Effects of Soybean Phosphate Transporter Gene GmPHT2 on Pi Transport and Plant Growth under Limited Pi Supply Condition
Abstract
1. Introduction
2. Results
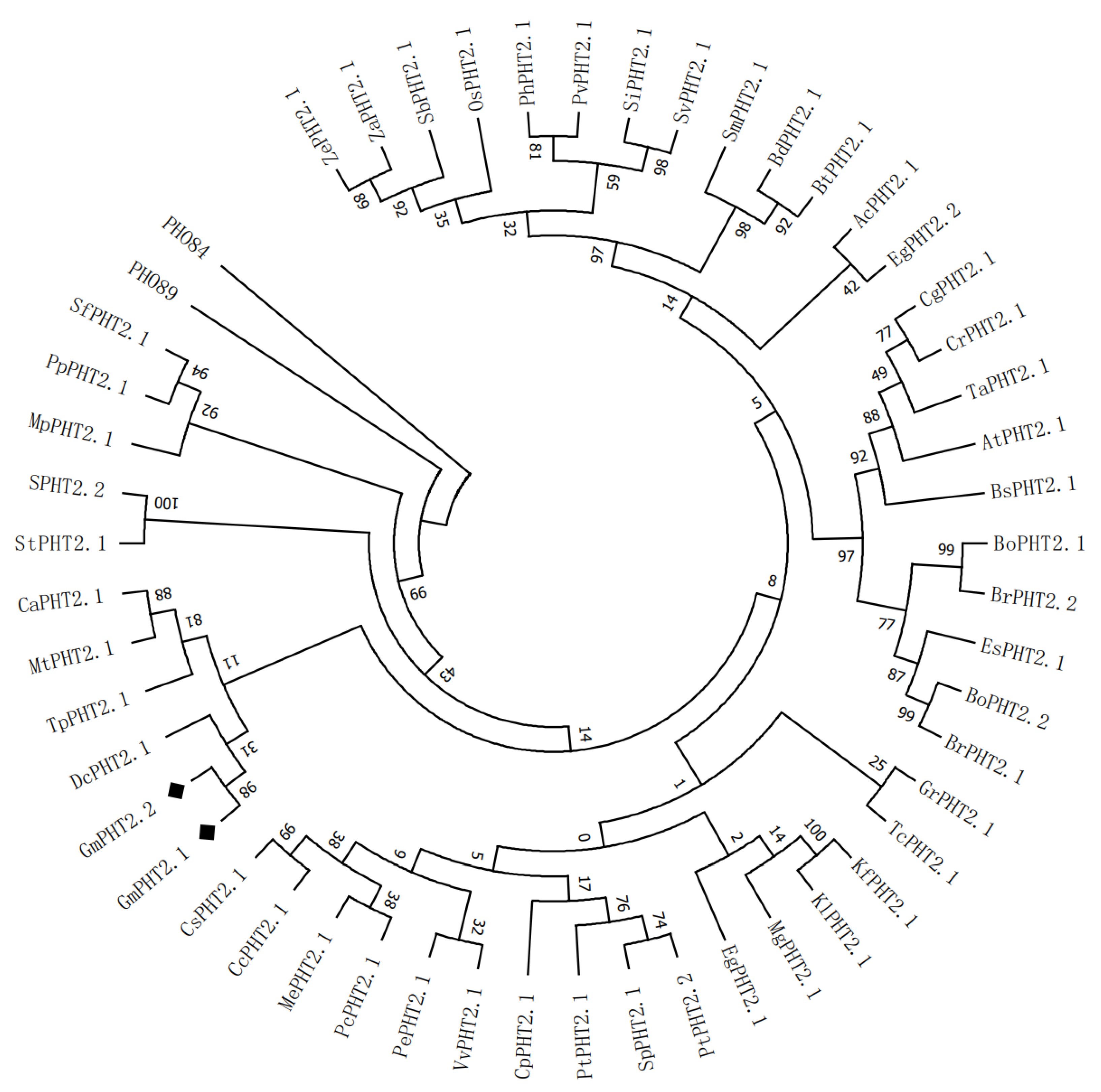
2.1. Phylogenetic Analysis of GmPHT2 Gene Family
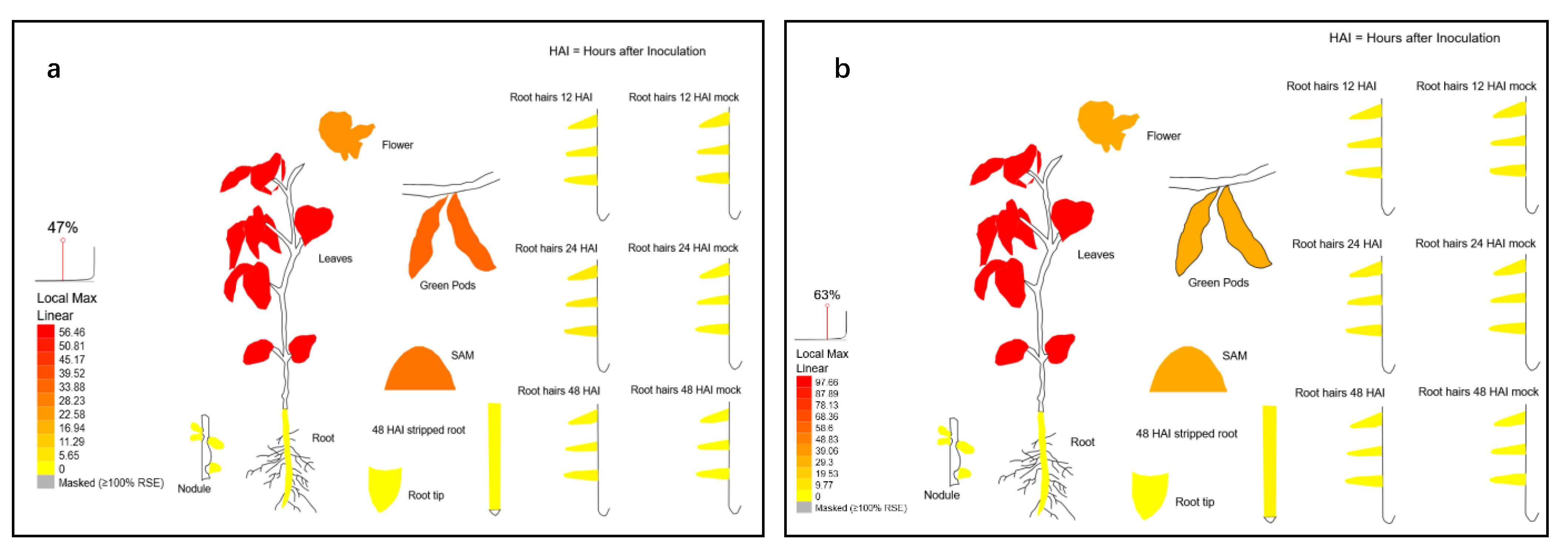
2.2. Expression Patterns of GmPHT2 Genes in Soybean
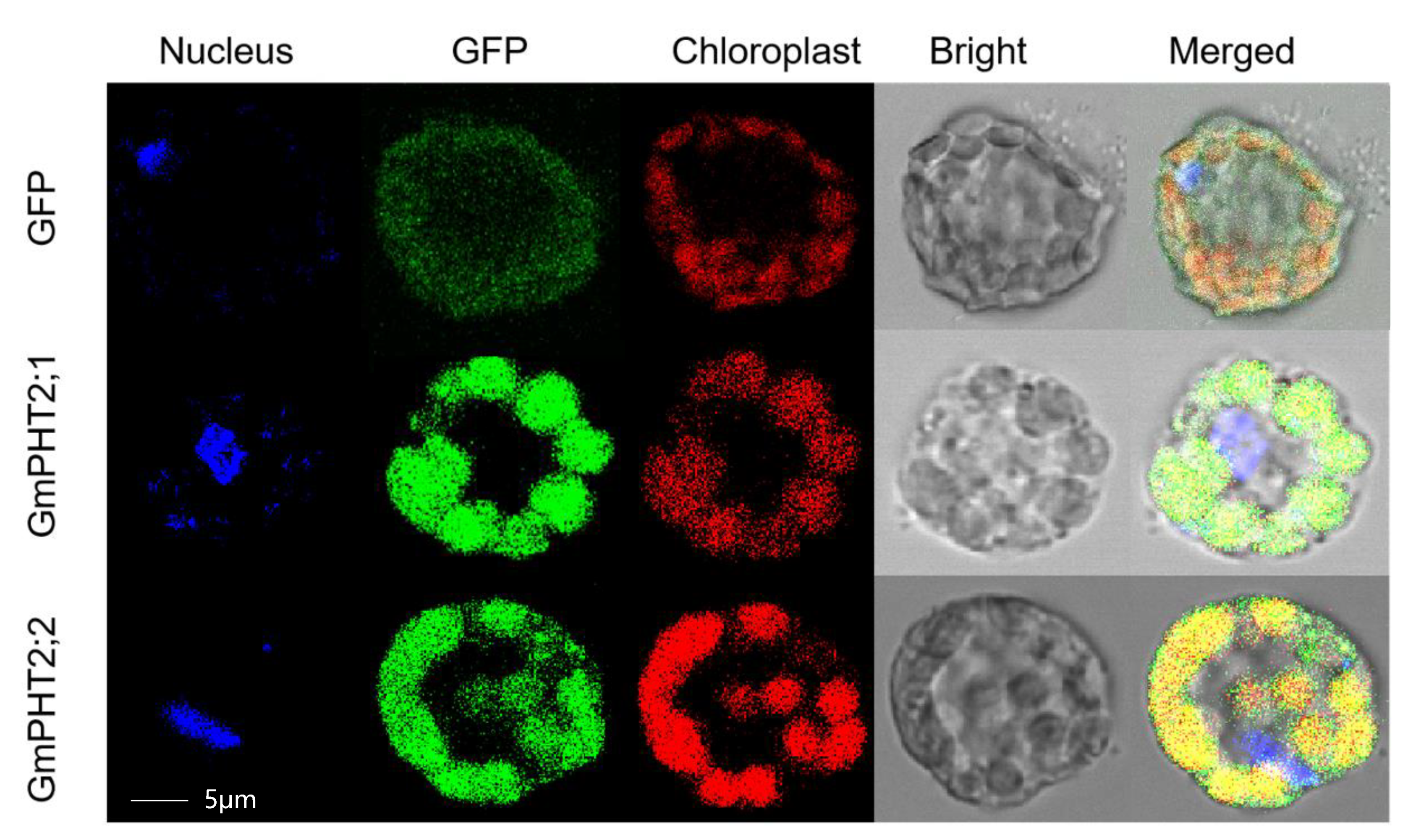
2.3. Subcellular Localization of Gmpht2 Family Member
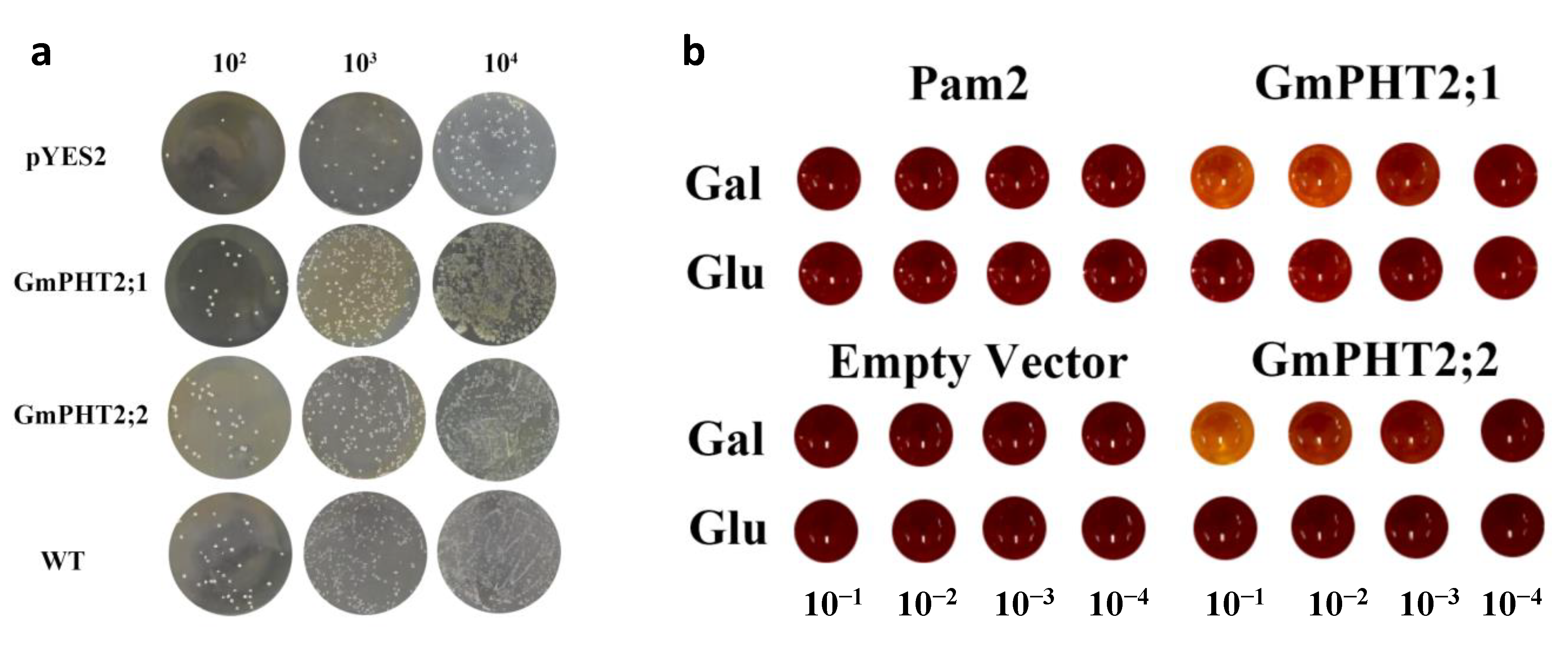
2.4. Pi Transport Activity of GmPHT2 Genes in Yeast
2.5. Expression Patterns of GmPHT2 Genes in Response to Different Treatments
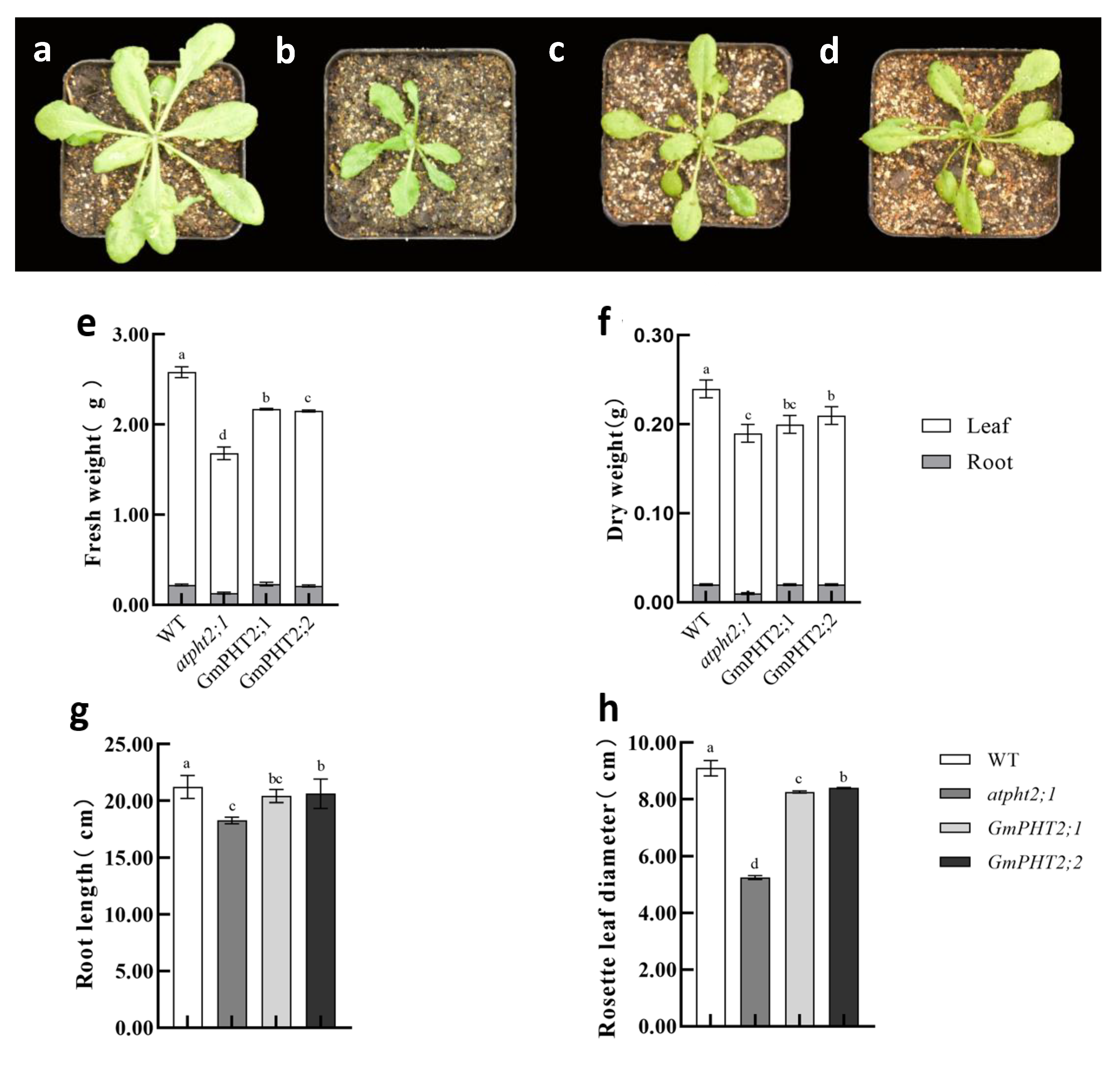
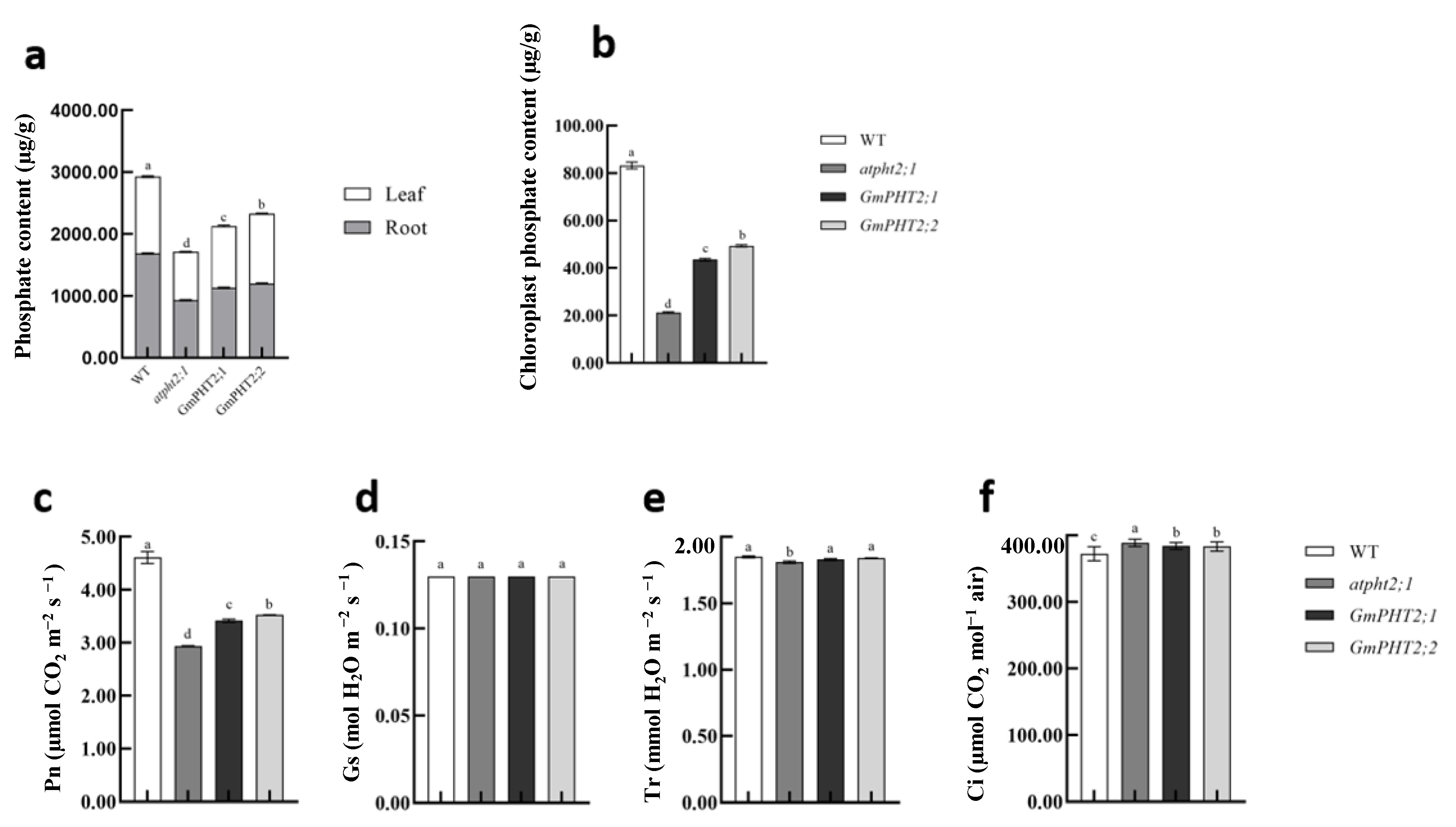
2.6. Heterologous Expression of GmPHT2 Enhanced Pi Uptake in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Bioinformatic and Phylogenetic Analysis of GmPHT2 Gene Family
4.3. RNA Isolation, Quantification, and First-Strand cDNA Synthesis
4.4. Quantitative Real-Time PCR (qRT-PCR)
4.5. Subcellular Localization of Genes
4.6. Complementation of Yeast Pi Transport Mutant by PHT2 Genes
4.7. Analysis of Promoters of GmPHT2 Family Genes
4.8. Vector Construction and Gene Transformation
4.9. Phenotypic Identification of Arabidopsis Plants
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Vance Carroll, P. Quantitative trait loci, epigenetics, sugars, and microRNAs: Quaternaries in phosphate acquisition and use. Plant Physiol. 2010, 154, 582–588. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Domagalski, J.; Lin, C.; Luo, Y.; Kang, J.; Wang, S.; Brown, L.R.; Munn, M.D. Eutrophication study at the Panjiakou-Daheiting Reservoir system, northern Hebei Province, People’s Republic of China: Chlorophyll—A model and sources of phosphorus and nitrogen. Agric. Water Manag. 2007, 94, 43–53. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 2000, 3, 182–187. [Google Scholar] [CrossRef]
- Yuan, H.; Liu, D. Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 2008, 50, 849–859. [Google Scholar] [CrossRef]
- Wei, X.; Fu, Y.; Yu, R.; Wu, L.; Wu, Z.; Tian, P.; Li, S.; Yang, X.; Yang, M. Comprehensive sequence and expression profile analysis of the phosphate transporter gene family in soybean. Sci. Rep. 2022, 12, 20883. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Xue, L.; Sun, M.; Yi, K.; Zhao, H. Functional Analysis of Phosphate Transporter OsPHT4 Family Members in Rice. Rice Sci. 2020, 27, 493–503. [Google Scholar]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef]
- Liu, T.; Huang, T.; Yang, S.; Hong, Y.; Huang, S.; Wang, F.; Chiang, S.; Tsai, S.; Lu, W.; Chiou, T. Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 2016, 7, 11095. [Google Scholar] [CrossRef] [PubMed]
- Mimura, T. Regulation of Phosphate Transport and Homeostasis in Plant Cells. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 1999; Volume 191. [Google Scholar]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008, 177, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Meng, S.; Li, M.; Zhao, Z. Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front. Plant Sci. 2016, 7, 1398. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chang, X.-J.; Ye, Y.; Xie, W.-B.; Wu, P.; Lian, X.-M. Comprehensive Sequence and Whole-Life-Cycle Expression Profile Analysis of the Phosphate Transporter Gene Family in Rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Piñeros, M.A.; Wang, Z.; Wang, W.; Li, C.; Wu, Z.; Kochian, L.V.; Wu, P. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 2014, 37, 1159–1170. [Google Scholar] [CrossRef]
- Teng, W.; Zhao, Y.Y.; Zhao, X.Q.; He, X.; Ma, W.Y.; Deng, Y.; Chen, X.-P.; Tong, Y.-P. Genome-wide Identification, Characterization, and Expression Analysis of PHT1 Phosphate Transporters in Wheat. Front. Plant Sci. 2017, 8, 543. [Google Scholar] [CrossRef]
- Preuss, C.P.; Huang, C.Y.; Gilliham, M.; Tyerman, S.D. Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol. 2010, 152, 1431–1441. [Google Scholar] [CrossRef]
- Nagy, R.; Vasconcelos, M.J.V.; Zhao, S.; McElver, J.; Bruce, W.; Amrhein, N.; Raghothama, K.G.; Bucher, M. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. 2006, 8, 186–197. [Google Scholar] [CrossRef]
- Ren, F.; Zhao, C.Z.; Liu, C.S.; Huang, K.L.; Guo, Q.Q.; Chang, L.L.; Xiong, H.; Li, X.-B. A Brassica napus PHT1 phosphate transporter, BnPht1;4, promotes phosphate uptake and affects roots architecture of transgenic Arabidopsis. Plant Mol. Biol. 2014, 86, 595–607. [Google Scholar] [CrossRef]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. Cell Mol. Biol. 2009, 57, 798–809. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef]
- He, J.; Jin, Y.; Du, Y.-L.; Wang, T.; Turner, N.C.; Yang, R.-P.; Siddique, K.H.M.; Li, F.-M. Genotypic Variation in Yield, Yield Components, Root Morphology and Architecture, in Soybean in Relation to Water and Phosphorus Supply. Front. Plant Sci. 2017, 8, 1499. [Google Scholar] [CrossRef]
- Zheng, H.F.; Chen, L.D.; Yu, X.Y.; Zhao, X.F.; Ma, Y.; Ren, Z.B. Phosphorus control as an effective strategy to adapt soybean to drought at the reproductive stage: Evidence from field experiments across northeast China. Soil Use Manag. 2015, 31, 19–28. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, X.; Gong, Z.; Ma, C.; Zhang, L. Effects of phosphorus nutrition on P absorption and yields of soybean. Plant Nutr. Fertil. Sci. 2012, 18, 670–677. [Google Scholar]
- Versaw Wayne, K.; Harrison Maria, J. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 2002, 14, 1751–1766. [Google Scholar] [CrossRef]
- Poirier, Y.; Bucher, M. Phosphate transport and homeostasis in Arabidopsis. Arab. Book 2002, 1, e0024. [Google Scholar] [CrossRef]
- Shi, S.; Wang, D.; Yan, Y.; Zhang, F.; Wang, H.; Gu, M.; Sun, S.; Xu, G. Function of phosphate transporter OsPHT2;1 in improving phosphate utilization in rice. Chin. J. Rice Sci. 2013, 27, 457–465. [Google Scholar]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef]
- Daram, P.; Brunner, S.; Rausch, C.; Steiner, C.; Amrhein, N.; Bucher, M. Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 1999, 11, 2153–2166. [Google Scholar] [CrossRef]
- Allen, J.F.; de Paula, W.B.; Puthiyaveetil, S.; Nield, J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011, 16, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K. The import and export business in plastids: Transport processes across the inner envelope membrane. Plant Physiol. 2011, 155, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Roessner, U.; Eickmeier, I.; Genc, Y.; Callahan, D.L.; Shirley, N.; Langridge, P.; Bacic, A. Metabolite profiling reveals distinct changes in carbon and nitrogen metabolism in phosphate-deficient barley plants (Hordeum vulgare L.). Plant Cell Physiol. 2008, 49, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Bao, Y.; Zhang, X.; Yang, M.; Zhang, X. Research progress of phosphorus transporter gene in plants. Ecol. Environ. Sci. 2017, 26, 342–349. [Google Scholar]
- Ayadi, A.; David, P.; Arrighi, J.F.; Chiarenza, S.; Thibaud, M.C.; Nussaume, L.; Marin, E. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Baumann, A.; Poirier, Y. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 2010, 152, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, K.; Takabatake, R.; Umehara, Y.; Kouchi, H.; Izui, K.; Hata, S. Cloning, functional expression, and mutational analysis of a cDNA for Lotus japonicus mitochondrial phosphate transporter. Plant Cell Physiol. 2002, 43, 1250–1253. [Google Scholar] [CrossRef]
- Kang, X.; Ni, M. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 2006, 18, 921–934. [Google Scholar] [CrossRef]
- Liu, T.Y.; Huang, T.K.; Tseng, C.Y.; Lai, Y.S.; Lin, S.I.; Lin, W.Y.; Chen, J.-W.; Chiou, T.J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168–2183. [Google Scholar] [CrossRef]
- Khan, G.A.; Bouraine, S.; Wege, S.; Li, Y.; de Carbonnel, M.; Berthomieu, P.; Poirier, Y.; Rouached, H. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1; H3 in Arabidopsis. J. Exp. Bot. 2014, 65, 871–884. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.H.; Shen, S.C.; Lee, L.Y.; Lee, S.H.; Chan, M.T.; Lin, C.S. Tape- Arabidopsis Sandwich—A simpler Arabidopsis protoplast isolation method. Plant Methods 2009, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.M.; Hall, A.; Millar, A.J.; Darrah, C.; Davis, S.J. Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 2009, 5, 1–7. [Google Scholar] [CrossRef]
- John, M.K. Colorimetric Determination of Phosphorus in Soil and Plant Materials with Ascorbic ACID. Soil Sci. 1970, 109, 214–220. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Xu, X.; Fu, Y.; Yang, X.; Wu, L.; Tian, P.; Yang, M.; Wu, Z. Effects of Soybean Phosphate Transporter Gene GmPHT2 on Pi Transport and Plant Growth under Limited Pi Supply Condition. Int. J. Mol. Sci. 2023, 24, 11115. https://doi.org/10.3390/ijms241311115
Wei X, Xu X, Fu Y, Yang X, Wu L, Tian P, Yang M, Wu Z. Effects of Soybean Phosphate Transporter Gene GmPHT2 on Pi Transport and Plant Growth under Limited Pi Supply Condition. International Journal of Molecular Sciences. 2023; 24(13):11115. https://doi.org/10.3390/ijms241311115
Chicago/Turabian StyleWei, Xiaoshuang, Xiaotian Xu, Yu Fu, Xue Yang, Lei Wu, Ping Tian, Meiying Yang, and Zhihai Wu. 2023. "Effects of Soybean Phosphate Transporter Gene GmPHT2 on Pi Transport and Plant Growth under Limited Pi Supply Condition" International Journal of Molecular Sciences 24, no. 13: 11115. https://doi.org/10.3390/ijms241311115
APA StyleWei, X., Xu, X., Fu, Y., Yang, X., Wu, L., Tian, P., Yang, M., & Wu, Z. (2023). Effects of Soybean Phosphate Transporter Gene GmPHT2 on Pi Transport and Plant Growth under Limited Pi Supply Condition. International Journal of Molecular Sciences, 24(13), 11115. https://doi.org/10.3390/ijms241311115





