgCYP716A47 | Cloning PgCYP716A47 gene into pUC57 | This study | Ampicillin |
| AtCPR | Cloning AtCPR gene into pUC57 | This study | Ampicillin |
| pEASY-Blunt | Cloning vector with multiple cloning sites | TransGen Biotech | Ampicillin |
| p-PgDDS | Cloning PPsad-PgDDS-TPsad cassette into pEASY-Blunt | This study | Ampicillin |
| p-PgCYP716A47 | Cloning PFDX-PgCYP716A47-TFDX cassette into pEASY-Blunt | This study | Ampicillin |
| p-AtCPR | Cloning PFDX-AtCPR-TFDX cassette into pEASY-Blunt | This study | Ampicillin |
| pOpt | Cloning vector with multiple cloning sites | Lauersen, et al. [39] | Zeocin |
| pOpt-Ble | Cloning vector with multiple cloning sites | This study | Zeocin |
| V1 | Cloning PPsad-PgDDS-TPsad and PFDX-PgCYP716A47-TFDX cassettes into pOpt2-Spec | This study | Spectinomycin |
| V2 | Cloning PPsad-PgDDS-TPsad, PFDX-PgCYP716A47-TFDX, and PFDX-AtCPR-TFDX cassettes into pOpt2-Ble | This study | Zeocin |
| Name | Description | Source |
|---|---|---|
| SV1 | Transformants that harbor V1 construct | This study |
| SV2 | Transformants that harbor V2 construct | This study |
| SV3 | Transformants that harbor V1 and V2 constructs | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.-L.; Li, X.-Y.; Lan, C.-X.; Yuan, Z.-L.; Zhao, J.-L.; Huang, Y.; Hu, Z.-L.; Jia, B. Promoting Photosynthetic Production of Dammarenediol-II in Chlamydomonas reinhardtii via Gene Loading and Culture Optimization. Int. J. Mol. Sci. 2023, 24, 11002. https://doi.org/10.3390/ijms241311002
Zhao M-L, Li X-Y, Lan C-X, Yuan Z-L, Zhao J-L, Huang Y, Hu Z-L, Jia B. Promoting Photosynthetic Production of Dammarenediol-II in Chlamydomonas reinhardtii via Gene Loading and Culture Optimization. International Journal of Molecular Sciences. 2023; 24(13):11002. https://doi.org/10.3390/ijms241311002
Chicago/Turabian StyleZhao, Mei-Li, Xiang-Yu Li, Cheng-Xiang Lan, Zi-Ling Yuan, Jia-Lin Zhao, Ying Huang, Zhang-Li Hu, and Bin Jia. 2023. "Promoting Photosynthetic Production of Dammarenediol-II in Chlamydomonas reinhardtii via Gene Loading and Culture Optimization" International Journal of Molecular Sciences 24, no. 13: 11002. https://doi.org/10.3390/ijms241311002





